Abstract
The complete genome sequence of Caulobacter crescentus was determined to be 4,016,942 base pairs in a single circular chromosome encoding 3,767 genes. This organism, which grows in a dilute aquatic environment, coordinates the cell division cycle and multiple cell differentiation events. With the annotated genome sequence, a full description of the genetic network that controls bacterial differentiation, cell growth, and cell cycle progression is within reach. Two-component signal transduction proteins are known to play a significant role in cell cycle progression. Genome analysis revealed that the C. crescentus genome encodes a significantly higher number of these signaling proteins (105) than any bacterial genome sequenced thus far. Another regulatory mechanism involved in cell cycle progression is DNA methylation. The occurrence of the recognition sequence for an essential DNA methylating enzyme that is required for cell cycle regulation is severely limited and shows a bias to intergenic regions. The genome contains multiple clusters of genes encoding proteins essential for survival in a nutrient poor habitat. Included are those involved in chemotaxis, outer membrane channel function, degradation of aromatic ring compounds, and the breakdown of plant-derived carbon sources, in addition to many extracytoplasmic function sigma factors, providing the organism with the ability to respond to a wide range of environmental fluctuations. C. crescentus is, to our knowledge, the first free-living α-class proteobacterium to be sequenced and will serve as a foundation for exploring the biology of this group of bacteria, which includes the obligate endosymbiont and human pathogen Rickettsia prowazekii, the plant pathogen Agrobacterium tumefaciens, and the bovine and human pathogen Brucella abortus.
Caulobacter crescentus, a Gram-negative bacterium that grows in dilute aquatic environments, is a member of the α-subdivision of proteobacteria. C. crescentus invariably differentiates and divides asymmetrically at each cell cycle. Asymmetric cell division and differentiation are recurring themes that underline cellular diversity in multicellular organisms. C. crescentus is a simple and highly manipulable single-celled model system to study cellular differentiation, asymmetric division, and their coordination with cell cycle progression (1, 2). Caulobacter does all that with less than 4,000 genes, allowing full genome-wide studies of a single differentiating cell.
This stalked bacterium adheres to solid surfaces via a holdfast at the tip of the stalk. It also has a motile swarmer cell stage during its life cycle. The stalked cell acts like a stem cell, continually giving rise to a new swarmer cell at each division (Fig. 1) (3). The production of a swarmer cell with an obligatory motile period minimizes competition during growth in a dilute environment by ensuring that the progeny cell will colonize in a new location (1). The swarmer cell is unable to initiate chromosome replication until it differentiates into a stalked cell. The regulation of cell cycle progression in C. crescentus occurs at several levels (4): temporally controlled transcriptional activation and repression, differential phosphorylation of two-component system regulatory proteins, and proteolysis of regulatory and structural proteins. The basic paradigm of cell cycle control used by eukaryotic cells, temporally controlled transcription, phosphorylation of regulatory factors, and targeted proteolysis, has been conserved in C. crescentus, although the proteins involved in these processes are different. Recent observations of regulatory proteins dynamically localized to defined cellular addresses at specific times in the C. crescentus cell cycle suggest that the three-dimensional organization of this cell adds yet another layer of control (5). The completion of the full genome sequence of this organism provides access to the complete signal transduction network that controls differentiation and cell cycle progression within the context of a unicellular organism growing in a dilute nutrient environment.
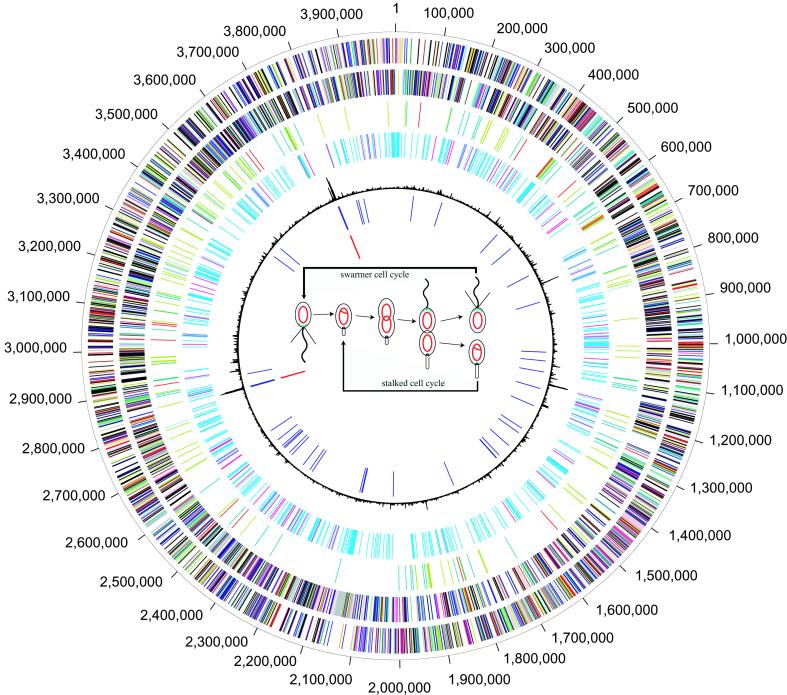
Figure 1.
