Summary
PTPMT1 was the first protein tyrosine phosphatase found localized to the mitochondria, but its biological function was unknown. Herein, we demonstrate that whole body deletion of Ptpmt1 in mice leads to embryonic lethality, suggesting an indispensable role for PTPMT1 during development. Ptpmt1-deficiency in mouse embryonic fibroblasts compromises mitochondrial respiration and results in abnormal mitochondrial morphology. Lipid analysis of Ptpmt1-deficient fibroblasts reveals an accumulation of phosphatidylglycerophosphate (PGP) along with a concomitant decrease in phosphatidylglycerol. PGP is an essential intermediate in the biosynthetic pathway of cardiolipin, a mitochondrial-specific phospholipid regulating the membrane integrity and activities of the organelle. We further demonstrate that PTPMT1 specifically dephosphorylates PGP in vitro. Loss of PTPMT1 leads to dramatic diminution of cardiolipin, which can be partially reversed by the expression of catalytic active PTPMT1. Our study identifies PTPMT1 as the mammalian PGP phosphatase and points to its role as a regulator of cardiolipin biosynthesis.
Introduction
Reversible phosphorylation is a major regulatory mechanism controlled by more than 500 kinases and 100 phosphatases (Alonso et al., 2004; Manning et al., 2002). Although the majority of kinases and phosphatases function on protein substrates, there are notable examples of kinases and phosphatases that regulate the production and breakdown of phospholipids. For instance, phosphoinositide 3-kinase (PI3K) and PTEN are two enzymes that regulate the abundance of key cellular phospholipids (Maehama and Dixon, 1998; Whitman et al., 1988). The importance of PTEN in tumor biology prompted us to identify additional phosphatases similar to PTEN (Pagliarini et al., 2004). We used the amino acid sequence surrounding the active site, HCKAGKGR, which includes two invariant basic amino acids (bold) along with the catalytically essential Cys-X5-Arg (CX5R) motif, to search for closely related phosphatases. To our surprise, we discovered a protein tyrosine phosphatase (PTP) that localized exclusively to the inner mitochondrial membrane (Pagliarini et al., 2005). This was the first CX5R phosphatase found in mitochondria, and because of its unique localization, we named it PTPMT1 (PTP localized to the Mitochondrion 1). This phosphatase is highly conserved and is present in animals, plants, and bacteria (Pagliarini et al., 2004).
Mitochondria are ubiquitous and dynamic organelles that participate in crucial cellular processes in eukaryotic organisms. They are responsible for the production of over 90% of cellular ATP (Voet, 2004). Mitochondria are also the sites of fatty acid oxidation, ketone body production, heme biosynthesis, cardiolipin metabolism, and coenzyme Q synthesis. In addition, mitochondria are a major source of reactive oxygen species. They harbor key steps of gluconeogenesis and the urea cycle, and are central to the mechanisms of apoptosis (Newmeyer and Ferguson-Miller, 2003; Voet, 2004).
Due to the unique localization of PTPMT1 in the inner mitochondrial membrane, we postulated that it plays important roles in oxidative phosphorylation or other mitochondrial functions. Therefore, we sought to elucidate the cellular function(s) and identify the endogenous substrate(s) of this phosphatase.
Results
Inactivation of the Ptpmt1 gene leads to embryonic lethality
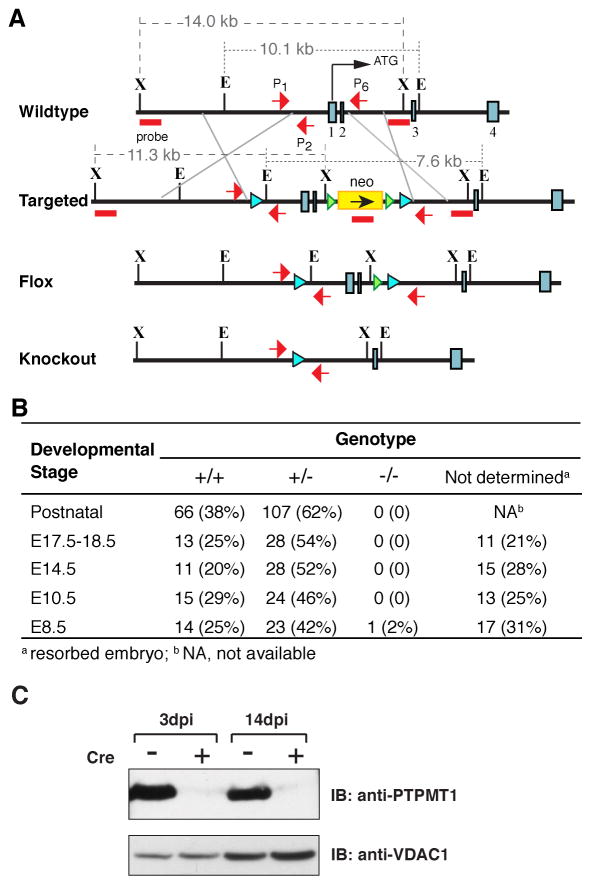
To study the physiological function(s) of PTPMT1, we genetically engineered mice in which the Ptpmt1 gene was ablated through homologous recombination (Figure 1A). Subsequently, we intercrossed Ptpmt1-heterozygous mice to examine the consequences of PTPMT1 deficiency. Out of 173 offspring born, 66 were wild type, 107 were heterozygous, and none were homozygous null (Figure 1B) (see Figure S1 for detailed characterization of mouse genotyping), indicating a Mendelian ratio typical of embryonic lethality. We next conducted timed mating experiments to determine the stage of the embryonic lethality. Embryos from E8.5, E10.5, E14.5, and E17.5-18.5 gestational stages were primarily wild type or heterozygous for the Ptpmt1 allele (Figure 1B). Further dissection identified an average of 26% resorption sites on the uterus lacking discernible embryos from all developmental stages (Figure 1B), suggesting that Ptpmt1−/− embryos implanted but died prior to E8.5. Hence, our results suggest that PTPMT1 plays an indispensable role in early development.
Figure 1. Inactivation of the Ptpmt1 gene leads to mouse embryonic lethality.
A) Gene targeting strategy. Exons (blue boxes), translation initiation site (ATG), selection gene (neomycin, neo), positions of 5′ and 3′ probes for Southern blot (red bars), PCR primer (red arrows), loxP sites ( ), Frt sites ( ), and restriction sites are indicated. X-XbaI, E-EcoRI. B) Genotype of offspring from Ptpmt1 heterozygous intercrosses at various developmental stages. C) Derivation of Ptpmt1-deficient mouse embryonic fibroblasts. Ptpmt1flox/flox MEFs were infected with recombinant adenoviruses encoding GFP(−) or GFP-Cre (+). Cells were harvested at the indicated time points post-infection for western blot analysis. dpi, days post infection. See also Figure S1.
Inhibition of mitochondrial respiration in cells lacking PTPMT1
To further investigate the physiological functions of PTPMT1, we generated a floxed allele of Ptpmt1 in mice for Cre-mediated conditional knockout (Ptpmt1+/flox) (Figure 1A and Figure S1B). We next derived Ptpmt1flox/flox mouse embryonic fibroblasts (MEFs) from E13.5 embryos obtained from intercrosses of Ptpmt1+/flox mice. The cells were then infected with control adenovirus or adenovirus expressing Cre recombinase. The expression of PTPMT1 protein was almost completely ablated in the Cre-treated (KO) cells 3 days after the infection and remained negligible for at least two weeks in culture (Figure 1C). The level of PTPMT1 mRNA transcripts was also greatly diminished in those cells (Figure S1C).
MEFs lacking PTPMT1 grow slowly. Three days after infection with Cre recombinase, the proliferation rate of KO cells was reduced 25% compared to that of control adenovirus-treated cells (Flox) (Figure S2A). Inhibition of cell growth was more pronounced in KO MEFs at day 6. A colony formation assay was also performed to examine the long-term effect of PTPMT1 deficiency on cell growth. Not surprisingly, KO MEFs formed fewer and smaller colonies than Flox cells (Figure S2B and S2C). It is noteworthy that Ptpmt1-null cells were not apoptotic or necrotic (data not shown) and had cell sizes similar to Flox MEFs (Figure S2D).
