Abstract
We performed a meta-analysis of 14 genome-wide association studies of coronary artery disease (CAD) comprising 22,233 cases and 64,762 controls of European descent, followed by genotyping of top association signals in 60,738 additional individuals. This genomic analysis identified 13 novel loci harboring one or more SNPs that were associated with CAD at P<5×10−8 and confirmed the association of 10 of 12 previously reported CAD loci. The 13 novel loci displayed risk allele frequencies ranging from 0.13 to 0.91 and were associated with a 6 to 17 percent increase in the risk of CAD per allele. Notably, only three of the novel loci displayed significant association with traditional CAD risk factors, while the majority lie in gene regions not previously implicated in the pathogenesis of CAD. Finally, five of the novel CAD risk loci appear to have pleiotropic effects, showing strong association with various other human diseases or traits.
It has been estimated that heritable factors account for 30–60% of the interindividual variation in the risk of coronary artery disease (CAD) 1. Recently, genome-wide association (GWA) studies have identified several common variants that associate with risk of CAD 2. However, in aggregate these variants explain only a small fraction of the heritability of CAD, probably partly due to the limited power of previous studies to discover effects of modest size. Recognizing the need for larger studies, we formed the transatlantic Coronary ARtery DIsease Genome wide Replication and Meta-analysis (CARDIoGRAM) consortium 3. We perfomed a meta-analysis of 14 GWA studies of CAD comprising 22,233 cases and 64,762 controls, all of European ancestry (Supplementary Table 1a and Additional Table 1a, see www.imbs-luebeck.de/imbs/sites/default/files/myfilemanager/500/AdditionalInformation.pdf). We then genotyped lead SNPs within the most promising novel loci as well as a subset of previously reported CAD loci in up to 60,738 additional subjects (approximately half cases and controls) (Supplementary Table 1b and Additional Table 1b, see www.imbs-luebeck.de/imbs/sites/default/files/myfilemanager/500/AdditionalInformation.pdf). Lastly, we explored potential mechanisms and intermediate pathways by which novel loci may mediate risk.
Nine of the 12 loci previously associated with CAD through individual GWA studies achieved genome-wide significance (P<5×10−8) in our initial meta-analysis (Table 1). We were, however, unable to test the previously reported association with a haplotype and a rare SNP in LPA in our GWA data,4–5 but observed robust association with the rare LPA variant in our replication samples through direct genotyping (Table 1). Thus, 10 of the 12 loci previously associated with CAD at a genome-wide significance level surpassed the same threshold of significance in CARDIoGRAM.
Table 1.
Association evidence in CARDIoGRAM for previously published loci for coronary disease (previously reported with genome-wide significance (P<5×10−8).
| Band | SNP | Gene(s) in region | n | Risk allele frequency (risk allele) |
CARDIoGRAM | reference | |
|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR | |||||
| 1p13.3 | rs599839* | SORT1 | 83,873 | 0.78 (A) | 1.11 (1.08; 1.15) | 2.89·10−10 | 1.29 (1.18; 1.40) 19 |
| 1p32.3 | rs11206510*** | PCSK9 | 102,352 | 0.82 (T) | 1.08 (1.05; 1.11) | 9.10·10−08 | 1.15 (1.10; 1.21) 8 |
| 1q41 | rs17465637**** | MIA3 | 25,197 | 0.74 (C) | 1.14 (1.09; 1.20) | 1.36·10−08 | 1.20 (1.12; 1.30) 19 |
| 2q33.1 | rs6725887* | WDR12 | 77,954 | 0.15 (C) | 1.14 (1.09; 1.19) | 1.12·10−09 | 1.16 (1.10; 1.22) 8 |
| 3q22.3 | rs2306374* | MRAS | 77,843 | 0.18 (C) | 1.12 (1.07; 1.16) | 3.34·10−08 | 1.15 (1.11; 1.19) 7 |
| 6p24.1 | rs12526453* | PHACTR1 | 83,050 | 0.67 (C) | 1.10 (1.06; 1.13) | 1.15·10−09 | 1.13 (1.09; 1.17) 8 |
| 6q25.3 | rs3798220** | LPA | 32,584 | 0.02 (C) | 1.54 (1.36; 1.74) | 9.62·10−12 | 1.92 (1.48; 2.49) 12 |
| 9p21.3 | rs4977574* | CDKN2A/B, ANRIL | 84,256 | 0.46 (G) | 1.29 (1.23; 1.36) | 1.35·10−22 | 1.25 (1.18; 1.31) - 1.37 (1.26; 1.48) 19-6 |
| 10q11.21 | rs1746048*** | CXCL12 | 136,416 | 0.87 (C) | 1.09 (1.07; 1.13) | 2.12·10−10 | 1.33 (1.20; 1.48) 19 |
| 12q24.12 | rs3184504* | SH2B3 | 67,746 | 0.44 (T) | 1.07 (1.04;1.10) | 6.35·10−06 | 1.13 (1.08; 1.18) 20 |
| 19p13.2 | rs1122608* | LDLR | 49,693 | 0.77 (G) | 1.14 (1.09; 1.18) | 9.73·10−10 | 1.14 (1.09; 1.19) 8 |
| 21q22.11 | rs9982601* | MRPS6 | 46,230 | 0.15 (T) | 1.18 (1.12; 1.24) | 4.22·10−10 | 1.19 (1.13; 1.27) 8 |
Data taken *from meta-analysis;
from replication;
from combined analysis,
only genotyped data from a subset of studies
We selected 23 novel loci with a significance level of P<5×10−6 in the meta-analysis for follow-up (see Online Methods and Supplementary Note for details). Taking the number of loci into consideration, our replication study had >90% power to detect effect sizes observed in the GWA meta-analysis. Of the 23 loci, 13 replicated using our a priori definition of a validated locus i.e., showing independent replication after Bonferroni correction and also achieving a P value of < 5 × 10−8 in the combined discovery and replication data (Table 2, Figure 1, Supplementary Figures 1 and 2). Results for all loci in the replication phase are shown in Supplementary Table 2.