Circular representation of the C. crescentus genome. Coordinate markers around the outside of the circle are in base pairs. First circle, predicted coding regions on the plus strand color coded by role category: violet, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups, and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; gold, DNA metabolism; light gray, energy metabolism; magenta, fatty acid and phospholipid metabolism; pink, protein synthesis/fate; orange, purines, pyrimidines, nucleosides, nucleotides; olive, regulatory functions; dark green, transcription; teal, transport and binding proteins; salmon, plasmid, phage, and transposon functions; blue, unknown function, hypothetical and conserved hypothetical proteins. Second circle, predicted coding regions on the minus strand color coded by role category. Third circle, genes involved in chemotaxis and motility color coded by role category: olive, two-component regulatory genes; red, methyl-accepting chemotaxis genes; dark green, extracellular function sigma factors; teal, TonB and the TonB-dependent receptors. Fourth circle, cell cycle-regulated genes (2). Fifth circle, atypical nucleotide composition curve. Sixth circle, tRNAs. Seventh circle, rRNAs. The center of the circle contains a schematic of the C. crescentus cell cycles. Within the cells, the red circles indicate the nonreplicating chromosome, and the red theta structures indicate replicating the chromosome.
Methods
ORF Prediction and Gene Identification.
ORFs were identified by using glimmer (6). Annotation of the identified ORFs was accomplished by manual curation of the outputs of a variety of similarity searches. Searches of the predicted coding regions were performed with blastp, as previously described (7). The protein–protein matches are aligned with blast_extend_repraze, a modified Smith–Waterman (8) algorithm that maximally extends regions of similarity across frameshifts. Gene identification is facilitated by searching against a database of nonredundant bacterial proteins (nraa) developed at tigr and curated from the public archives GenBank, Genpept, PIR, and SwissProt. Searches matching entries in nraa have the corresponding role, gene common name, percent identity and similarity of match, pairwise sequence alignment, and taxonomy associated with the match assigned to the predicted coding region and stored in the database. ORFs were also analyzed with two sets of hidden Markov models (HMMs) constructed for a number of conserved protein families from pfam (9) and tigrfam (10). Regions of the genome without ORFs and ORFs without a database match are reevaluated by using blastx as the initial search, and ORFs are extrapolated from regions of alignment. Finally, each putatively identified gene is assigned to one of 113 role categories adapted from Riley (11).
Construction of Paralogous Families.
Paralogous families were built in stages. First, for each pfam HMM scoring above the cutoff to two or more proteins, an alignment of matching regions was constructed. In some cases, the alignment was discarded, trimmed, or used to generate a new global HMM and improved hit region alignment. Second, all peptide sequence outside of the accepted HMM hit regions was subject to automated domain clustering and alignment by mkdom (12). Several mkdom cluster alignments were rejected as insignificant or trimmed. The 678 nonoverlapping paralogous domain alignments include 259 supported by pfam HMMs and 419 created by mkdom. The alignments include 2,893 regions from 1,801 different proteins of 3,767.
Dinucleotide Signatures Analysis.
The data for dinucleotide signature analysis were computed by the method described by Karlin et al. (13) with a window size of 100,000 bp and a granularity of 100 bp. χ2 analysis: the distribution of all 64 trinucleotides (3 mers) was computed for the complete genome in all 6 reading frames, followed by the 3-mer distribution in 2,000 bp windows. Windows overlapped by 1,000 bp. For each window, the χ2 statistic on the difference between its 3-mer content and that of the whole genome was computed.
General Features of the Genome.
