Abstract
4’-Ethynyl-2-fluoro-2’-deoxyadenosine (EFdA) is the most potent inhibitor of HIV reverse transcriptase (RT). We have recently named EFdA a Translocation Defective RT Inhibitor (TDRTI) because after its incorporation in the nucleic acid it blocks DNA polymerization, primarily by preventing translocation of RT on the template/primer that has EFdA at the 3’-primer end (T/PEFdA). The sugar ring conformation of EFdA may also influence RT inhibition by a) affecting the binding of EFdA triphosphate (EFdATP) at the RT active site and/or b) by preventing proper positioning of the 3’-OH of EFdA in T/PEFdA that is required for efficient DNA synthesis. Specifically, the North (C2’-exo/C3’-endo), but not the South (C2’-endo/C3’-exo) nucleotide sugar ring conformation is required for efficient binding at the primer-binding and polymerase active sites of RT. In this study we use nuclear magnetic resonance (NMR) spectroscopy experiments to determine the sugar ring conformation of EFdA. We find that unlike adenosine nucleosides unsubstituted at the 4’-position, the sugar ring of EFdA is primarily in the North conformation. This difference in sugar ring puckering likely contributes to the more efficient incorporation of EFdATP by RT than dATP. In addition, it suggests that the 3’-OH of EFdA in T/PEFdA is not likely to prevent incorporation of additional nucleotides and thus it does not contribute to the mechanism of RT inhibition. This study provides the first insights into how structural attributes of EFdA affect its antiviral potency through interactions with its RT target.
Keywords: EFdA, Translocation Defective Reverse Transcriptase Inhibitors, Sugar Ring Conformation, Reverse Transcriptase, HIV, Antivirals
INTRODUCTION
HIV-1 reverse transcriptase (RT) is the most targeted viral protein by approved anti-HIV drugs due to its critical role in replication of the virus (24, 14, 29, 5, 32, 8). These inhibitors, which are either nucleoside reverse transcriptase inhibitors (NRTIs) or non-nucleoside reverse transcriptase inhibitors (NNRTIs), interfere with the enzyme’s ability to synthesize the viral DNA. In particular, NRTIs mimic the natural dNTP substrate of the enzyme and bind to the 3’-primer terminus in the polymerase active site. Once incorporated into the primer, the NRTI prevents further elongation of the DNA by acting as a chain terminator. All currently approved NRTIs lack a 3’-OH moiety, which has long been considered a requirement for inhibitors to be successful chain terminators. Although this lack of a 3’-OH group promotes effective chain termination, it imparts a negative effect on the potency of the NRTI, including a diminished binding affinity for the RT target and decreased ability to be activated by cellular kinases (12).
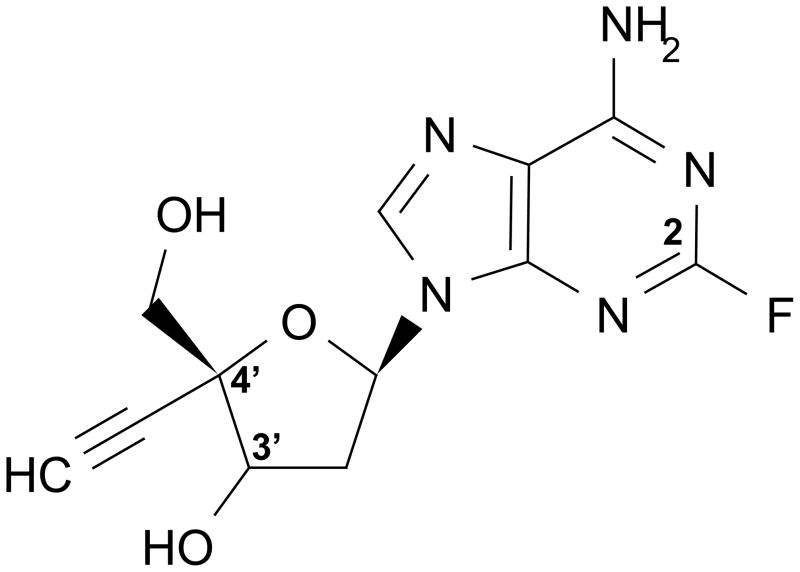
We reported previously that a group of NRTIs with 4’-substitutions and a 3’-OH are very effective at inhibiting both wild-type (WT) and multi-drug resistant strains of HIV (18). The most potent compound in this collection is 4’-ethynyl-2-fluoro-2’-deoxyadenosine (EFdA), an adenosine analog containing a 4’-ethynyl group on the deoxyribose ring and a 2-fluoro group on the adenine base (Figure 1). EFdA is able to inhibit both WT and multi-drug resistant strains of HIV several orders of magnitude more efficiently than all other currently approved NRTIs (22). Moreover, clinically-observed drug resistant HIV strains are sensitive (38, 21), and in some cases hypersensitive (17), to EFdA. Recently, we have shown that EFdA acts primarily as a chain terminator because it prevents translocation of RT on the EFdA-terminated primer after incorporation. Antiviral compounds demonstrating this novel mechanism of inhibition have been termed Translocation Defective Reverse Transcriptase Inhibitors (TDRTIs) (22).
