Abstract
The deletion end-points of a number of type I (less than monomeric) plasmid deletants obtained by transforming recA+ or recA- E. coli with linear pBR322 DNA were determined by DNA sequencing. In both monodirectional and bidirectional deletions the recyclization point was normally characterized by recombination between directly repeated sequences of between 4 and 10 bp present on each arm of the linearized pBR322 molecule. Frequently, short tracts of uninterrupted homology involved in recombinational recircularization were embedded in regions of relative non-homology. A model predicting the probability of matching sequences in either end of a linear plasmid molecule is presented. It is proposed that exonucleolytic processing of the exposed termini of linear plasmid molecules generates substrates for subsequent recombinational recyclization and deletion. The activity of host recombination and repair functions in recircularizing linear DNA molecules explains the generation of many of the aberrant recombinant DNA constructs obtained during gene cloning procedures.
Full text
PDF




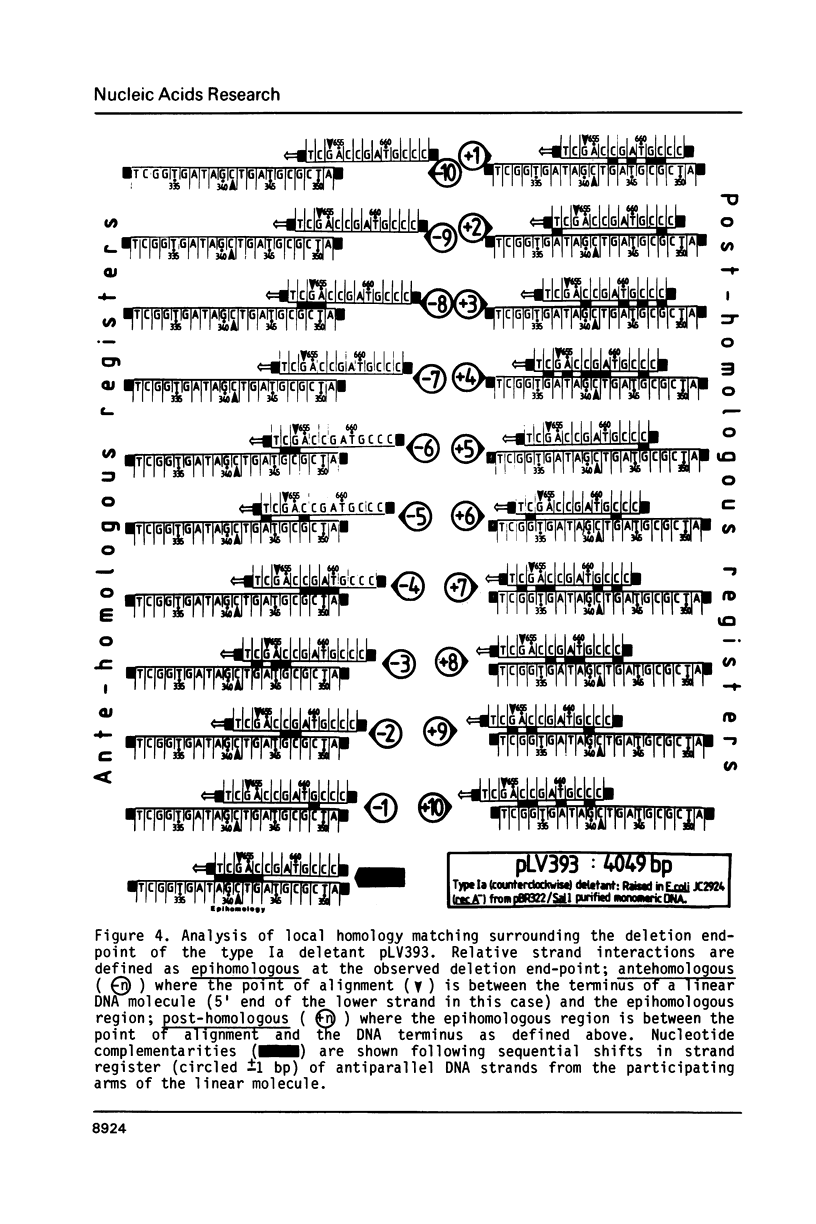
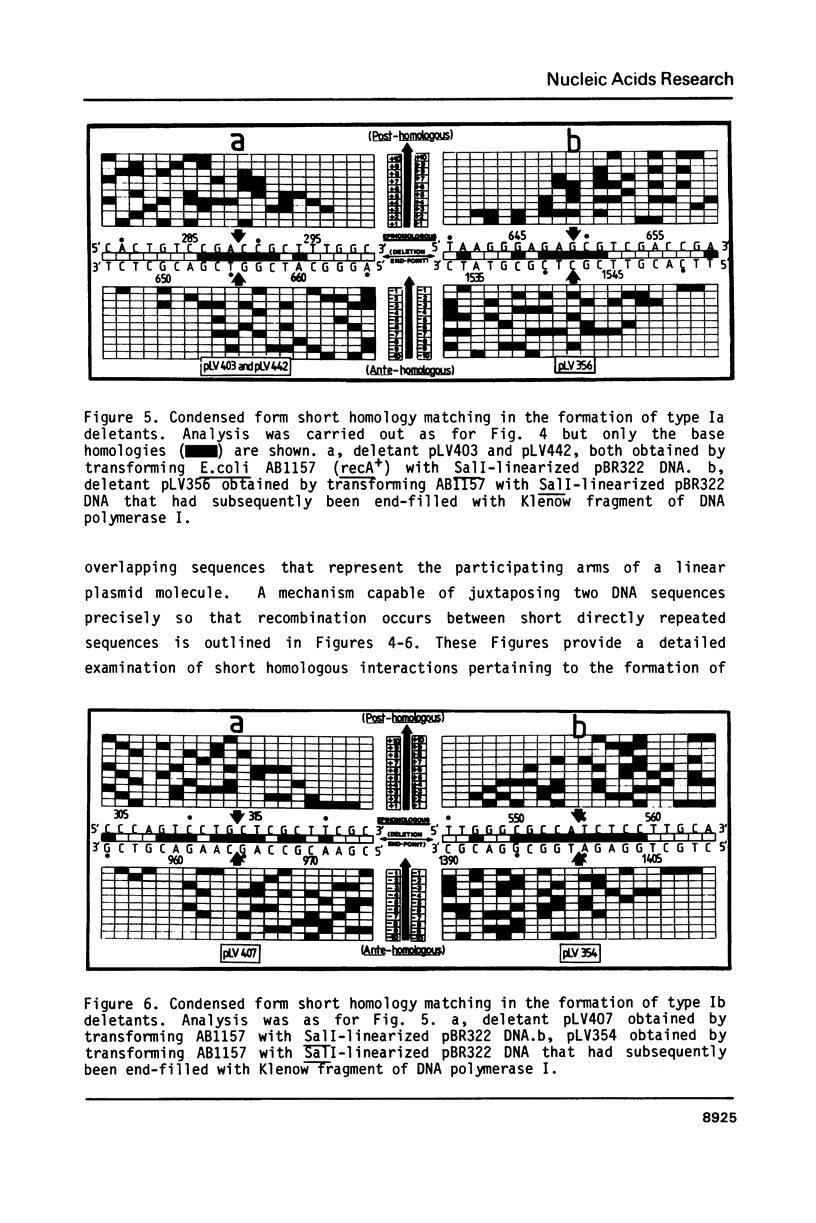


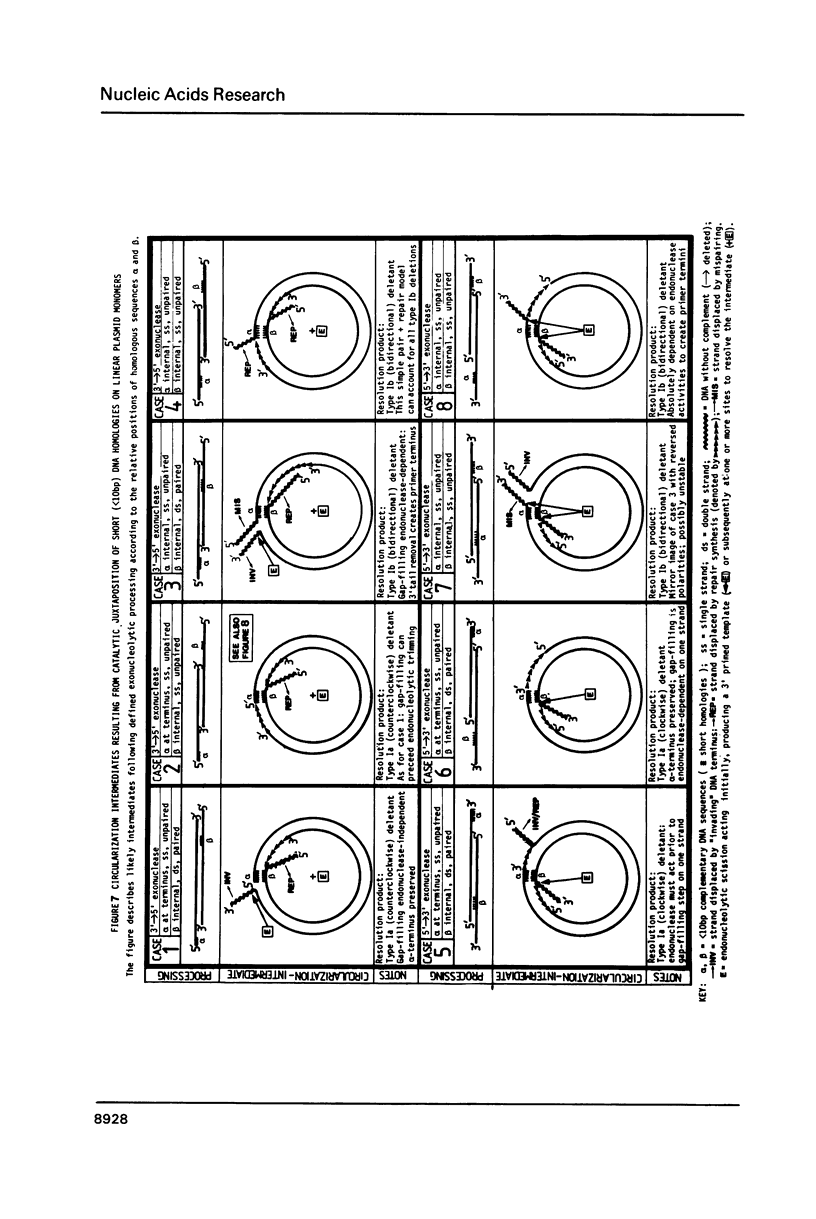

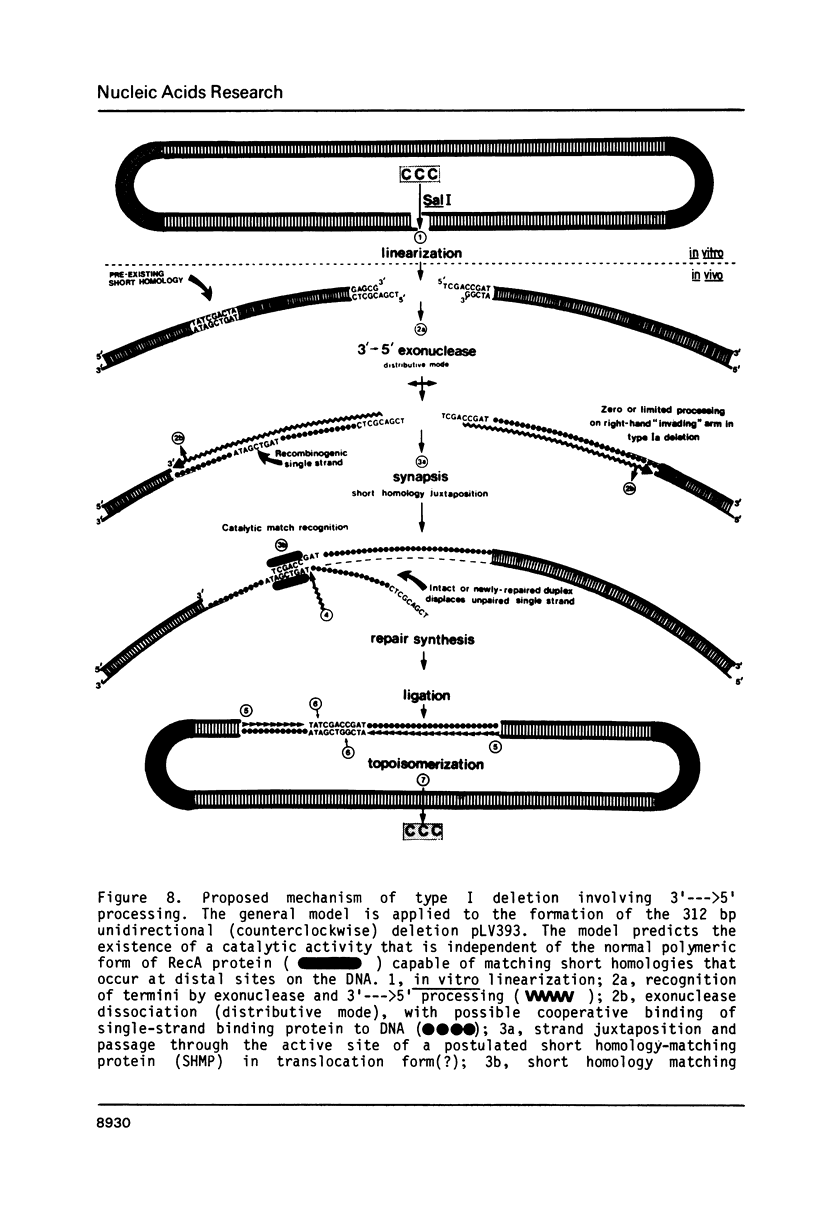