The genome sequence of C. crescentus CB15 was determined by the whole genome random sequencing method (14). The genome consists of a circular chromosome of 4,016,942 bp with an average G + C content of 67.2%. A total of 3,767 predicted ORFs were identified, of which 2,030 (53.9%) are assigned putative functions, 725 (19.2%) have matches to hypothetical proteins, and 1,012 (26.9%) have no database match (Fig. 4, published as supplemental data on the PNAS web site, www.pnas.org). Coding regions comprise 90.6% of the chromosome. Approximately 1/2 of the proteins (1,801) are members of 678 paralogous families. The largest protein families are response regulators (71 proteins); TonB-dependent outer membrane channels (65 proteins); histidine kinases (61 proteins); and ATP-binding cassette domain transporters (45 proteins) (7). Base pair 1 of the chromosome was assigned within the experimentally determined origin of replication (15), which was also revealed by GC skew analysis (G-C/G + C) (16). Two nontandem ribosomal RNA operons and 51 tRNAs representing all 20 amino acids are present (Table 1).
Table 1.
General Features of the C. crescentus genome
| Size, bp | 4,016,942 |
| G+C percent | 67.2 |
| Total no. ORFs | 3,767 |
| ORF size, bp | 969 |
| Percent coding | 90.6 |
| No. rRNA operons (16S-23S-5S) | 2 |
| No. tRNA | 51 |
| No. similar to known proteins | 2,030 (53.9%) |
| No. similar to proteins of unknown function | 721 (19.2%) |
| No. hypothetical proteins | 1,012 (26.9%) |
| No. of ORFs in paralogous families | 1,801 (47.8%) |
The genome contains 23 insertion sequences, which consist of 5 multicopy and 3 single-copy elements. Two of the multicopy elements, IS511 and IS298, have been previously described (17); the remaining elements, to our knowledge, are novel (Table 2). There are two nontandem identical 2.2-kb regions containing N-acetylglucosamine phosphotransferase system (PTS) components. The two PTS systems presumably reflect a very recent gene duplication event. The 5′ portion of the DNA damage repair gene radC is split in the genome from the 3′ portion by a 60-kb insert. This insert contains genes for an extracytoplasmic function sigma factor, a TonB-dependent channel, a putative methyl-accepting chemotaxis homologue, and a metal ion efflux protein. It also contains genes for transposases, conjugal transfer proteins, and several hypothetical proteins that are concentrated toward the outer boundaries of the insertion. Trinucleotide skew analysis identified these outer portions of the insert as different from the genome at large, suggesting that the disrupted radC gene resulted from a plasmid insertion and subsequent recombination events.
Table 2.
Multicopy insertion sequences
| Element/family copies | Length/DR* | Structure | Similarity/species | Terminal inverted repeats (5′-3′) | ||||
|---|---|---|---|---|---|---|---|---|
| IS511 | IS3 | 4 | 1266 | 4 | orfA/orfB | self† | C. crescentus | TGACCTGCCCCTGATTTTTT |
| TGACCTGCCTCTGATCTTTC | ||||||||
| IS298 | IS5 | 4 | 845 | 4 | orfA/orfB | self‡ | C. crescentus | GTGGTGTGGACTCTAAGGAT |
| GCGGTGTGGACACTTATCGC | ||||||||
| ISCc1 | IS5 | 5§ | 848 | 4 | orfA/orfB | IS298 | C. crescentus | GCCGTAGTGACGATTTAGGA |
| GTGGCGGTGACCATTTAGCT | ||||||||
| ISCc2 | IS110 | 4¶ | 1140 | 2 | orfA | IS492 | Pseudomonas atlantica | TATCTGGATTGCAGCGCCAT |
| TGTCTGGATCGTCAAGCGGC | ||||||||
| ISCc3 | IS3 | 3 | 1514 | 2 | orfA/orfB | ISD1 | Desulfovibrio vulgaris | TGTCCGCCGTCAGCGCCAAT |
| TGTACGTCGTCAGAAGTTTT | ||||||||
Size in base pairs of the element (Length) and the direct repeat (DR) generated by insertion into the chromosomal target site.
IS298 gi/4836363/gb/AF117124.1/AF117124[4836363].
One copy of ISCc1 is truncated.
All four copies of ISCc2 occur at the same position in an abundant DNA repeat and may represent a single insertion event.
Cell Cycle.