Figure 1.
The chemical structure of EFdA.
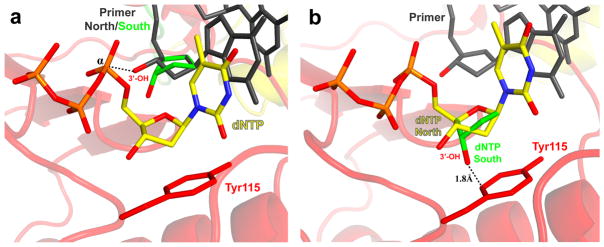
It has been demonstrated that the conformation of the sugar ring affects the biological activity of NRTIs (16, 30, 27, 20, 25, 4, 31, 2, 3). In solution, the structure of the deoxyribose ring of nucleosides exists in a dynamic equilibrium between the C2’-exo/C3’-endo (North) and C2’-endo/C3’-exo (South) conformations. It has previously been shown that the sugar ring conformation of NRTIs is important for recognition by RT at both the primer and dNTP binding sites. For efficient DNA polymerization to occur, both the nucleotide at the 3’-end of the primer and the incoming dNTP or NRTI are required to be in the North conformation. In the North conformation, the 3’-OH of the nucleotide at the 3’-primer terminus is properly positioned for in-line nucleophilic attack on the α-phosphate of the incoming dNTP or NRTI (Figure 2a). The North conformation is also important for the incoming dNTP or NRTI, because if the sugar ring were in the South conformation, the 3’-OH would be very close to Tyr115 of RT (d = 1.8 Å), creating unfavorable steric interactions between the substrate and enzyme (Figure 2b) (20, 23, 2).
Figure 2.
The effect of sugar ring conformation in the HIV-1 RT polymerase active site. The sugar ring conformation at the 3’-primer end is required to be in the North (2’-exo/3’-endo) conformation (2a, dark gray) for successful in-line nucleophilic attack of the α-phosphate of the incoming dNTP or NRTI. The South (2’-endo/3’-exo) (2a, green) conformation of the sugar ring at the primer terminus positions the 3’-OH away from the α-phosphate and thus DNA polymerization is not as efficient. The preferred conformation of the sugar ring of the incoming dNTP or NRTI is the North conformation (2b, yellow). The South conformation of the sugar ring of the incoming dNTP or NRTI (2b, green) would result in a very short distance between the 3’-OH and the aromatic ring of Tyr115 (red), causing steric interactions with the enzyme. Coot (10) was used to perform simple modeling of the 2’-endo/3’-exo sugar ring conformations of the primer and dNTP using structural coordinates from PDB Code 1RTD. Images were generated using PyMOL (7).