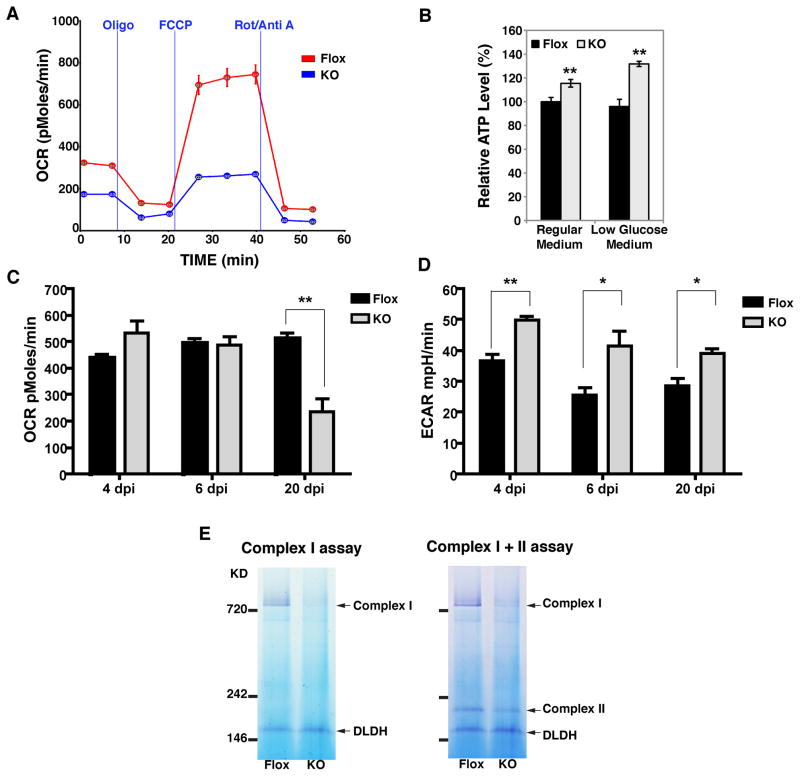
The position of PTPMT1 on the inner mitochondrial membrane places it within close proximity to electron transport chain complexes and enzymes of the tricarboxylic acid cycle. Therefore, we investigated the effects of PTPMT1 deletion on mitochondrial respiration and oxidative phosphorylation. Intact cell oxygen consumption rates (OCR) were measured at 13 days after deletion of Ptpmt1 (Figure 2A). The basal respiration rates were substantially lower in Ptpmt1 KO cells relative to that of Flox MEFs. We also assessed State 4 rates (a measurement of proton leakage across the inner membrane) by the addition of the ATP synthase inhibitor oligomycin, and maximal respiratory rates by the addition of the uncoupler, carbonylcyanide-ρ-trifluoromethoxyphenylhydrazone (FCCP). A dramatic reduction in maximal respiratory rates as well as state 4 rates was observed in the KO cells (Figure 2A), suggesting that PTPMT1 is required for optimal mitochondrial respiration. In contrast, when we concomitantly measured the cellular ATP content, an increase in ATP level was observed in Ptpmt1-deficient MEFs. The increase was more evident after pre-incubation of cells with low glucose medium (Figure 2B), consistent with our previous report that suppression of PTPMT1 expression by small-interfering RNA (siRNA) markedly enhances ATP levels in INS-1 832/13 cells in response to low glucose exposure (Pagliarini et al., 2005). Mitochondrial oxidative phosphorylation is a major source of cellular ATP production; however, it is known that cells can modulate the balance between aerobic and anaerobic (glycolytic) ATP production (Warburg, 1956) and that lowering extracellular glucose enhances glucose transport and utilization (Kletzien and Perdue, 1975). The diminished mitochondrial respiratory capacity in the presence of an elevated ATP level prompted us to hypothesize that glycolysis is increased in Ptpmt1-deficient cells. Therefore, we carried out a time course analysis of mitochondrial respiration versus glycolysis following low glucose incubation. Four and 6 days after Ptpmt1 deletion, the maximal oxygen consumption rates in Ptpmt1-deficient cells were not statistically different than those in Flox cells (Figure 2C). However, significant reduction of maximal respiration was observed in Ptpmt1-deficient cells 20 days later. Similar results were obtained in cells cultured in regular growth media (data not shown). To determine the glycolytic capacity of KO cells, we measured the rates of extracellular acidification as an indicator of glycolytic lactate production in Flox and KO MEFs after low glucose incubation (Parce et al., 1989; Wu et al., 2007). Indeed, a substantial increase of extracellular acidification rates (ECAR) was observed in KO MEFs at all time points, even as early as 4 days after the deletion of Ptpmt1 (Figure 2D). Additionally, when we measured cellular lactate production levels directly using a colorimetric assay, KO MEFs displayed a marked increase of lactate at day 13 (Figure S2E), when the inhibition of mitochondrial respiration was apparent (Figure 2A). Together, our results show that prolonged loss of PTPMT1 expression leads to a profound reduction of mitochondrial maximal respiration, concomitant with an increase in cellular ATP content. The increase of ATP levels in KO cells may result from an augmentation of glycolysis, although we cannot rule out the possibility that the utilization of ATP was also diminished, particularly since KO MEFs grow at a much slower rate than Flox cells (Figure S2, panels A–C).
Figure 2. Inhibition of mitochondrial respiration in Ptpmt1-deficient cells.
A) Oxygen consumption rates (OCR) of intact MEFs cultured in regular medium (25mM glucose) at 13dpi. The basal rate, the State 4 rate (with 1uM oligomycin, Oligo), and the maximal uncoupler stimulated rate (in the presence of FCCP) are shown. Rotenone (Rot) and antimycin A (Anti A) were used to block all electron flow through Complex I and III, demonstrating that non-mitochondrial oxygen consumption in these cells is negligible. Mean ± SEM from four replicates. B) Whole cell ATP contents in MEFs (13dpi) cultured in regular medium or pre-incubated with 3mM glucose containing medium overnight. Mean ± S.D., n=3, **p<0.01. Maximal uncoupler stimulated respiration rates (C) and extracellular acidification rate (ECAR) rates (D) of Ptpmt1-Flox and KO MEFs preincubated with low glucose (3mM) media overnight are obtained at the indicated time post adenoviral infection. Mean ± SEM from four replicates. **p<0.01, *p<0.05. E) At 13dpi, crude mitochondria were isolated from Flox or KO MEFs, solubilized in β-dodecylmaltoside, subjected to BN-PAGE, and followed by consecutive Complex I and Complex II in-gel activity assays. The activities of each complex are indicated by the appearance of a pink colored band. See also Figure S2.
To further understand the mechanism by which PTPMT1 reduces mitochondrial respiratory capacity, we isolated mitochondria from both Flox and KO MEFs at 13 days after deletion of the Ptpmt1 gene. The electron transport chain complexes were separated by blue-native polyacrylamide gel electrophoresis (BN-PAGE) followed by in-gel activity assays. A NADH: Nitrotetrazolium Blue (NTB) reductase assay was first carried out to examine the functional integrity of Complex I (Figure 2E, left panel). Under the same assay conditions, a lower band was also observed, which is likely to be dihydrolipoamide dehydrogenase (DLDH) (Yan and Forster, 2009), a component of multiple mitochondrial dehydrogenases. A profound reduction of Complex I activity was observed in cells lacking PTPMT1, whereas the DLDH activity remained unchanged. Subsequently, we determined the activity of Complex II by performing a succinate: NTB reductase assay on the same gel strip. In Ptpmt1-null mitochondria, Complex II activity was slightly decreased. (Figure 2E, right panel). Our results demonstrate that PTPMT1 deletion profoundly inhibits the activity of Complex I, consistent with the decreased respiratory rates we observe in those cells. Together, our results indicate that PTPMT1 is required for mitochondrial respiration, likely through its regulation of electron transport chain complexes.