The control of cell cycle progression in C. crescentus has been shown to depend, in large measure, on the differential availability and activation by phosphorylation of the two-component system response regulator CtrA and the CckA histidine kinase. Both of these regulators are essential for viability and control the time of chromosome replication initiation, DNA methylation mediated by the CcrM DNA methyltransferase, cell division, and flagella and pili biogenesis (18, 19). Another essential response regulator, DivK, functions via two nonessential histidine kinases, DivJ and PleC, to coordinate cell division with polar differentiation events (20–22). Critical to cell cycle progression is the proteolysis of CtrA∼P at the G1-S transition. Access to the C. crescentus genome sequence has now allowed a global approach to the regulatory mechanisms that include targeted proteolysis, signaling protein activation, DNA methylation, and differential gene transcription that allows cell differentiation within the context of the cell cycle.
Proteolysis.
Previous work in C. crescentus has demonstrated that bacteria, like eukaryotes, regulate proteolysis of specific proteins to control cell cycle progression and morphogenesis (23, 24). For both the response regulator CtrA and the chemoreceptor McpA, residues at or near the C terminus are necessary, although not sufficient, for proper cell cycle-dependent turnover. CtrA ends in a double alanine (AA), and McpA ends in a string of small hydrophobic residues followed by WEEF. Searching the predicted proteome of C. crescentus reveals that nearly 20% of all response regulators, and more than 50% of all cell cycle-regulated response regulators and histidine kinases, have C-terminal residues of AA, IA, or VA. The chemoreceptors McpB, McpC, McpD, McpE, like McpA, are all cell cycle-regulated and end in a short string of hydrophobic amino acids such as alanines and valines followed by WEEF. Those hydrophobic residues have recently been found to be essential for turnover (I.P. and M.R.K.A., unpublished results). Of the 12 predicted chemoreceptors lacking the WEEF motif, 8 end in AA or VA. Additionally, 8 of the 23 other predicted chemotaxis genes end in AA or VA. The similarity of C-terminal residues in large families of proteins for which proteolysis has been shown to play a role in cell cycle-regulated turnover suggests that regulated proteolysis is a significant component of the progression of the cell cycle.
Two-Component Signal Transduction Proteins.
C. crescentus has the largest number of signal transduction proteins of any sequenced bacterium when adjusted for genome size. In light of the role played by these signal transduction proteins in the control of cell differentiation during cell cycle progression (20) and growth in a dilute aquatic environment, the large number of newly identified members of this family of proteins provides a valuable resource for understanding the complete signaling pathways that control these processes. Analysis of the genome sequence revealed 34 histidine protein kinase (HPK) genes, 44 response regulator (RR) genes, and 27 hybrid HPK/RR genes. Of these, 22 HPKs, 21 RRs, and 26 hybrids were newly identified by the sequence analysis. Approximately 1/3 of the HPK genes are located adjacent to RR genes on the chromosome and are likely to be functional pairs involved in responses to environmental changes. The role of these cognate pairs contrasts with that of the dispersed HPKs and RRs, many of which function in cell cycle regulation (4, 20). A number of these cell cycle-regulated HPKs and RRs are essential for cell viability (4, 18–21). The transcription of at least 35 of the 105 genes encoding two-component signal transduction proteins has recently been shown to be temporally regulated during the cell cycle and to include all those that are essential except DivL (2). Further, of the four studied by fluorescence microscopy in living cells, all were found to be dynamically localized during the cell cycle (19, 22, 23). The signaling proteins whose genes were found to be cell cycle-regulated but are otherwise uncharacterized are thus candidates for having regulatory roles in cell cycle progression.
Eleven of the hybrid HPK/RR proteins are predicted to be cytoplasmic, as are 13 of the nonhybrid HPKs. Surprisingly, many of these cytoplasmic kinases (14 proteins) contain PAS domains, which are often involved in sensing changes in cellular energy levels, oxygen levels, or redox potential. In contrast, few PAS domains are found in the membrane-associated kinases. Thus, it appears that C. crescentus relies heavily on molecular networks that sense and respond to intracellular oxygen and redox state.
DNA Methylation.
Chromosome methylation on the N-6 adenine of the sequence GAnTC is catalyzed by the CcrM DNA methyltransferase (25). The transcription of the ccrM gene is under tight cell cycle control; the CcrM protein is present only in the predivisional cell, when it is available to bring the two newly replicated chromosomes from the hemi- to the full methylation state. CcrM is essential for viability, and its expression at inappropriate times in the cell cycle causes defects in cell division and DNA replication. Thus, temporally regulated methylation of GAnTC site is a component of cell cycle progression. We therefore examined the number of GAnTC sites in the genome and their location with respect to coding sequences. Given the size of the C. crescentus genome, if the occurrence of these sites were random, we would expect there to be approximately 12,000 GAnTC sites. In fact, there are only 4,496 of these sites in the genome, and 22% are located between the ORFs that comprise 90.6% of the genome, supporting the argument that the methylation of these sites plays an important regulatory role. Knowledge of the genome position of these sites now opens the way for understanding their function in the regulation of the cell cycle.