If EFdA were in the South conformation after incorporation into the primer, then the 3’-OH would not be properly positioned for in-line nucleophilic attack on the α-phosphate of the next incoming nucleotide, therefore contributing to inefficient catalysis and inhibition of DNA synthesis. In order to evaluate the influence of these structural attributes on the antiviral properties of EFdA, we carried out nuclear magnetic resonance (NMR) spectroscopy experiments using EFdA and the natural substrate dA. The results of this study allow us to compare the sugar ring puckering of EFdA and other nucleoside analogs, such as 2’-fluoro-2’,3’-dideoxynucleoside 5’-triphosphates (23), AZT, ddI, ddA (26), and bicyclo[3.1.0]hexene nucleosides (28), and determine if its structure 1) is in the proper conformation for optimal recognition by RT and 2) if it contributes to the inhibition mechanism of HIV RT.
MATERIALS AND METHODS
Chemicals
The compound dA was purchased from Sigma-Aldrich (St. Louis, MO). EFdA was provided by Yamasa Corporation (Chiba, Japan). d6-Dimethyl sulfoxide (DMSO) was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA). All other materials were purchased from Fisher Scientific (Pittsburgh, PA).
NMR Spectroscopy
One-dimensional 1H NMR spectra were collected in 10 °C increments from 20 to 50 °C on a Bruker Avance DRX500 Spectrometer equipped with a 5mm HCN cryoprobe. Both dA and EFdA were dissolved in d6-DMSO to final concentrations of 2–4 mM. Spectra used for coupling constant analysis were acquired with 64 scans and 33K data points with a sweep width of 4960 Hz and a relaxation delay of 4.3 s. Spectra were processed with a line broadening of 0.3 Hz. Coupling constants were read directly from the spectra for first-order resonances. Complex multiplets were analyzed by spectral simulation using SpinWorks 3.0 (19). Simulations were performed for the spin systems on the deoxyribose ring only. RMS deviations of calculated and experimental coupling constants for both compounds were < 0.05 Hz.
Pseudorotational Analysis
Sugar ring conformations of dA and EFdA were determined using the program PSEUROT 6.3 (13, 6, 36) (acquired from Dr. Cornelis Altona, University of Leiden, Netherlands). This version utilizes an improved generalized Karplus equation, as described by Donders et al. (9), which uses experimental data as a basis for the iterative calculation of coupling constants. In the case of dA, the puckering amplitude of the less favored conformation ( φm(N)) was held constant during calculations. For EFdA, the puckering amplitudes of both conformations were fixed during calculation. RMS deviations of calculated and experimental coupling constants for both compounds were < 0.2 Hz.
Molecular Modeling
A model of the ternary complex of HIV-1 RT / DNA / EFdA triphosphate (EFdATP) was built using the coordinates of the crystal structure of the HIV-1 RT-DNA-tenofovir diphosphate (TFVDP) complex (35). The triphosphate of the EFdATP was built using the corresponding atoms of TFVDP in the structure from PDB code 1T05 and of dTTP in PDB code 1RTD (15). The structure of the EFdATP was assembled from its components using the sketch module of SYBYL 7.3 (34), and minimized by the semi-empirical quantum chemical method PM3 (33). After removing the TFVDP from 1T05, the PM3 charges and the docking module of SYBYL 7.0 were used to dock the EFdATP at the RT dNTP binding site to give the ternary complex of HIV-1 RT / DNA / EFdATP. The final complex structure was minimized for 100 cycles using the AMBER force field with Coleman united charges on the protein and DNA molecules.
RESULTS
NMR Spectroscopy and Pseudorotational Analysis
To evaluate the sugar ring conformation of dA and EFdA, one-dimensional 1H NMR spectra were collected over a range of temperatures for both compounds. Coupling constants determined from each spectrum were used to calculate the structural parameters of the deoxyribose ring. The changes in the value of each coupling constant with temperature were examined for both compounds, and these changes were used to calculate the pseudorotational phase angle, P, and the degree of maximum ring pucker, φm, using PSEUROT 6.3 (Table 1) (1).
Table 1.
Summary of pseudorotational analysis.
| Compound | PN, deg | PS, deg | φm(N), deg | φm(S), deg | % N | RMS error (Hz)a |
|---|---|---|---|---|---|---|
| dA | 18.7 | 169.1 | 39.0b | 31.1 | 23 | 0.034 |
| EFdA | 38.7 | 146.5 | 39.0b | 39.0b | 75 | 0.197 |
Root mean square deviation between experimental and calculated coupling constants.
Parameter fixed during calculations.