PTPMT1 maintains mitochondrial membrane integrity
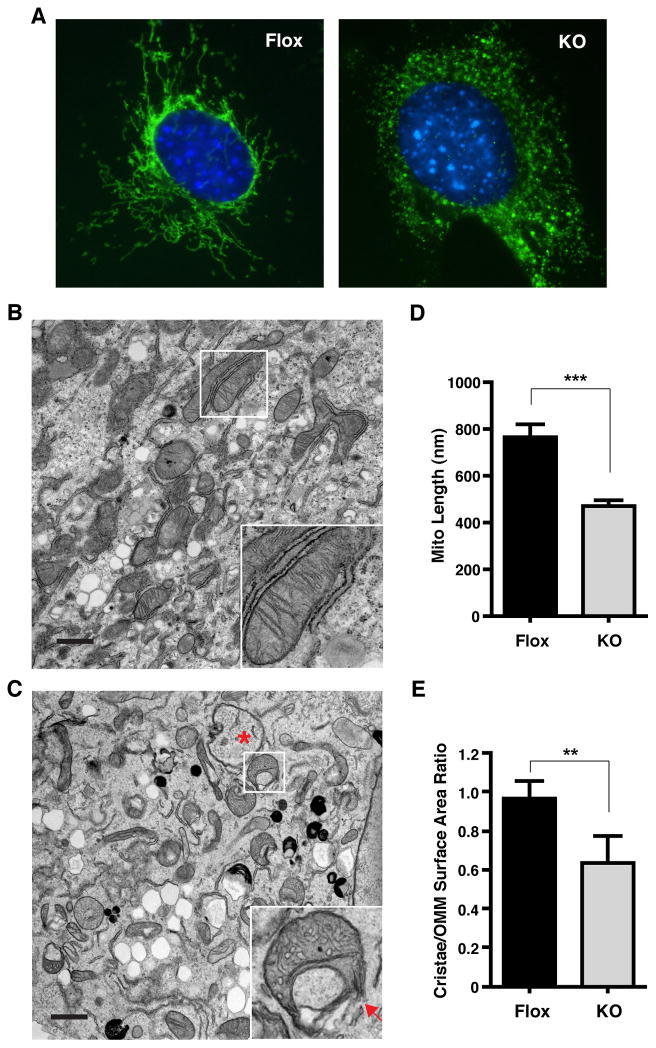
We next examined the effect of Ptpmt1 deletion on mitochondrial morphology using immunocytochemistry. Cytochrome c staining of Flox MEFs revealed a typical reticular mitochondrial network that distributed in a radial manner (Figure 3A, left panel). In contrast, KO MEFs exhibited a distinct punctate staining pattern, suggesting fragmentation of mitochondria (Figure 3A, right panel).
Figure 3. PTPMT1 maintains mitochondrial membrane integrity.
A) Ptpmt1-deficient MEFs have fragmented mitochondria. Two weeks after deletion of Ptpmt1, cells were stained with anti-cytochrome c antibody and visualized by immunofluorescence microscopy. DNA was stained blue with 4′,6-diamidino-2-phenylindole (DAPI). B) Electron micrographs of Flox MEFs. Normal mitochondria with lamellar cristae are enlarged in the inset. Scale bar, 1μm. C) EM images of defective mitochondria in KO cells. Inset shows a vesicular mitochondrion and rupture of the outer mitochondrial membrane/OMM (red arrow). Red asterisk, loss of cristae and matrix components. D) Morphometric analysis of mitochondrial length. Mean ± SEM, n=80. E) Morphometric analysis of cristae/OMM surface area ratio. Mean ± SEM, n=52. ***p<0.001, **p<0.01.
In order to further characterize this phenotype, the ultrastructure of the mitochondria was examined in Flox and KO MEFs by transmission electron microscopy. As shown in the electron micrograph in Figure 3B, Flox cells had many normal, elongated mitochondria with abundant cristae in a lamellar architecture (inset). In contrast, deletion of Ptpmt1 produced many smaller mitochondria with a variety of morphological abnormalities (Figure 3C), such as vesicular matrix compartments (inset) with ruptured outer mitochondrial membranes (red arrow), and degradation of cristae and matrix components (red asterisk). Mitochondrial length was subsequently determined by morphometric analysis of electron micrograph images. Not surprisingly, mitochondria were much shorter in KO cells than those in Flox cells (Figure 3D), which further supports that loss of PTPMT1 resulted in mitochondrial fragmentation. In addition, the loss of cristae in KO MEFs was confirmed by morphometric analysis comparing the ratio of cristae/outer mitochondrial membrane (OMM) surface area in Flox and KO cells (Figure 3E). Matrix vesicular formation has been observed in other paradigms of mitochondrial damage, such as apotosis (Sun et al., 2007). However, the distortion and deficiency of cristae in KO MEFs indicated that PTPMT1 is essential for the integrity of the inner mitochondrial membrane.
Accumulation of PGP in Ptpmt1-deficient cells
A full understanding of PTPMT1’s function requires identification of the endogenous substrate(s) of this phosphatase. Although initially characterized as a phosphoinositide phosphatase with a preference for phosphatidylinositol 5-phosphate (PI(5)P) in vitro, our results indicated that PI(5)P is not likely to be the physiological substrate of PTPMT1 (Pagliarini et al., 2004, 2005). In addition, the poor in vitro activity of PTPMT1 toward proteinaceous substrates prompted us to employ an unbiased search for a lipid substrate by comparing cellular phospholipid profiles of Flox and KO MEFs.
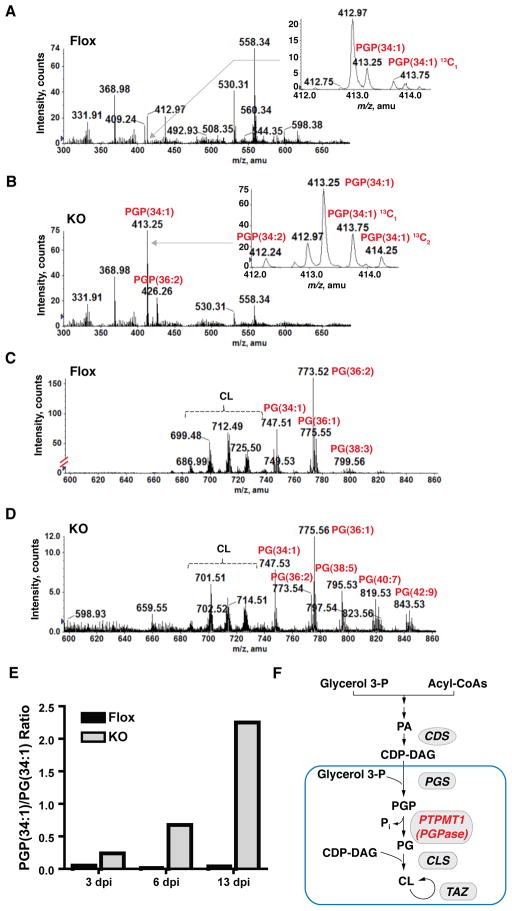
Total lipids from both cells were extracted and subjected to liquid chromatography followed by tandem mass spectrometry (MS/MS) analysis. Levels of phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol were comparable in both Flox and KO MEFs (data not shown). The abundance of the major phosphatidylglycerophosphate (PGP) species (34:1) as well as minor forms of PGP were markedly elevated in KO cells. In contrast, levels of all PGP species were barely detectable in Flox MEFs (Figure 4A and 4B, see Figure S3B for the identification of 34:1 PGP species by MS/MS analysis).
Figure 4. Loss of PTPMT1 leads to the accumulation of phosphatidylglycerophosphate in vivo.