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Bergsma D. J., Olive D. M., Hartzell S. W., Byrne B. J., Subramanian K. N. Cyclization of linear chimeric plasmids in vivo by a novel end-to-end joining reaction or by intramolecular recombination: one of the products contains a 147-bp perfect palindrome stable in Escherichia coli. Gene. 1982 Dec;20(2):157–167. doi: 10.1016/0378-1119(82)90034-8. [DOI] [PubMed] [Google Scholar]
- Conley E. C., Saunders J. R. Recombination-dependent recircularization of linearized pBR322 plasmid DNA following transformation of Escherichia coli. Mol Gen Genet. 1984;194(1-2):211–218. doi: 10.1007/BF00383519. [DOI] [PubMed] [Google Scholar]
- Conley E. C., Saunders V. A., Saunders J. R. Deletion and rearrangement of plasmid DNA during transformation of Escherichia coli with linear plasmid molecules. Nucleic Acids Res. 1986 Nov 25;14(22):8905–8917. doi: 10.1093/nar/14.22.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund T., Normark S. Recombination between short DNA homologies causes tandem duplication. Nature. 1981 Jul 16;292(5820):269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Garaev M. M., Bobkov A. F., Bobkova A. F., Kalinin V. N., Smirnov V. D., Khudakov YuE, Tikchonenko T. I. The site-specific deletion in plasmid pBR322. Gene. 1982 Apr;18(1):21–28. doi: 10.1016/0378-1119(82)90052-x. [DOI] [PubMed] [Google Scholar]
- Gonda D. K., Radding C. M. By searching processively RecA protein pairs DNA molecules that share a limited stretch of homology. Cell. 1983 Sep;34(2):647–654. doi: 10.1016/0092-8674(83)90397-5. [DOI] [PubMed] [Google Scholar]