Coupling constants between most of the hydrogen atoms on the deoxyribose ring required spectral simulation in order to be resolved. The pseudorotational parameters listed in Table 1 were calculated with PSEUROT 6.3, which assumes a two-state approximation (North and South) and utilizes a generalized Karplus equation combined with a non-linear Newton-Raphson minimization process to examine the conformational flexibility of five-membered rings. An iterative approach was used to determine the optimal pseudorotational parameters PN, PS, φm(N), and φm(S) in which some of these parameters were fixed during refinement.
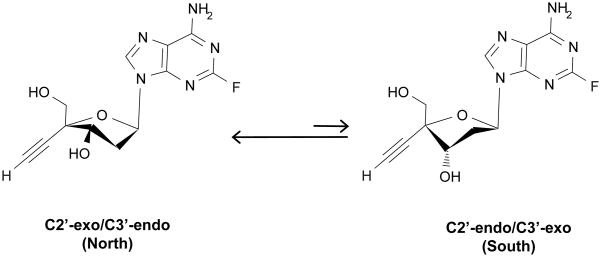
The results of the conformational analysis of dA demonstrate that the sugar ring heavily favors the South conformation over the North conformation in solution. This result is in agreement with previous conformational studies of dA (37, 11). On the other hand, the results of the conformational analysis of EFdA show that the sugar ring favors the North conformation in solution (Figure 3). The value of PN is 38.7°, which is just slightly outside of the range observed for most typical nucleosides (PN = 0–36°) but still in the Northern hemisphere (1). This observation is similar to that previously reported for 4’-ethynyl-2’,3’-dideoxycytidine (31). The value of PS is 146.5°, which falls in the range commonly observed for traditional nucleosides (PS = 144–180°). The values of φm(N) and φm(S) were fixed during calculations to 39°, the average value commonly observed for purine-base nucleosides (1). This result suggests that either the 4’-ethynyl, 2-fluoro, or both in combination greatly influence the sugar ring conformation of EFdA compared to the natural substrate, dA.
Figure 3.
Dynamic equilibrium between the North and South sugar ring conformations of EFdA in solution.
Molecular Modeling
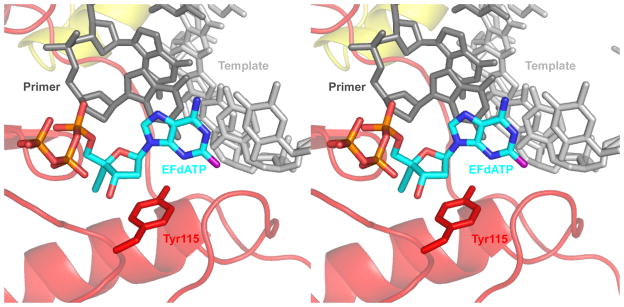
To understand the molecular basis of importance of the sugar ring conformation for RT recognition, we used our pre-catalytic RT / DNA / EFdATP ternary complex to assess the effect of EFdA sugar ring conformation on the binding interactions of the inhibitor with the polymerase active site of the enzyme (Figure 4). In our molecular model, the 4’-ethynyl group is stabilized in a hydrophobic pocket formed by RT residues Ala114, Tyr115, Phe160, Met184, and the aliphatic segment of Asp185. With the 3’-carbon in the North conformation, the 3’-OH is able to fit nicely in the RT polymerase active site. However, when the 3’-carbon of EFdATP is placed in the South conformation, the 3’-OH interacts sterically with the aromatic ring of Tyr115 (short distance of 1.8 Å).
Figure 4.
Stereo view of EFdATP (cyan) modeled in the RT active site. The 3’-OH of EFdATP is in the North conformation and is free of steric interactions with Tyr115 (red sticks). The primer is in dark gray, the template in light gray, the connection subdomain in yellow, and the palm subdomain in red. The fingers subdomain was removed for clarity. Image generated using PyMOL (7).