Mass spectra of PGP in Flox (panel A) and KO (panel B) MEFs 13 days after deletion of Ptpmt1. Insets of panel A) and B) identified [M-2H]2− ions of 34:1 PGP (exact mass m/z 413.239) at m/z 413.25, its 13C1-isotope at m/z 413.75, 13C2-isotope at m/z 414.25, and 34:2 PGP at m/z 412.24. The total number of carbons and unsaturations in the fatty acyl chains are listed to indicate the molecular species of each ion. See also Figure S3. C) and D) Mass spectra [M-H]− ions of PG and [M-2H]2− ions cardiolipin (CL) in Flox and KO MEFs, respectively. The scales of panel C and D are set at different levels to facilitate the observation of PG and cardiolipin ions in KO cells. E) Ratio of 34:1 PGP to 34:1 PG at the indicated time after Ptpmt1 deletion. F) Schematic diagram of cardiolipin biosynthesis pathway. PA, phosphatidic acid; CDS, CDP-DAG synthase; PGS, PGP synthase; CLS, cardiolipin synthase; TAZ, cardiolipin remodeling enzyme, Tafazzin. Enzymatic reactions that take place in mitochondrion are enclosed within the blue rectangle.
In the 1950’s and 1960’s, Eugene Kennedy and his co-workers characterized the biosynthetic pathways for numerous cellular phospholipids including cardiolipin (Vance and Vance, 2008). In eukaryotes, glycerol-3-phosphate is first acylated to form phosphatidic acid and then converted to cytidinediphosphate diacylglycerol (CDP-DAG) by the enzyme CDP-DAG synthase. Subsequent steps in cardiolipin biosynthesis take place in the inner membrane of mitochondria. These steps involve the conversion of CDP-DAG to phosphatidylglycerol (PG) by the sequential action of PGP synthase and a PGP phosphatase. Cardiolipin is subsequently produced via condensation of PG with CDP-DAG by cardiolipin synthase (Figure 4F). Most of the biosynthetic enzymes in this pathway have been extensively studied except the PGP phosphatase whose identity has remained elusive in mammals. Our phospholipid profiling analyses show that PGP is markedly elevated in KO MEFs, indicating that PTPMT1 is the mammalian enzyme responsible for the conversion of PGP to PG. If PTPMT1 was the phosphatase that dephosphorylates PGP, we would expect to find PG levels decreased in KO cells. As predicted, while several molecular forms of PG were identified in the Flox cells (Figure 4C), we observed a substantial reduction in all PG species in cells lacking PTPMT1 (Figure 4D). To further analyze the change in PGP and PG levels induced by loss of PTPMT1, MEFs were harvested at 3, 6, and 13 days after Ptpmt1 ablation. The ratio of 34:1 PGP/PG was determined by mass spectrometry analysis (Figure 4E). Notably, an increase in the PGP/PG ratio was found in KO MEFs as early as 3 days after the deletion of Ptpmt1 (Figure 1C). Moreover, the PGP/PG ratio was augmented in a time-dependent manner in KO cells as compared to that of Flox MEFs. A similar accumulation of PGP and a reduction of PG were observed in C2C12 mouse myoblasts in which PTPMT1 was knocked down by small interfering RNA in a separate lipid analysis (data not shown). These results indicate that PTPMT1 specifically catalyzes the conversion of PGP to PG in multiple cell types.
PTPMT1 dephosphorylates PGP in vitro
To determine whether PTPMT1 possesses PGP phosphatase activity in vitro, we enzymatically synthesized radiolabeled PGP. Briefly, phosphatidyl-sn-[U-14C]glycerol phosphate (14C-PGP) was produced by incubating CDP-DAG with sn-[U-14C] glycerol 3-phosphate in the presence of purified PGP synthase (PGS) from E. coli (Dowhan, 1992). An overnight incubation resulted in the quantitative conversion of starting materials to 14C-PGP as assessed by thin layer chromatography (TLC) (data not shown).
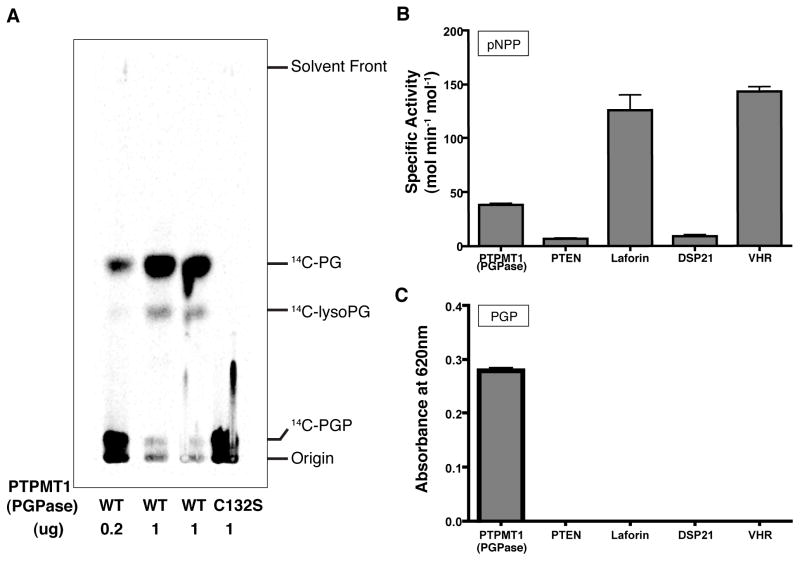
The 14C-PGP was then incubated with purified recombinant PTPMT1. The radioactive reaction products were extracted and separated by TLC analysis. 14C-labeled PGP remained near the origin, whereas 14C-PG migrated to the middle of the TLC plate (Figure 5A), as indicated by the co-migration of non-radiolabeled PG standards (data not shown). While recombinant PTPMT1 efficiently converted PGP to PG in a dose-dependent manner, the catalytically inactive mutant of PTPMT1 (C132S) failed to catalyze the conversion. Our results demonstrate that PTPMT1 dephosphorylates PGP in vitro. To rule out the possibility that the PGP phosphatase activity is a general feature of the PTP superfamily, we compared the activity of PTEN, laforin, DSP21, and VHR against PGP to that of PTPMT1. The specific activity of each enzyme using pNPP as a substrate is consistent with previously reported values (Figure 5B) (Denu et al., 1995; Maehama and Dixon, 1998; Rardin et al., 2008; Worby et al., 2006). PTPMT1 was more efficient at removing phosphate from 14C-PGP than the other phosphatases (Figure 5C), suggesting that the PGP phosphatase activity is not a common property shared by PTP superfamily members. Taken together, our results strongly suggest that PGP is the physiological substrate of PTPMT1.
Figure 5. PTPMT1 dephosphorylates phosphatidylglycerophosphate in vitro.
A) Conversion of 14C-labeled PGP to 14C-PG by recombinant wild type (WT) versus catalytically inactive (C132S) PTPMT1. A trace amount of radiolabeled lysoPG was generated during the reaction. B) The specific activities of PTEN, Laforin, DSP21, and VHR are compared with that of PTPMT1 using pNPP as the substrate. Mean ± S.D., n=3. C) PGP phosphatase activity is not a general feature of protein tyrosine phosphatases. The activity of each enzyme using PGP as the substrate is indicated by the absorbance at 620nm using malachite green reagents to measure the release of inorganic phosphate (Mean ± S.D., n=3).
PTPMT1 is required for cardiolipin biosynthesis
As the conversion of PGP to PG is an integral part of the de novo cardiolipin biosynthetic pathway, we predict that loss of PTPMT1 would lead to a reduction in cardiolipin content in the cells. Cardiolipin is one of the major components of the mitochondrial inner membrane (Daum, 1985). It optimizes mitochondrial respiration through its interactions with electron transport chain complexes and multiple carrier proteins including the adenine nucleotide transporter, as well as stabilizing the supercomplex formation of respiratory enzymes (Hatch, 1998; Houtkooper and Vaz, 2008; Schlame et al., 2000). As such, it is understandable that Ptpmt1-deficient cells would have profound defects in mitochondrial respiratory capacity and inner mitochondrial membrane morphology due to cardiolipin deficiency.