Transcription.
There are 16 putative RNA polymerase sigma factors in the C. crescentus genome, only three of which, rpoD (26), rpoH (27, 28), and rpoN (29), have been previously identified. The 13 new sigma factors revealed by the genome analysis are all extracytoplasmic function (ECF) sigma factors, which typically act to couple periplasmic or extracellular stimuli to changes in gene expression. Two of these new ECF sigma factors, SigT and SigU, are specifically transcribed at the swarmer-to-stalked cell transition and are components of the genetic network that controls cell cycle progression (2). Thus, the newly identified complement of sigma factors will clearly contribute to understanding the control of the 19% of C. crescentus genes whose transcription has been shown to be cell cycle regulated (2). The RNA polymerase holoenzyme containing RpoN (sigma 54) is used in C. crescentus for the transcription of genes involved in cell differentiation events, such as flagellar biogenesis (30). Accessory factors that control transcriptional activation, such as those required to work in concert with sigma-54, are candidate mediators of differential gene expression during the cell cycle. There are only four putative RpoN activators in the C. crescentus genome, compared with 12 sigma-54 dependent activators in Escherichia coli (31), and at least one of these four C. crescentus RpoN activators is temporally regulated (2, 30).
Adaptation to Dilute Aquatic Conditions.
Genome analysis identified a large number of genes that would enable utilization of dilute carbon sources and provides a comprehensive picture of the strategies used by C. crescentus for survival in nutrient-limiting conditions. Unlike E. coli and Vibrio cholerae, C. crescentus has no OmpF-type outer membrane porins that allow the passive diffusion of hydrophilic substrates across the outer membrane. However, it does possess 65 members of the family of TonB-dependent outer membrane channels that catalyze energy-dependent transport across the outer membrane. This is more than any other organism thus far characterized, with the next highest being 34 in Pseudomonas aeruginosa (32), and with no other sequenced proteobacteria possessing more than 10. C. crescentus has substantially fewer cytoplasmic membrane transporters relative to genome size than either E. coli or V. cholerae (33). Given C. crescentus ' low nutrient habitat, it is surprising that PTS or ATP-binding cassette domain transporters, which usually have high affinity for their substrates, are not overly represented compared with low-substrate affinity transporters. This transporter configuration, energy-gated outer membrane channels for specific substrates and lower-affinity cytoplasmic membrane transporters, may be essential for C. crescentus nutrient scavenging, compared with other organisms that use passive diffusion of substrates across their outer membrane by using nonspecific porins and high-affinity inner membrane transporters.
A variety of efflux systems are predicted in C. crescentus. This bacterium displays a paracrystalline S-layer on its outer surface composed of a single protein, and this system has been exploited for the heterologous expression of proteins and peptide fragments (34). The S-layer protein is secreted by an ATP-binding cassette (ABC) domain transporter, together with membrane fusion proteins (MFP) and outer membrane factor (OMF) family constituents (35). At least one other ABC-type protein secretion system and a PTS complex polysaccharide extrusion system are present in C. crescentus, which may also play roles in secreting cell adhesion products. Representatives are also present from all known prokaryotic families of multidrug efflux systems, as well as four amino acid efflux systems and four resistance-nodulation-cell division- (RND) type metal ion efflux systems. Thus, C. crescentus is equipped to carry out active scavenging and secretion processes when growing in extreme dilute environments.
C. crescentus possesses a large number of genes for sensing and responding to environmental substrates (Fig. 1). Approximately 2.5% of the genome is devoted to swarmer cell motility (chemotaxis and flagellum related genes). Before the determination of the genome sequence, 44 sequenced genes were implicated in assembly or activity of the C. crescentus flagellum (30). Nine additional genes are present in the genome whose predicted products are similar to proteins with roles in flagella biogenesis. There are two chemotaxis operons in C. crescentus. The previously characterized mcpA operon (McpA, CheX, CheYI, CheAI, CheWI, CheRI, CheBI, CheYII, CheD, CheU, CheYIII, and CheE) is essential for chemotaxis (36, 37) and is closely related to the chemotaxis operons from other α-proteobacteria such as Rhodobacter sphaeroides (38) and Sinorhizobium meliloti (39). The newly revealed second chemotaxis operon (McpK, CheAII, CheWII, CheYIV, CheBII, and CheRII) is most similar to that in the α-proteobacterium, Rhodospirillum centenum. In addition to the two defined chemotaxis operons, a large number of mcp genes and cheY genes are scattered throughout the genome. The genome contains 16 unlinked genes encoding chemoreceptors (MCPs), indicating that Caulobacter has the ability to respond to a wide variety of compounds. Six of the MCPs lack membrane-spanning domains and may be involved in sensing cytoplasmic substrates linked to cell cycle events.