DISCUSSION
Our results show that EFdA has a dramatically different sugar ring conformation than dA. The equilibrium between the North and South sugar ring conformations is shifted from the preferred South conformation for the natural substrate dA to the North conformation for EFdA. It is not clear if the structural changes are due to the 2- or 4’-substitutions in EFdA. However, it is likely that steric interactions between the 4’-ethynyl and the 3’-OH disfavor the South conformation of EFdA. This is because the North conformation of the sugar ring positions the 4’-ethynyl and the 3’-OH groups farther apart than they would be in the South conformation. However, even in the absence of the 3’-OH, the sugar ring in the crystal structure of 4'-ethynyl-2',3’-dideoxycytidine is also in the North conformation (31). Hence, it appears that in addition to steric interactions between the 3’-OH and 4’-ethynyl, other factors contribute to the propensity of 4’-ethynyl-substituted NRTIs to have a sugar ring in the North conformation.
The NMR experiments confirm that EFdA is primarily in the North conformation, rendering its structure (and presumably the structure of its active metabolite EFdATP) in the optimal conformation for binding at the polymerase active site of RT. While the natural substrate, dA, favors the South conformation, the energy barrier required to convert the sugar ring of dA to the North conformation is very small, approximately 1 kcal/mol (11). EFdA (and presumably EFdATP) already favors the North conformation and does not have to overcome this energy barrier in order to bind to the primer. This is likely a contributing factor in the more efficient incorporation of EFdATP into the primer strand than dATP, contributing to the high potency of the inhibitor as reported (22).
Molecular modeling studies were performed with EFdATP to gain further insight into the structural effects of the sugar ring conformation on RT recognition. When EFdATP is positioned in the nucleotide binding site prior to incorporation into the primer, the importance of the sugar ring puckering becomes apparent. With the sugar ring of EFdATP positioned in the North conformation, the 3’-OH is free of unfavorable interactions with RT (particularly from steric interactions with Tyr115) and fits perfectly in the active site. In addition, the sugar ring geometry of EFdA allows favorable interactions of the 4'-ethynyl into a hydrophobic pocket defined by RT residues A114, Y115, F160, M184 and the aliphatic chain of D185. These interactions are thought to contribute both to enhanced RT utilization of EFdATP and difficulty in translocation of 3'-terminal EFdAMP primers (22).
Upon incorporation into the primer strand, the position of the 3’-OH of the EFdAMP in the North conformation is in perfect alignment for in-line nucleophilic attack on the α-phosphate of the next incoming dNTP. Because the North conformation of the sugar ring is heavily favored over the South conformation for EFdA, this is evidence that misalignment of the 3’-OH for nucleophilic attack is not a contributing factor to the chain termination mechanism of inhibition of EFdA. While the sugar ring conformation of the 3’-terminal EFdAMP is in the North conformation, which favors incorporation of the next dNTP, this step cannot occur because of the inability of RT to translocate from the nucleotide binding site. This further supports the observation that the inability of RT to translocate after EFdA incorporation is the primary mechanism of inhibition (22).
Slight changes in the chemical composition of adenosine analogs have a pronounced effect on the efficiency of TDRTIs in blocking HIV replication. In particular, substitutions at the 4’-position of the deoxyribose ring and the 2-position of the adenine base favor structural conformations of EFdA that improve its interactions with the RT target, thereby enhancing its antiviral potency.
Acknowledgments
We would like to thank Dr. Wei Wycoff for assistance with variable temperature NMR data collection. The purchase of the 500 MHz NMR spectrometer was partially supported by NSF grant CHE-89-08304, and NIH/NCRR grant S10 RR022341-01 (cold probe). This work was supported in part by NIH grants AI076119, AI076119-S1, AI076119-02S1, and AI094715 (to S.G.S.) and AI079801 (to M.A.P.). We also acknowledge support from GeneMatrix and the Korea Food & Drug Administration and the Ministry of Knowledge and Economy, Bilateral International Collaborative R&D Program. B.M. is a recipient of the amfAR Mathilde Krim Fellowship. We are grateful to Yamasa Corporation for providing EFdA.