Notably, our mass spectrometry analysis of phospholipids revealed a marked decrease in cardiolipin (CL) content in KO MEFs 13 days after deletion of Ptpmt1 (Figure 4C and 4D). Moreover, both the content and the acyl chain composition of cardiolipin were altered in KO cells. To further quantify the effect of PTPMT1 on cardiolipin metabolism, we next determined the steady state phospholipid composition of Flox versus KO MEFs by labeling intact cells with 32P-orthophosphate. Two weeks after Cre recombinase infection, cardiolipin and PG levels in KO MEFs were reduced to 36% and 39% of those in Flox cells, respectively (Table 1), indicating that PTPMT1 is required for the biosynthesis of both lipids. Notably, the abundance and composition of cardiolipin remained indistinguishable in Flox and KO MEFs at 3 and 6 days after Ptpmt1 deletion (Figure S3, panels C–D), which in turn explained why the mitochondrial oxygen consumption rates were not reduced in KO cells at early time points (Figure 2C). The time-dependent loss of cardiolipin in KO cells may reflect the slow turnover rate of this mitochondrial lipid, which may be influenced by a combination of factors including degradation and recycling (Schlame, 2008). It is important to note that the relative abundance of phosphatidic acid, phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol were not affected by the loss of PTPMT1 (Table 1). The level of sphingomyelin was decreased to a lesser extent in the KO MEFs. The physiological significance of this reduction remains unclear. Collectively, our data demonstrate that PTPMT1 is required for cardiolipin biosynthesis, consistent with its function as the PGP phosphatase.
Table 1. Phospholipid Composition of Ptpmt1 Flox and KO MEFs.
After 13 days of adenoviral infection, cells were cultured in the medium containing 32Pi (1uCi/ml) for 4 days. Total cellular phospholipids were extracted and analyzed as described under the Experimental Procedures. Data shown are the Mean ± S.E.M. for three independent experiments and were statistically analyzed using a Student’s t-test.
| Flox | KO | |
|---|---|---|
| % of total phospholipids | ||
| Cardiolipin | 2.8 ± 0.2 | 1.0 ± 0.3*** |
| Phosphatidylglycerol | 1.8 ± 0.3 | 0.7 ± 0.2** |
| Phosphatidic acid | 1.6 ± 0.3 | 1.5 ± 0.4 |
| Phosphatidylcholine | 39.9 ± 2.0 | 45.1 ± 2.4 |
| Phosphatidylethanolamine | 28.1 ± 0.6 | 28.1 ± 0.5 |
| Phosphatidylserine | 11.0 ± 1.0 | 10.3 ± 1.4 |
| Phosphatidylinositol | 11.2 ± 0.5 | 10.7 ± 0.3 |
| Sphingomyelin | 3.7 ± 0.3 | 2.6 ± 0.4* |
p < 0.001,
p < 0.01,
p< 0.05 between Flox and KO MEFs
The catalytic activity of PTPMT1 is essential for cardiolipin synthesis
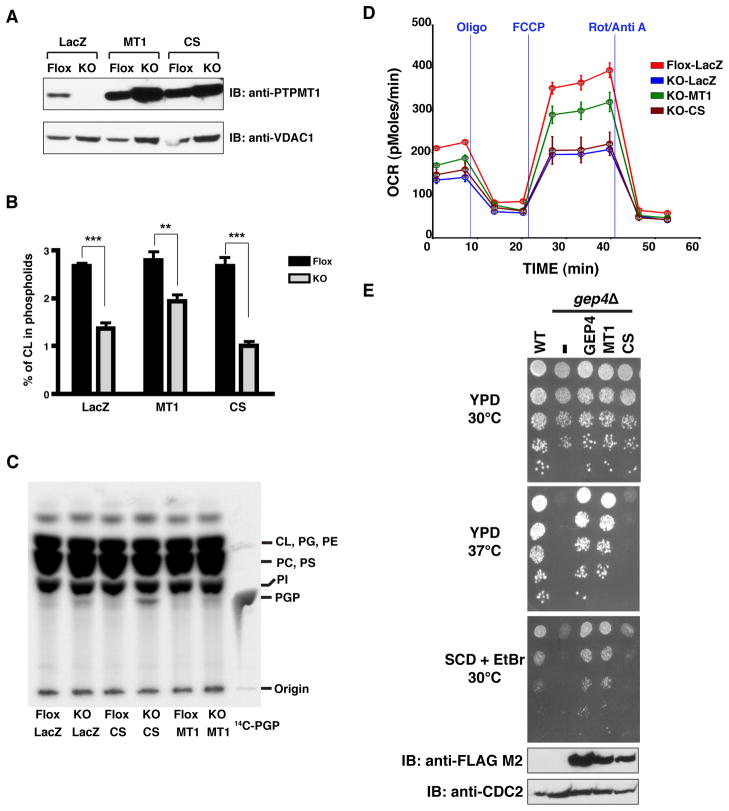
To verify that the catalytic activity of PTPMT1 is required for cardiolipin biosynthesis, we next examined whether wildtype or a phosphatase inactive mutant (C132S) of PTPMT1 is able to rescue the mitochondrial defects in KO cells. One week after deletion of Ptpmt1, MEFs were infected with recombinant adenoviruses encoding β-galactosidase (LacZ), wildtype or mutant PTPMT1. Cardiolipin content and mitochondrial respiration were subsequently analyzed after rescuing for 7 days. Both wildtype and mutant PTPMT1 efficiently expressed in the cells (Figure 6A), but expression of the CS mutant severely impaired the cell growth of KO MEFs (data not shown). Nonetheless, we examined steady state phospholipid composition. The abundance of cardiolipin was greatly diminished in KO MEFs expressing LacZ, compared to that of the LacZ-expressing Flox cells (Figure 6B), consistent with our earlier observation (Table 1). Interestingly, overexpression of wildtype PTPMT1 partially restored the cardiolipin content of KO MEFs. In contrast, the cardiolipin level was further decreased in KO cells expressing the CS mutant, suggesting that the catalytic activity of PTPMT1 is important for cardiolipin biosynthesis. Given the dramatic alteration of mitochondrial morphology in KO cells, it is conceivable that overexpression of wildtype PTPMT1 may not be able to fully reverse the structural defect and cardiolipin deficiency in the time frame of our experiment. Moreover, the abundance of PGP was simultaneously examined by TLC analysis (Figure 6C). PGP was barely detectable in Flox cells, whereas a distinct PGP band was found in KO MEFs expressing LacZ, as indicated by the co-migration of 14C-labeled PGP. As expected, wildtype PTPMT1 alleviated the accumulation of PGP in KO cells. In contrast, overexpression of the PTPMT1 CS mutant in Ptpmt1-null cells substantially augmented cellular PGP content, suggesting this mutant protein may function as a trap to accumulate PGP. Similar “substrate-trapping” mutants have been studied in many members of PTP family to stabilize and identify their endogenous targets (Tonks and Neel, 2001). In contrast, the CS mutant had a minimal effect on the levels of cardiolipin and PGP in Flox cells, likely because the endogenous PTPMT1 can efficiently convert PGP to PG. To rescue the mitochondrial defects in Ptpmt1-deficient cells, we next overexpressed either wildtype or mutant PTPMT1 and measured mitochondrial respiration rates of those cells (Figure 6D). As anticipated, intact cell oxygen consumption rates were much slower in LacZ-expressing KO MEFs, compared to that of Flox cells. Wildtype PTPMT1 partially reversed the inhibition of mitochondrial respiration in Ptpmt1-null cells, whereas CS mutants failed to rescue the phenotype. Together, our results indicate that the catalytic activity of PTPMT1 is indeed essential for cardiolipin synthesis and mitochondrial function.
Figure 6. Wildtype but not an active site mutant PTPMT1 rescues cardiolipin deficiency.
A) The expression of β-galactosidase (LacZ), wildtype PTPMT1 (MT1), or catalytically inactive PTPMT1 mutant (CS). B) After labeling MEFs with 32P-orthophosphate, phospholipids were extracted and the percentage of cardiolipin (CL) among total phospholipids was determined by TLC analysis. Mean ± S.E.M. from three independent experiments, one-way ANOVA with a post hoc F test, ***p<0.001, **p<0.01. C) The accumulation of PGP in Ptpmt1-deficient cells. For each sample, 50,000cpm of 32P-labeled phospholipids were applied onto TLC plates and visualized by phosphoimager. D) Oxygen consumption rates of intact MEFs cultured in regular medium. The basal rate, the State 4 rate (Oligo), and the maximal uncoupler stimulated rate (FCCP) are shown as described in Figure 2. Mean ± SEM from four replicates. E) Wildtype but not mutant PTPMT1 complements the growth deficiency of yeast PGP phosphatase GEP4-null (gep4Δ) cells in a serial dilution spotting assay. The expression levels of FLAG-tagged GEP4 and PTPMT1 are indicated by western blot analysis. The level of CDC2 is shown as control. WT, wildtype strain. (−) empty vector. SCD, synthetic complete medium with dextrose. EtBr, Ethidium Bromide. See also Figure S4 and Table S1.