Phylogeny.
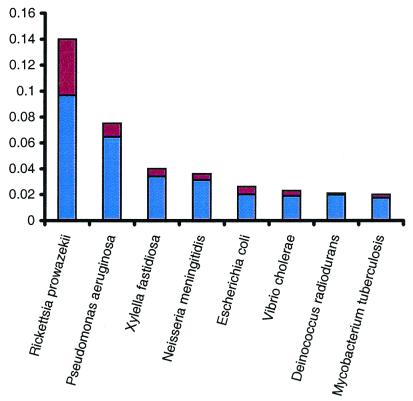
Comparison of the C. crescentus proteome to those of all other organisms for which the complete genome sequence is available demonstrated the close relationship between C. crescentus and the nonfree-living endosymbiont, Rickettsia prowazekii, the only other sequenced α-proteobacterium (40). Analysis of the genome sequence of Rickettsia indicated that the obligate endosymbiosis of this bacterium has led to a dramatic reduction of its genome size and the elimination of large numbers and sets of genes (40). We predict that those genes critical to cell cycle progression in Rickettsia are less likely to have been lost during its reductive evolution. This prediction is supported by two comparative global analyses. First, of the C. crescentus ORFs whose predicted protein matched closest to a Rickettsia homolog, more than 30% were cell cycle regulated in C. crescentus (2) (Fig. 2). That same percentage was only 22% for E. coli, 17% for Neisseria meningitidis, and less than 15% for all others. Second, we generated a complete list of cell cycle-regulated genes from C. crescentus that had homologs in the Rickettsia genome. This list of around 150 genes included genes known to be critical for cell cycle progression in C. crescentus, such as ctrA, parAB, and recA, but did not include genes required only for extraneous cell cycle processes such as flagellar biogenesis. This type of global comparison between the C. crescentus genome and the reduced genome of Rickettsia may ultimately help to discriminate between the “core” cell cycle genes required for proper progression of the cell cycle and the “peripheral” genes that are cell cycle-regulated but not needed for full viability in C. crescentus.
Figure 2.
Comparison of the C. crescentus strain CB15 ORFs to those of other completely sequenced organisms. The sequences of all proteins from each completely sequenced genome were retrieved from the National Center for Biotechnology Information and tigr databases. All C. crescentus ORFs were searched against the ORFs from all other genomes with FASTA3. The number of ORFs with the highest similarity (P < 10–5) to an ORF from a given species is shown as a proportion of the total number of ORFs in that species. The red portion of each organism's bar represents the percentage of genes that were also found to be cell cycle-regulated in C. crescentus (2). Only the organisms with the most hits, after adjustment for genome size, are presented.