ABBREVIATIONS
- dA
2’-Deoxyadenosine
- dATP
2’-Deoxyadenosine triphosphate
- EFdA
4’-Ethynyl-2-fluoro-2’-deoxyadenosine
- EFdAMP
4’-Ethynyl-2-fluoro-2’-deoxyadenosine monophosphate
- EFdATP
4’-Ethynyl-2-fluoro-2’-deoxyadenosine triphosphate
- NRTI
Nucleoside reverse transcriptase inhibitor
- RT
Reverse transcriptase
- T/PEFdA
Template/Primer terminated 4’-ethynyl-2-fluoro-2’-deoxyadenosine
- TDRTI
Translocation defective reverse transcriptase inhibitor
Footnotes
The abstract of this article was presented at the World Conference on Cellular and Molecular Biology in Indore, India, November 2 – 6, 2009.
References
- 1.Altona C, Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides: A new description using the concept of pseudorotation. J Am Chem Soc. 1972;94:8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- 2.Boyer PL, Julias JG, Marquez VE, Hughes SH. Fixed conformation nucleoside analogs effectively inhibit excision-proficient HIV-1 reverse transcriptases. J Mol Biol. 2005;345:441–450. doi: 10.1016/j.jmb.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 3.Boyer PL, Julias JG, Ambrose Z, Siddiqui MA, Marquez VE, Hughes SH. The nucleoside analogs 4'C-methyl thymidine and 4'C-ethyl thymidine block DNA synthesis by wild-type HIV-1 RT and excision proficient NRTI resistant RT variants. J Mol Biol. 2007;371:873–882. doi: 10.1016/j.jmb.2007.05.043. [DOI] [PubMed] [Google Scholar]
- 4.Choi Y, George C, Comin MJ, Barchi JJ, Jr, Kim HS, Jacobson KA, Balzarini J, Mitsuya H, Boyer PL, Hughes SH, Marquez VE. A conformationally locked analogue of the anti-HIV agent stavudine. An important correlation between pseudorotation and maximum amplitude. J Med Chem. 2003;46:3292–3299. doi: 10.1021/jm030116g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Clercq E. Anti-HIV drugs. Verh K Acad Geneeskd Belg. 2007;69:81–104. [PubMed] [Google Scholar]
- 6.De Leeuw FAAM, Altona C. Computer assisted pseudorotational analysis of 5-membered rings by means of 3JHH coupling constants: program PSEUROT. J Comp Chem. 1983;4:428–437. [Google Scholar]
- 7.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. http://www.pymol.org. [Google Scholar]
- 8.Deval J. Antimicrobial strategies: inhibition of viral polymerases by 3'-hydroxyl nucleosides. Drugs. 2009;69:151–166. doi: 10.2165/00003495-200969020-00002. [DOI] [PubMed] [Google Scholar]
- 9.Donders LA, De Leeuw FAAM, Altona C. Relationship between proton-proton NMR coupling constants and substituent electronegativities. IV. An extended Karplus equation accounting for interactions between subtituents and its application to coupling constant data calculated by the extended Hueckel method. Magn Reson Chem. 1989;27:556–563. [Google Scholar]
- 10.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 11.Ford H, Jr, Dai F, Mu L, Siddiqui MA, Nicklaus MC, Anderson L, Marquez VE, Barchi JJ., Jr Adenosine deaminase prefers a distinct sugar ring conformation for binding and catalysis: kinetic and structural studies. Biochemistry. 2000;39:2581–2592. doi: 10.1021/bi992112c. [DOI] [PubMed] [Google Scholar]
- 12.Gallois-Montbrun S, Schneider B, Chen Y, Giacomoni-Fernandes V, Mulard L, Morera S, Janin J, Deville-Bonne D, Veron M. Improving nucleoside diphosphate kinase for antiviral nucleotide analogs activation. J Biol Chem. 2002;277:39953–39959. doi: 10.1074/jbc.M206360200. [DOI] [PubMed] [Google Scholar]