PTPMT1 functionally replaces yeast PGP phosphatase GEP4
Putative PTPMT1 orthologs have been identified in a variety of evolutionarily distinct species, except the budding yeast S. cerevisiae (Pagliarini et al., 2004). Osman et al recently reported that GEP4 functions as a PGP phosphatase in yeast mitochondria (Osman et al., 2010). GEP4 is a member of the haloacid dehydrogenase superfamily and is present only in fungi and plants. Therefore, we sought to determine whether PTPMT1 serves as a functional equivalent of Gep4 in yeast.
FLAG-tagged GEP4, wildtype PTPMT1, or mutant PTPMT1 was first transformed into GEP4-null strain and the expression was confirmed by western blot analysis (Figure 6E, bottom two panels). PTPMT1 contains an N-terminal mitochondrial targeting sequence (Pagliarini et al., 2005), which we hypothesized, would also direct PTPMT1 to the yeast mitochondria. Indeed, our subcellular fractionation analysis indicated that PTPMT1 preferentially accumulated in the yeast mitochondria similar to GEP4 (data not shown). Consistent with the observation by Osman et al., the yeast strain that lacks GEP4 did not grow at 37°C on glucose-containing medium (Figure 6E, second panel). Both GEP4 and wildtype PTPMT1 efficiently rescued the temperature sensitive growth defects. However, the PTPMT1 CS mutant failed to complement the loss of GEP4. In addition, gep4Δ cells were unable to grow in the presence of ethidium bromide, an agent that induces loss of mitochondrial DNA and defects in cell wall (Janitor and Subik, 1993; Zhong et al., 2005). Notably, overexpression of wildtype PTPMT1 greatly improved the growth of gep4Δ cells in ethidium bromide containing medium, similar to that of GEP4-expressing cells (Figure 6E, third panel). Our lipid analysis indicated that wildtype, but not mutant PTPMT1, rescued the cardiolipin deficiency of gep4Δ cells (Figure S3C). Together, our results demonstrate that PTPMT1 complements the loss of GEP4 in yeast and this rescue is dependent on the phosphatase activity of PTPMT1.
Discussion
Over 100 human genes have been identified belonging to the PTP superfamily, all of which contain a highly conserved CX5R motif in the active site for catalysis (Alonso et al., 2004; Tonks, 2006). Most members of the PTP family are key mediators of signal transduction through their dephosphorylation of proteins and phosphoinositides (Tonks, 2006). The unique subcellular localization of PTPMT1 prompted us to investigate whether it plays an important role in mitochondrial signaling. In the present study, we discovered that PTPMT1 dephosphorylates PGP, a glycerophospholipid, and is essential for mitochondrial function through its regulation of cardiolipin biosynthesis. Our findings strongly suggest that phosphatases in the PTP family can be involved in cellular processes other than signal transduction. It will be of interest to determine whether some of the less characterized PTP family members target unorthodox substrates like PTPMT1.
The enzyme reaction catalyzed by the PGP phosphatase was first characterized by Kennedy et al. in 1963 using rat and chicken liver mitochondria (Kiyasu et al., 1963). This reaction, which is conserved in bacteria, is catalyzed by PgpA, PgpB, and the recently identified PgpC, three bacterial enzymes that display no homology to PTPMT1 (Icho and Raetz, 1983; Lu et al., 2011). Osman et al recently reported that GEP4 dephosphorylates PGP in yeast (Osman et al., 2010). GEP4 is a member of the haloacid dehydrogenase superfamily which contains a DxDx(T/V) motif in the active site and utilizes a unique Asp-based catalytic mechanism (Kerk et al., 2008). GEP4 orthologs exist in fungi and plant, but are not found in higher eukaryotes, providing little insight into the identity of the mammalian PGP phosphatase. On the other hand, PTPMT1 is one of the most widely distributed and highly conserved PTPs, with over 60 orthologs identified in Animalia, Plantae, Protista, and Eubacteria (Pagliarini et al., 2004), which raises the possibility that PTPMT1 orthologs also utilize PGP as their substrates. Although a PTPMT1-like sequence is not present in Saccharomyces cerevisiae, our results demonstrate that mammalian PTPMT1 serves as a functional equivalent of GEP4 in yeast (Figure 6E). PTPMT1, GEP4, PgpA, PgpB, and PgpC are structurally and evolutionarily distinct enzymes with distinct regulatory mechanisms for their activities (Table S2) (Kerk et al., 2008; Kumaran et al., 2006; Pagliarini et al., 2004; Stukey and Carman, 1997). Why nature has utilized such divergent phosphatases to remove the phosphate from PGP is presently unclear, but it may reflect fundamental differences among organisms to regulate cardiolipin synthesis and possibly energy metabolism.
Substantial accumulation of PGP was observed in cells lacking PTPMT1 (Figure 4B). Notably, PGP exists in cells at low abundance (Kiyasu et al., 1963; MacDonald and McMurray, 1980). Little is known about the function of PGP in cells other than it is an intermediate in the cardiolipin biosynthetic pathway. However, we found that depletion of PTPMT1 not only inhibited cell growth (Figure S2, panels A–C), but also led to an increase in cellular ATP content and glycolysis (Figure 2B and 2D). This phenomenon was observed even before the onset of cardiolipin deficiency (Figure S3, panels C–D) and inhibition of mitochondrial respiratory capacity (Figure 2C), implying a possible role for PGP or other intermediates of the pathway in the regulation of aerobic and anaerobic metabolism. It will be interesting to determine whether deletion of Ptpmt1 alters the activity of glycolytic enzymes or the abundance of glycolytic intermediates as a result of PGP accumulation. Indeed, several enzymes of the glycolytic pathway have been found associated with mitochondrial membranes, such as hexokinase on the outer membrane (Wilson, 2003) and FAD-linked glycerol-3-phosphate dehydrogenase on the inner membrane (Klingenberg, 1970). In addition, it is likely that the rate of energy utilization is diminished in the slower growing KO cells, which in turn may contribute to the overall increase of cellular ATP content. Further investigation of glycolytic intermediates, glycolytic rates, and ATP consumption in both Flox and KO MEFs is clearly warranted and is ongoing in our lab.
Our results found that Ptpmt1−/− embryos implanted but died prior to E8.5, suggesting an essential role for the Ptpmt1 gene during early embryonic development. However, the cause of embryonic lethality is not fully understood. It is noteworthy that the demand for mitochondria varies at different embryonic developmental stages. In the early embryo, mitochondria are maternally inherited and their population remains relatively constant until after implantation, when rapid mitochondrial biogenesis and mitochondrial DNA replication resume in response to the increasing demand for energy production during organogenesis (Cerritelli et al., 2003; Van Blerkom, 2009; Hance et al., 2005; Larsson et al., 1998). Therefore, it is possible that PTPMT1 is required for the increased mitochondrial biogenesis that occurs in post-implantation embryos.
Identifying the mammalian PGP phosphatase is of interest not only because it is a missing step of lipid biosynthesis, but also due to its potential implication in many cardiolipin deficiency-related diseases, such as ischemia/reperfusion injury, heart failure, aging, and neurodegeneration (Chicco and Sparagna, 2007; Houtkooper and Vaz, 2008). It is noteworthy that the reactive cysteine in PTP family phosphatases is highly susceptible to oxidative stress (Tonks, 2005). PTPMT1 is tethered to the inner mitochondrial membrane, where its activity could be regulated by mitochondrial reactive oxygen species (ROS). Loss of two copies of the Ptpmt1 gene in all tissues is embryonic lethal; however, a single copy of a functional Ptpmt1 gene could result in an organism being highly susceptible to episodes of oxidative stress. Furthermore, loss of cardiolipin content in mitochondria has been well documented in cardiac ischemia/reperfusion injury (Chicco and Sparagna, 2007), but the underlying molecular mechanism is poorly understood. Hence, our identification of a potentially redox-sensitive enzyme in the cardiolipin synthesis pathway raises the question of whether inactivation of PTPMT1 by mitochondrial ROS contributes to cardiolipin deficiency under those conditions.