Table 2.
Novel loci for coronary disease.
| Meta-analysis | Replication | Combined analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Band | SNP | Gene(s) in region | Risk allele frequency (risk allele) |
P | n | P | n | OR (95% CI) | P |
| 1p32.2 | rs17114036 | PPAP2B | 0.91 (A) | 1.43·10−08 | 80,870 | 3.18·10−12 | 52,356 | 1.17 (1.13; 1.22) | 3.81·10−19 |
| 6p21.31 | rs17609940 | ANKS1A | 0.75 (G) | 2.21·10−06 | 83,997 | 1.18·10−03 | 53,415 | 1.07 (1.05; 1.10) | 1.36·10−08 |
| 6q23.2 | rs12190287 | TCF21 | 0.62 (C) | 4.64·10−11 | 78,290 | 3.25·10−04 | 52,598 | 1.08 (1.06; 1.10) | 1.07·10−12 |
| 7q32.2 | rs11556924 | ZC3HC1 | 0.62 (C) | 2.22·10−09 | 80,011 | 7.37·10−10 | 54,189 | 1.09 (1.07; 1.12) | 9.18·10−18 |
| 9q34.2 | rs579459 | ABO | 0.21 (C) | 1.16·10−07 | 77,138 | 7.02·10−08 | 46,840 | 1.10 (1.07; 1.13) | 4.08·10−14 |
| 10q24.32 | rs12413409 | CYP17A1, CNNM2, NT5C2 | 0.89 (G) | 1.47·10−06 | 80,940 | 1.38·10−04 | 48,801 | 1.12 (1.08; 1.16) | 1.03·10−09 |
| 11q23.3 | rs964184 | ZNF259, APOA5-A4-C3-A1 | 0.13 (G) | 8.02·10−10 | 82,562 | 2.20·10−09 | 52,930 | 1.13 (1.10; 1.16) | 1.02·10−17 |
| 13q34 | rs4773144 | COL4A1, COL4A2 | 0.44 (G) | 4.15·10−07 | 77,113 | 1.31·10−03 | 37,618 | 1.07 (1.05; 1.09) | 3.84·10−09 |
| 14q32.2 | rs2895811 | HHIPL1 | 0.43 (C) | 2.67·10−07 | 63,184 | 4.59·10−05 | 51,054 | 1.07 (1.05; 1.10) | 1.14·10−10 |
| 15q25.1 | rs3825807 | ADAMTS7 | 0.57 (A) | 9.63·10−06 | 80,849 | 1.39·10−08 | 48,803 | 1.08 (1.06; 1.10) | 1.07·10−12 |
| 17p11.2 | rs12936587 | RASD1, SMCR3, PEMT | 0.56 (G) | 4.89·10−07 | 76,952 | 1.35·10−04 | 52,648 | 1.07 (1.05; 1.09) | 4.45·10−10 |
| 17p13.3 | rs216172 | SMG6, SRR | 0.37 (C) | 6.22·10−07 | 57,235 | 2.11·10−04 | 54,303 | 1.07 (1.05; 1.09) | 1.15·10−09 |
| 17q21.32 | rs46522 | UBE2Z, GIP, ATP5G1, SNF8 | 0.53 (T) | 3.57·10−06 | 83,867 | 8.88·10−04 | 53,766 | 1.06 (1.04; 1.08) | 1.81·10−08 |
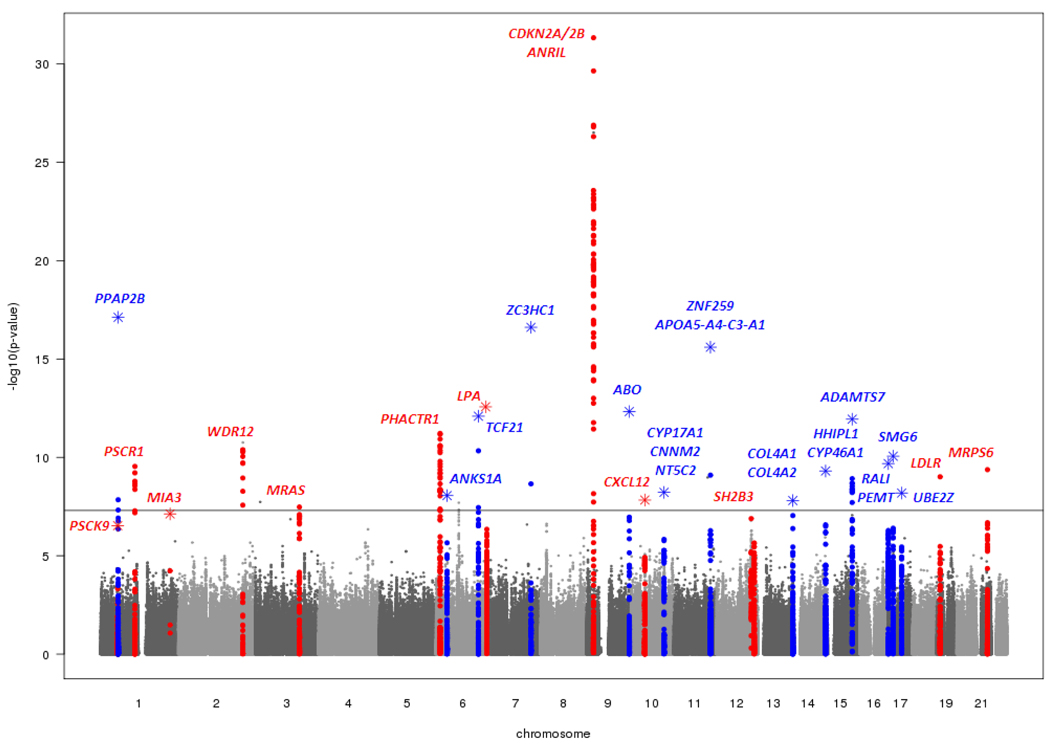
Figure 1. Graphical summary (Manhattan plot) of genome-wide association results.
The x-axis represents the genome in physical order; the y-axis shows -log10 P values for all SNPs. Data from the discovery phase are shown in circles and data from the combined discovery and replication phases in stars. Genes at the significant loci are listed above the signals. Known loci are shown in red and novel loci are shown in blue.
The 13 novel loci displayed risk allele frequencies ranging from 0.13 to 0.91 and were associated with a 6 to 17 percent increase in the risk of CAD per allele (Table 2). Out of the 13 novel loci the additive model appeared most appropriate for six while the recessive model performed best at 5 and the dominant model at 2 loci (Additional Table 2, see www.imbs-luebeck.de/imbs/sites/default/files/myfilemanager/500/AdditionalInformation.pdf).