- 13.Haasnoot CAG, De Leeuw FAAM, De Leeuw HPM, Altona C. The relationship between proton-proton NMR coupling constants and substituent electronegativities II-Conformational analysis of the sugar ring in nucleosides and nucleotides in solution using a generalized Karplus equation. Org Magn Reson. 1981;15:43–52. [Google Scholar]
- 14.Hammer SM, Saag MS, Schechter M, Montaner JS, Schooley RT, Jacobsen DM, Thompson MA, Carpenter CC, Fischl MA, Gazzard BG, Gatell JM, Hirsch MS, Katzenstein DA, Richman DD, Vella S, Yeni PG, Volberding PA. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society--USA panel. Top HIV Med. 2006;14:827–843. [PubMed] [Google Scholar]
- 15.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 16.Jagannadh B, Reddy DV, Kunwar AC. 1H NMR study of the sugar pucker of 2',3'-dideoxynucleosides with anti-human immunodeficiency virus (HIV) activity. Biochem Biophys Res Commun. 1991;179:386–391. doi: 10.1016/0006-291x(91)91382-m. [DOI] [PubMed] [Google Scholar]
- 17.Kawamoto A, Kodama E, Sarafianos SG, Sakagami Y, Kohgo S, Kitano K, Ashida N, Iwai Y, Hayakawa H, Nakata H, Mitsuya H, Arnold E, Matsuoka M. 2'-deoxy-4'-C-ethynyl-2-halo-adenosines active against drug-resistant human immunodeficiency virus type 1 variants. Int J Biochem Cell Biol. 2008;40:2410–2420. doi: 10.1016/j.biocel.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Kodama EI, Kohgo S, Kitano K, Machida H, Gatanaga H, Shigeta S, Matsuoka M, Ohrui H, Mitsuya H. 4'-Ethynyl nucleoside analogs: potent inhibitors of multidrug-resistant human immunodeficiency virus variants in vitro. Antimicrob Agents Chemother. 2001;45:1539–1546. doi: 10.1128/AAC.45.5.1539-1546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marat K. SpinWorks 3.0, released April 11, 2008. University of Manitoba; Winnipeg, Manitoba, Canada: http://www.umanitoba.ca/chemistry/nmr/spinworks/index.html. [Google Scholar]
- 20.Marquez VE, Ezzitouni A, Russ P, Siddiqui MA, Ford H, Jr, Feldman RJ, Mitsuya H, George C, Barchi JJ., Jr HIV-1 Reverse Transcriptase Can Discriminate Between Two Conformationally Locked Carbocyclic AZT Triphosphate Analogues. J Am Chem Soc. 1998;120:2780–2789. [Google Scholar]
- 21.Mascolini M, Larder BA, Boucher CA, Richman DD, Mellors JW. Broad advances in understanding HIV resistance to antiretrovirals: report on the XVII International HIV Drug Resistance Workshop. Antivir Ther. 2008;13:1097–1113. [PubMed] [Google Scholar]
- 22.Michailidis E, Marchand B, Kodama EN, Singh K, Matsuoka M, Kirby KA, Ryan EM, Sawani AM, Nagy E, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. Mechanism of inhibition of HIV-1 reverse transcriptase by 4'-ethynyl-2-fluoro-2'-deoxyadenosine triphosphate, a translocation defective reverse transcriptase inhibitor. J Biol Chem. 2009;284:35681–35691. doi: 10.1074/jbc.M109.036616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mu L, Sarafianos SG, Nicklaus MC, Russ P, Siddiqui MA, Ford H, Jr, Mitsuya H, Le R, Kodama E, Meier C, Knispel T, Anderson L, Barchi JJ, Jr, Marquez VE. Interactions of conformationally biased north and south 2'-fluoro-2', 3'-dideoxynucleoside 5'-triphosphates with the active site of HIV-1 reverse transcriptase. Biochemistry. 2000;39:11205–11215. doi: 10.1021/bi001090n. [DOI] [PubMed] [Google Scholar]
- 24.Parniak MA, Sluis-Cremer N. Inhibitors of HIV-1 reverse transcriptase. Adv Pharmacol. 2000;49:67–109. doi: 10.1016/s1054-3589(00)49024-1. [DOI] [PubMed] [Google Scholar]