- Howard-Flanders P., West S. C., Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984 May 17;309(5965):215–219. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- Jones I. M., Primrose S. B., Ehrlich S. D. Recombination between short direct repeats in a recA host. Mol Gen Genet. 1982;188(3):486–489. doi: 10.1007/BF00330053. [DOI] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. Rapid assay for extrachromosomal homologous recombination in monkey cells. Mol Cell Biol. 1985 Mar;5(3):529–537. doi: 10.1128/mcb.5.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D. W., Taylor A. F., Smith G. R. Escherichia coli RecBC pseudorevertants lacking chi recombinational hotspot activity. J Bacteriol. 1983 Aug;155(2):664–680. doi: 10.1128/jb.155.2.664-680.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R., Schultz D. W., Crasemann J. M. Generalized recombination: nucleotide sequence homology between Chi recombinational hotspots. Cell. 1980 Mar;19(3):785–793. doi: 10.1016/s0092-8674(80)80054-7. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Morrison P., Kolodner R. Intramolecular recombination of linear DNA catalyzed by the Escherichia coli RecE recombination system. J Mol Biol. 1985 Dec 5;186(3):515–525. doi: 10.1016/0022-2836(85)90126-3. [DOI] [PubMed] [Google Scholar]
- Thompson R., Achtman M. The control region of the F sex factor DNA transfer cistrons: physical mapping by deletion analysis. Mol Gen Genet. 1979 Jan 16;169(1):49–57. doi: 10.1007/BF00267544. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Walker J. N., Dean P. D., Saunders J. R. Identification and characterisation of PmaCI an endonuclease of novel specificity from Pseudomonas maltophila. Nucleic Acids Res. 1986 Feb 11;14(3):1293–1301. doi: 10.1093/nar/14.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Suggs S. V., Miyoshi K., Bhatt R., Itakura K. A set of synthetic oligodeoxyribonucleotide primers for DNA sequencing in the plasmid vector pBR322. Gene. 1981 Dec;16(1-3):21–26. doi: 10.1016/0378-1119(81)90057-3. [DOI] [PubMed] [Google Scholar]
- Watt V. M., Ingles C. J., Urdea M. S., Rutter W. J. Homology requirements for recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4768–4772. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H., Weissmann C. Formation of genes coding for hybrid proteins by recombination between related, cloned genes in E. coli. Nucleic Acids Res. 1983 Aug 25;11(16):5661–5669. doi: 10.1093/nar/11.16.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C., Howard-Flanders P. Duplex-duplex interactions catalyzed by RecA protein allow strand exchanges to pass double-strand breaks in DNA. Cell. 1984 Jun;37(2):683–691. doi: 10.1016/0092-8674(84)90401-x. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]