In conclusion, our work has identified the substrate of PTPMT1 by comparing phospholipid analyses coupled with mass spectrometry of lipids extracted from wild type versus knock out cells. These analyses provided us with the key to understanding the biological functions of PTPMT1 within mitochondria. This approach can be applied to understanding the molecular mechanisms underlying other alterations in phospholipid metabolism. Finally, in addition to the discovery of a lipid biosynthetic enzyme, we demonstrated that PTPMT1 functions as an important regulator of mitochondrial physiology, which may provide perspectives into the etiology of many mitochondria deficiency-related diseases.
Experimental Procedures
Cell culture and reagents
MEFs and C2C12 mouse myoblast cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 100 units/ml penicillin, 100 mg/ml streptomycin, 2 mM glutamine, and cultured at 37°C in a humidified incubator with 5% CO2. The expression and purification of recombinant PTPMT1, PTEN, laforin, DSP21, and VHR were previously described (Denu et al., 1995; Maehama and Dixon, 1998; Pagliarini et al., 2004; Rardin et al., 2008; Worby et al., 2006). 1,2-dioleoyl-sn-glycerol-3-cytidine diphosphate (ammonium salt) and 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL), DL-α-glycerol phosphate dipotassium salt was from Sigma, sn-[U-14C] glycerol 3-phosphate disodium salt was from American Radiolabeled Chemicals, Inc. (Saint Louis, MO).
Mice
Generation of Ptpmt1 knockout mice is described in the Supplemental Information. All procedures were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of University of California San Diego.
Derivation and adenoviral infection of MEFs
MEFs from E13.5 embryos from intercrossed of Ptpmt1+/flox mice were isolated, cultured, genotyped, and immortalized. For transduction of Ptpmt1flox/flox fibroblasts with the Cre recombinase, cells were infected with adenoviruses encoding either GFP or GFP-Cre (Gene transfer vector core, University of Iowa). Adenoviral vectors containing untagged wildtype or C132S mutant of murine PTPMT1 were constructed into pAd/CMV/V5-DEST vector (Invitrogen) and adenoviruses were generated and purified. pAd/CMV/V5-GW/lacZ (Invitrogen) was used as the control.
Western blot and Immunofluorescence Analysis
The western blots were performed using antibodies against PTPMT1 (Pagliarini et al., 2005), VDAC1 (Calbiochem), FLAG M2 (Sigma), and CDC2 (Santa Cruz). For immunofluorescent staining, MEFs were plated on glass coverslips, fixed with 3.7% formaldehyde and then incubated with anti-cytochrome c (BD Biosciences) antibody and Alexa Fluor 488 goat anti-mouse-conjugated secondary antibody (Molecular Probes). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Molecular Probes).
Liquid Chromatography/Tandem Mass Spectrometry Analysis (LC/MS/MS)
Total cellular phospholipids were extracted (Bligh and Dyer, 1959) and quantified using normal phase Liquid Chromatography coupled to a QSTAR XL tandem mass spectrometer (Applied Biosystems, Foster City, CA) equipped with an electrospray source (see the Supplemental Information for details). Data acquisition and analysis were performed using the instrument’s Analyst QS software.
PGP Phosphatase Assay
14C-labeled PGP was synthesized as described (Dowhan, 1992). The PGP phosphatase activities of recombinant PTPMT1, PTEN, Laforin, DSP21, and VHR were determined by TLC or malachite green analysis (see Supplemental Information).
Phospholipid Quantification
Cellular phospholipids were extracted (Bligh and Dyer, 1959), and separated by TLC using Chloroform/Methanol/ammonium hydroxide/water (130:75:2:6). For the separation of PGP, ethylacetate/2-propanol/ethanol/6% ammonia (3:9:3:9) was used as TLC solvent (Osman et al., 2010). Individual species were identified by co-migration of nonradiolabeled standards. Their relative intensities were analyzed using the Typhoon 9410 Variable Mode Imager and ImageQuant TL software (GE Healthcare Life Science).
Mitochondrial Functional Analyses
Oxygen consumption (OCR) and extracellular acidification rates (ECAR) were measured using a Seahorse Extracellular Flux Analyzer XF24 (Seahorse Bioscience, North Billerica, MA) (Wu et al., 2007). 4X104 cells were plated into each well prior to the assay. OCR and ECAR were normalized by the DNA content. (See Supplemental Information for details). The data was presented as the means ± SEM for four replicates. Qualitatively similar results were obtained in three separate experiments. Differences in rates were analyzed by one-way ANOVA with a post hoc F test. For measurement of cellular ATP content, MEFs were plated and grown overnight in regular or low glucose (3mM) media and whole cell ATP content was measured by CellTiter-Glo Luminescent Cell Viability/ATP Assay kit (Promega), and normalized by DNA content. Statistical analysis was performed using one-way ANOVA with a post hoc F test. Cellular lactate was measured by a colorometric assay kit (Abcam) and normalized by total protein amount. Differences between Flox and KO MEFs were examined for statistical significance using a Student’s t-test.
Blue-native PAGE
Subconfluent MEFs were harvested and crude mitochondria were isolated by differential centrifugation (Pallotti and Lenaz, 2001). 20ug of mitochondria were solubilized on ice with 1% β-dodecylmaltoside (Invitrogen) for 30 min and further separated by 4–16% Native PAGE gel (Invitrogen) (Schagger and von Jagow, 1991; McKenzie et al., 2007). To assess the activity of Complex I, the gel was incubated in 2mM Tris/HCl, pH7.4 containing 0.1mg/ml NADH and 2.5mg/ml nitrotetrazolium blue (Zerbetto et al., 1997). Subsequently, the gel was incubated in 2mM Tris/HCl, pH7.4 buffer in the presence of 20mM sodium succinate, 2mM phenazine methosulfate, and 2.5mg/ml nitrotetrazolium blue for the Complex II in-gel activity assay (Wittig et al., 2007).
Transmission Electron Microscopy and Morphometric Analysis
Cells were grown on 35 mm glass bottom culture dishes (MatTek, Ashland, MA). Sample preparation and conventional EM analysis were carried out as described in the Supplemental Information. Mitochondrial lengths and cristae/OMM surface area ratio were measured using ImageJ (NIH). Differences between Flox and KO MEFs were examined for statistical significance using a Student’s t-test.
Yeast Complementation Assays
Yeast strains used in this study are derivatives of S288C (Brachmann et al., 1998). GEP4 knockout strains are obtained from Open Biosystems and verified by gene-specific PCR primer pairs. The plasmids for complementation assays in yeast were constructed by inserting the PCR products of GEP4, M. musculus PTPMT1 (wildtype), mutant PTPMT1 (C132S) with C-terminal FLAG tag into pRS415-GPD vector (ATCC). Subsequently, gep4Δ cells were transformed with plasmids containing GEP4, PTPMT1, or empty vector. Cells were grown in the appropriate selective medium at 30°C to mid-log phase, counted and followed by 5-fold serial dilution. Each dilution was spotted onto either YPD plates or Synthetic Completed Medium with Dextrose (SCD) in the presence of 25ug/ml ethidium bromide, and cultured at the indicated temperature for 2 days.
Full Experimental Procedures and associated references are enclosed in Supplementary Information.
Supplementary Material
Highlights.