- 25.Poznanski J, Bretner M, Kulikowski T, Balzarini J, Van Aerschot A, De Clercq E. Synthesis, solution conformation and anti-HIV activity of novel 3'-substituted-2',3'-dideoxy-5-hydroxymethyluridines and their 4,5-substituted analogues. Antivir Chem Chemother. 2003;14:127–138. doi: 10.1177/095632020301400302. [DOI] [PubMed] [Google Scholar]
- 26.Reddy DV, Jagannadh B, Kunwar AC. NMR study of dideoxynucleotides with anti-human immunodeficiency virus (HIV) activity. J Biochem Biophys Methods. 1996;31:113–121. doi: 10.1016/0165-022x(95)00027-o. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez JB, Marquez VE, Nicklaus MC, Mitsuya H, Barchi JJ., Jr Conformationally locked nucleoside analogues. Synthesis of dideoxycarbocyclic nucleoside analogues structurally related to neplanocin C. J Med Chem. 1994;37:3389–3399. doi: 10.1021/jm00046a024. [DOI] [PubMed] [Google Scholar]
- 28.Russ PL, Gonzalez-Moa MJ, Vu BC, Sigano DM, Kelley JA, Lai CC, Deschamps JR, Hughes SH, Marquez VE. North- and south-bicyclo[3.1.0]hexene nucleosides: the effect of ring planarity on anti-HIV activity. ChemMedChem. 2009;4:1354–1363. doi: 10.1002/cmdc.200900153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schinazi RF, Hernandez-Santiago BI, Hurwitz SJ. Pharmacology of current and promising nucleosides for the treatment of human immunodeficiency viruses. Antiviral Res. 2006;71:322–334. doi: 10.1016/j.antiviral.2006.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siddiqui MA, Driscoll JS, Marquez VE, Roth JS, Shirasaka T, Mitsuya H, Barchi JJ, Jr, Kelley JA. Chemistry and anti-HIV properties of 2'-fluoro-2',3'-dideoxyarabinofuranosylpyrimidines. J Med Chem. 1992;35:2195–2201. doi: 10.1021/jm00090a008. [DOI] [PubMed] [Google Scholar]
- 31.Siddiqui MA, Hughes SH, Boyer PL, Mitsuya H, Van QN, George C, Sarafinanos SG, Marquez VE. A 4'-C-ethynyl-2',3'-dideoxynucleoside analogue highlights the role of the 3'-OH in anti-HIV active 4'-C-ethynyl-2'-deoxy nucleosides. J Med Chem. 2004;47:5041–5048. doi: 10.1021/jm049550o. [DOI] [PubMed] [Google Scholar]
- 32.Sluis-Cremer N, Tachedjian G. Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors. Virus Res. 2008;134:147–156. doi: 10.1016/j.virusres.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart JJP. Optimization of parameters for semiempirical methods I. Method. J Comput Chem. 1989;10:209–220. [Google Scholar]
- 34.SYBYL 7.3. Tripos International; 1699 South Hanley Rd., St. Louis, Missouri, 63144, USA: [Google Scholar]
- 35.Tuske S, Sarafianos SG, Clark AD, Jr, Ding J, Naeger LK, White KL, Miller MD, Gibbs CS, Boyer PL, Clark P, Wang G, Gaffney BL, Jones RA, Jerina DM, Hughes SH, Arnold E. Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nat Struct Mol Biol. 2004;11:469–474. doi: 10.1038/nsmb760. [DOI] [PubMed] [Google Scholar]
- 36.Van Wijk J, Huckriede BD, Ippel JH, Altona C. Furanose sugar conformations in DNA from NMR coupling constants. Methods Enzymol. 1992;211:286–306. doi: 10.1016/0076-6879(92)11017-d. [DOI] [PubMed] [Google Scholar]
- 37.Westhof E, Plach H, Cuno I, Ludemann HD. Proton magnetic resonance studies of 2'-, 3'-, and 5'-deoxyadenosine conformations in solution. Nucelic Acids Res. 1977;4:939–953. doi: 10.1093/nar/4.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White KL, Chen JM, Feng JY, Margot NA, Ly JK, Ray AS, Macarthur HL, McDermott MJ, Swaminathan S, Miller MD. The K65R reverse transcriptase mutation in HIV-1 reverses the excision phenotype of zidovudine resistance mutations. Antivir Ther. 2006;11:155–163. doi: 10.1177/135965350601100209. [DOI] [PubMed] [Google Scholar]