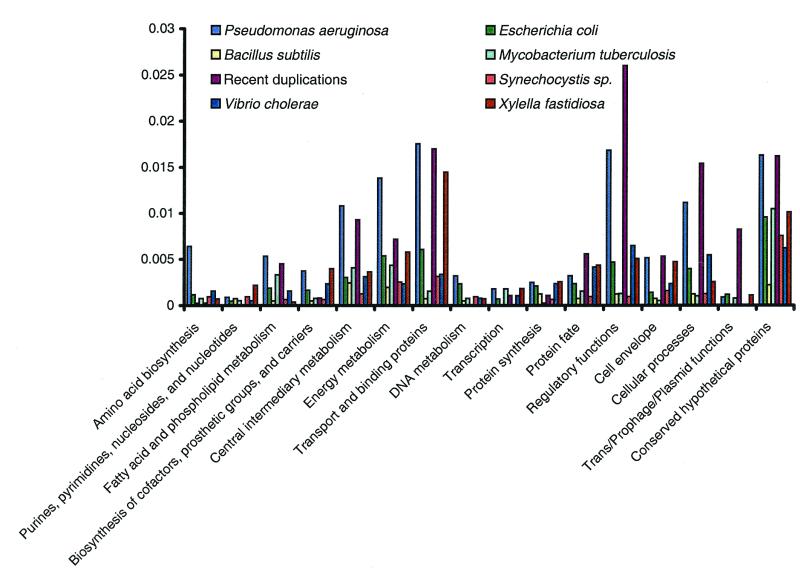
Comparison of the Caulobacter proteome to that of other species reveals that there are more matches to the P. aeruginosa proteome, when scaled for genome size, than to any species other than R. prowazekii (Fig. 2). There are approximately twice as many best matches to P. aeruginosa than to other γ-proteobacteria (Fig. 3). An explanation for this observation is the existence of a shared biology between members of these genera and the opportunity for gene transfer between these two lineages. This is supported by the nonuniform distribution of best matching proteins across role categories; functions such as transcription and translation are underrepresented, whereas peripheral metabolic functions are overrepresented. The presence in C. crescentus of a 20-gene cluster for the metabolism of aromatic compounds, a pathway extensively characterized only in soil bacteria including Pseudomonas and Streptomyces species (41), highlights a shared biology between this aquatic species and various species of soil bacteria. It also suggests that C. crescentus may be exposed to diverse substrates of terrestrial origin in its natural habitat. As revealed by comparative genome analysis, this shared biology between C. crescentus and soil organisms extends to other cellular processes. The conservation of gene order and the sequence similarity of genes involved in intermediary metabolism again suggests that gene transfer between these species has taken place. Consistent with this concept, it has been experimentally demonstrated that C. crescentus is able to integrate, retain, and efficiently express plasmid encoded degradative pathway genes from Pseudomonas putida (42). The presence of genes for the breakdown of numerous plant polysaccharides, including cellulose, xylan, lignin, glucan, and pectin, as well as transporter systems for the import of the resulting sugars, suggests that, unexpectedly, plant polymers are a significant source of metabolites for the central intermediary metabolism of this organism.
Figure 3.
Comparison of the C. crescentus strain CB15 ORFs to those of other completely sequenced organisms by major biological role categories. The number of ORFs in C. crescentus that are most similar to other completed genomes was size adjusted, and only those organisms with over 100 significant (P < 10–5) most similar hits are presented. ORFs with best hits to R. prowazekii are not included to allow detection of the lower number of similarities not caused by the shared common lineage.
Conclusion
Caulobacters are the most prevalent organisms adapted solely for survival in nutrient-poor aquatic and marine environments. The completion of the genomic sequence now lays the foundation for understanding, on a molecular level, how this bacterium's obligate differentiation and asymmetric division enable it to thrive in such dilute habitats. Furthermore, the tools developed for genetic manipulation of C. crescentus make it an attractive organism for development as a bioremediation agent (J.S., unpublished results).
With the completion of the annotated sequence of the C. crescentus genome, a full description of the genetic network that controls its cell differentiation, cell growth, and cell cycle progression is within reach. Cell cycle analysis of global transcription patterns, proteomics of stable and unstable proteins, genetic and biochemical analysis of phosphotransfer to regulatory proteins, and time-lapse fluorescent imaging for spatial tracking of regulatory proteins will generate a comprehensive map of the C. crescentus cell cycle genetic circuitry.
Supplementary Material
Acknowledgments
This work was supported by U.S. Department of Energy Office of Biological and Environmental Research Cooperative Agreement DEFC0295ER61962.
Abbreviations
- HMM
hidden Markov model
- HPK
histidine protein kinase
- RR
response regulator
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AE005673).