Inactivation of PTPMT1 results in early embryonic lethality in mice
Loss of PTPMT1 inhibits mitochondrial respiration and alters mitochondrial structure
PTPMT1 dephosphorylates phosphatidylglycerophosphate
PTPMT1 is essential for cardiolipin biosynthesis
Acknowledgments
We thank Ju Chen and Matthew Rardin for strains and reagents and Gregory Taylor, David Pagliarini, Matthew Gentry, Mikhail Bogdanov, as well as members of the Dixon laboratory for technical assistance. We thank Claudia Kent, Vincent Tagliabracci, and Seema Matto for critically reading the manuscript. This work was supported by grants from the National Institutes of Health (DK18024 and DK18849 to J.E.D., DK054441 to A.N.M., GM56389 to W.D., GM069338 to the LIPID MAPS Large Scale Collaborative Grant, and RR04050 to the National Center for Microscopy and Imaging Research) and the Larry L. Hillblom Foundation postdoctoral fellowship to J. Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol Cell Physiol. 2007;292:C33–44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- Daum G. Lipids of mitochondria. Biochim Biophys Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- Denu JM, Zhou G, Wu L, Zhao R, Yuvaniyama J, Saper MA, Dixon JE. The purification and characterization of a human dual-specific protein tyrosine phosphatase. J Biol Chem. 1995;270:3796–3803. doi: 10.1074/jbc.270.8.3796. [DOI] [PubMed] [Google Scholar]
- Dowhan W. Phosphatidylglycerophosphate synthase from Escherichia coli. Methods Enzymol. 1992;209:313–321. doi: 10.1016/0076-6879(92)09039-6. [DOI] [PubMed] [Google Scholar]
- Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum Mol Genet. 2005;14:1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- Hatch GM. Cardiolipin: biosynthesis, remodeling and trafficking in the heart and mammalian cells (Review) Int J Mol Med. 1998;1:33–41. doi: 10.3892/ijmm.1.1.33. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Vaz FM. Cardiolipin, the heart of mitochondrial metabolism. Cell Mol Life Sci. 2008;65:2493–2506. doi: 10.1007/s00018-008-8030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icho T, Raetz CR. Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J Bacteriol. 1983;153:722–730. doi: 10.1128/jb.153.2.722-730.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janitor M, Subik J. Molecular cloning of the PEL1 gene of Saccharomyces cerevisiae that is essential for the viability of petite mutants. Curr Genet. 1993;24:307–312. doi: 10.1007/BF00336781. [DOI] [PubMed] [Google Scholar]
- Kerk D, Templeton G, Moorhead GB. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 2008;146:351–367. doi: 10.1104/pp.107.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyasu JY, Pieringer RA, Paulus H, Kennedy EP. The biosynthesis of phosphatidylglycerol. J Biol Chem. 1963;238:2293–2298. [PubMed] [Google Scholar]
- Kletzien RF, Perdue JF. Induction of sugar transport in chick embryo fibroblasts by hexose starvation. Evidence for transcriptional regulation of transport. J Biol Chem. 1975;250:593–600. [PubMed] [Google Scholar]
- Klingenberg M. Localization of the glycerol-phosphate dehydrogenase in the outer phase of the mitochondrial inner membrane. Eur J Biochem. 1970;13:247–252. doi: 10.1111/j.1432-1033.1970.tb00924.x. [DOI] [PubMed] [Google Scholar]
- Kumaran D, Bonanno JB, Burley SK, Swaminathan S. Crystal structure of phosphatidylglycerophosphatase (PGPase), a putative membrane-bound lipid phosphatase, reveals a novel binuclear metal binding site and two “proton wires”. Proteins. 2006;64:851–862. doi: 10.1002/prot.21039. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Lu YH, Guan Z, Zhao J, Raetz CR. Three Phosphatidylglycerol-phosphate Phosphatases in the Inner Membrane of Escherichia coli. J Biol Chem. 286:5506–5518. doi: 10.1074/jbc.M110.199265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald PM, McMurray WC. Partial purification and properties of mammalian phosphatidylglycerophosphatase. Biochim Biophys Acta. 1980;620:80–89. doi: 10.1016/0005-2760(80)90187-3. [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Analysis of mitochondrial subunit assembly into respiratory chain complexes using Blue Native polyacrylamide gel electrophoresis. Anal Biochem. 2007;364:128–137. doi: 10.1016/j.ab.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Osman C, Haag M, Wieland FT, Brugger B, Langer T. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 2010;29:1976–1987. doi: 10.1038/emboj.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Wiley SE, Kimple ME, Dixon JR, Kelly P, Worby CA, Casey PJ, Dixon JE. Involvement of a mitochondrial phosphatase in the regulation of ATP production and insulin secretion in pancreatic beta cells. Mol Cell. 2005;19:197–207. doi: 10.1016/j.molcel.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Worby CA, Dixon JE. A PTEN-like phosphatase with a novel substrate specificity. J Biol Chem. 2004;279:38590–38596. doi: 10.1074/jbc.M404959200. [DOI] [PubMed] [Google Scholar]
- Pallotti F, Lenaz G. Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol. 2001;65:1–35. doi: 10.1016/s0091-679x(01)65002-7. [DOI] [PubMed] [Google Scholar]
- Parce JW, Owicki JC, Kercso KM, Sigal GB, Wada HG, Muir VC, Bousse LJ, Ross KL, Sikic BI, McConnell HM. Detection of cell-affecting agents with a silicon biosensor. Science. 1989;246:243–247. doi: 10.1126/science.2799384. [DOI] [PubMed] [Google Scholar]
- Rardin MJ, Wiley SE, Murphy AN, Pagliarini DJ, Dixon JE. Dual specificity phosphatases 18 and 21 target to opposing sides of the mitochondrial inner membrane. J Biol Chem. 2008;283:15440–15450. doi: 10.1074/jbc.M709547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res. 2008;49:1607–1620. doi: 10.1194/jlr.R700018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M, Rua D, Greenberg ML. The biosynthesis and functional role of cardiolipin. Prog Lipid Res. 2000;39:257–288. doi: 10.1016/s0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- Stukey J, Carman GM. Identification of a novel phosphatase sequence motif. Protein Sci. 1997;6:469–472. doi: 10.1002/pro.5560060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun MG, Williams J, Munoz-Pinedo C, Perkins GA, Brown JM, Ellisman MH, Green DR, Frey TG. Correlated three-dimensional light and electron microscopy reveals transformation of mitochondria during apoptosis. Nat Cell Biol. 2007;9:1057–1065. doi: 10.1038/ncb1630. [DOI] [PubMed] [Google Scholar]
- Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- Tonks NK, Neel BG. Combinatorial control of the specificity of protein tyrosine phosphatases. Curr Opin Cell Biol. 2001;13:182–195. doi: 10.1016/s0955-0674(00)00196-4. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J. Mitochondria in early mammalian development. Semin Cell Dev Biol. 2009;20:354–364. doi: 10.1016/j.semcdb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Vance D, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. Amsterdam: Elsevier; 2008. pp. 59–96.pp. 213–244. [Google Scholar]
- Voet D, Voet JG. Biochemistry. Vol. 1. Hoboken, NJ: Wiley; 2004. pp. 797–842. [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- Wilson JE. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol. 2003;206:2049–2057. doi: 10.1242/jeb.00241. [DOI] [PubMed] [Google Scholar]
- Wittig I, Karas M, Schagger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics. 2007;6:1215–1225. doi: 10.1074/mcp.M700076-MCP200. [DOI] [PubMed] [Google Scholar]
- Worby CA, Gentry MS, Dixon JE. Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- Yan LJ, Forster MJ. Resolving mitochondrial protein complexes using nongradient blue native polyacrylamide gel electrophoresis. Anal Biochem. 2009;389:143–149. doi: 10.1016/j.ab.2009.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbetto E, Vergani L, Dabbeni-Sala F. Quantification of muscle mitochondrial oxidative phosphorylation enzymes via histochemical staining of blue native polyacrylamide gels. Electrophoresis. 1997;18:2059–2064. doi: 10.1002/elps.1150181131. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Gvozdenovic-Jeremic J, Webster P, Zhou J, Greenberg ML. Loss of function of KRE5 suppresses temperature sensitivity of mutants lacking mitochondrial anionic lipids. Mol Biol Cell. 2005;16:665–675. doi: 10.1091/mbc.E04-09-0808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.