References
- 1.Brun Y V, Janakiraman R. In: Prokaryotic Development. Brun Y V, Shimkets L J, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 297–317. [Google Scholar]
- 2.Laub M T, McAdams H, Feldblyum T, Fraser C, Shapiro L. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 3.Stove J L, Stanier R Y. Nature (London) 1962;196:1189–1192. [Google Scholar]
- 4.Hung D, McAdams H, Shapiro L. In: Prokaryotic Development. Brun Y V, Shimkets L, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 361–378. [Google Scholar]
- 5.Shapiro L, Losick R. Cell. 2000;100:89–98. doi: 10.1016/s0092-8674(00)81686-4. [DOI] [PubMed] [Google Scholar]
- 6.Salzberg S, Delcher A, Kasif S, White O. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–408. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 8.Waterman M S. Methods Enzymol. 1988;164:765–793. doi: 10.1016/s0076-6879(88)64083-3. [DOI] [PubMed] [Google Scholar]
- 9.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haft D H, Loftus B J, Richardson D L, Yang F, Eisen J A, Paulsen I T, White O. Nucleic Acids Res. 2001;29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riley M. Microbiol Rev. 1993;57:862–952. doi: 10.1128/mr.57.4.862-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouzy J, Eugene P, Greene E A, Kahn D, Corpet F. Comput Appl Biosci. 1997;13:601–608. doi: 10.1093/bioinformatics/13.6.601. [DOI] [PubMed] [Google Scholar]
- 13.Karlin S, Campbell A M, Mrazek J. Annu Rev Genet. 1998;32:185–196. doi: 10.1146/annurev.genet.32.1.185. [DOI] [PubMed] [Google Scholar]
- 14.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Kickey E K, Peterson J D, Umayam L, et al. Nature (London) 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marczynski G T, Shapiro L. J Mol Biol. 1992;226:959–977. doi: 10.1016/0022-2836(92)91045-q. [DOI] [PubMed] [Google Scholar]
- 16.Lobry J R. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 17.Ohta M, Mullin D A, Tarleton J, Ely B, Newton A. J Bacteriol. 1990;172:236–242. doi: 10.1128/jb.172.1.236-242.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quon K, Marczynski G, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs C, Domian I, Maddock J R, Shapiro L. Cell. 1997;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 20.Ohta N, Grebe T W, Newton A. In: Prokaryotic Development. Brun Y V, Shimkets L J, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 341–359. [Google Scholar]
- 21.Hecht G B, Lane T, Ohta N, Sommer J N, Newton A. EMBO J. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler R, Shapiro L. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- 23.Domian I J, Quon K C, Shapiro L. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 24.Wright R J, Stephens C M, Zweiger G, Shapiro L, Alley M R K. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- 25.Reisenauer A, Kahng L S, McCollum S, Shapiro L. J Bacteriol. 1999;181:5135–5139. doi: 10.1128/jb.181.17.5135-5139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malakooti J, Ely B. J Bacteriol. 1995;177:6854–6860. doi: 10.1128/jb.177.23.6854-6860.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reisenauer A, Mohr C D, Shapiro L. J Bacteriol. 1996;178:1919–1927. doi: 10.1128/jb.178.7.1919-1927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J, Newton A. J Bacteriol. 1996;178:2094–2101. doi: 10.1128/jb.178.7.2094-2101.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brun Y V, Shapiro L. Genes Dev. 1992;6:2395–2408. doi: 10.1101/gad.6.12a.2395. [DOI] [PubMed] [Google Scholar]
- 30.Gober J W, England J C. In: Prokaryotic Development. Brun Y V, Shimkets L, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 319–339. [Google Scholar]
- 31.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 32.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, et al. Nature (London) 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 33.Paulsen I T, Nguyen L, Rabus R, Saier M H., Jr J Mol Biol. 2000;301:75–101. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 34.Bingle W H, Nomellini J F, Smit J. Mol Microbiol. 1997;26:277–288. doi: 10.1046/j.1365-2958.1997.5711932.x. [DOI] [PubMed] [Google Scholar]
- 35.Awram P, Smit J. J Bacteriol. 1998;180:3062–3069. doi: 10.1128/jb.180.12.3062-3069.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alley M R K, Gomes S L, Alexander W, Shapiro L. Genetics. 1991;129:333–341. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai J W, Alley M R. J Bacteriol. 2000;182:504–507. doi: 10.1128/jb.182.2.504-507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward M J, Bell A W, Hamblin P A, Packer H L, Armitage J P. Mol Microbiol. 1995;7:357–366. doi: 10.1111/j.1365-2958.1995.mmi_17020357.x. [DOI] [PubMed] [Google Scholar]
- 39.Greck M, Platzer J, Sourjik V, Schmitt R. Mol Microbiol. 1995;15:989–1000. doi: 10.1111/j.1365-2958.1995.tb02274.x. [DOI] [PubMed] [Google Scholar]
- 40.Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. Nature (London) 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 41.Iwagami S G, Yang K, Davies J. Appl Environ Microbiol. 2000;66:1499–1508. doi: 10.1128/aem.66.4.1499-1508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chatterjee D K, Chatterjee P. J Bacteriol. 1987;169:2962–2966. doi: 10.1128/jb.169.7.2962-2966.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.