In sub-group analyses, 20 out of 22 loci with P<5×10−8 (known and novel loci combined; for one locus age subgroups were not available) had higher odds ratios for early-onset than for late onset CAD (P=1.2×10−4 for observed vs. expected, Supplementary Table 3). The CAD loci showed consistent associations irrespective of case definition, although the odds ratios for most individual single nucleotide polymorphisms (SNPs) tended to be slightly greater for cases with angiographically proven CAD than for cases with unknown angiographic status (P=0.019 for observed vs. expected). In contrast, sub-group analyses in males and females revealed no sex specific effects for any risk alleles (Supplementary Table 3) or for their observed vs. expected pattern of association (P=0.4).
Among 7,523 controls and 7,637 CAD cases for whom we had individual-level genotype data, the minimum and maximum number of risk alleles observed per individual was 15 and 37, respectively, when considering 23 CAD susceptibility loci. The mean weighted risk score was significantly higher for cases than for controls (P <10−20). Furthermore, being in the top 10th percentile or lowest 10th percentile of the weighted score was associated with an odds ratio for CAD of 1.88 (95% confidence interval, 1.67 to 2.11) and 0.55 (95% confidence interval, 0.48 to 0.64), respectively, compared to the 50th percentile. The change in odds ratio for CAD across a broader spectrum of categories of the weighted score is shown in Supplementary Figure 3.
Three of the novel risk alleles were associated with differences in traditional CAD risk factors (Table 3 and Supplementary Table 4). The risk allele on chromosome 11q23.3 (rs964184, ZNF259, APOA5-A4-C3-A1 gene region) was associated with increased LDL cholesterol and decreased HDL cholesterol (and previously, with increased triglycerides) 6. The risk allele on chromosome 9q34.2 (rs579459, ABO) was associated with increased LDL and total cholesterol, in a direction consistent with the association of these SNPs with CAD risk (Table 3). The variant rs12413409 on chromosome 10q24.32 representing the CYP17A1/CNNM2/NT5C2 gene region was associated with hypertension.
Table 3.
Effects of novel CAD loci on traditional risk factors in combined analysis of ARIC and KORA F3/F4 (n = 13,171).
| SNP | Band | Gene(s) in region | Phenotype | β̂ (95% CI)* | P |
|---|---|---|---|---|---|
| rs579459 | 9q34.2 | ABO | Total cholesterol | 1.720 mg/dl (0.554; 2.885) | 0.0038 |
| LDL cholesterol | 1.538 mg/dl (0.468; 2.608) | 0.0049 | |||
| rs12413409 | 10q24.32 | CYP17A1, CNNM2, NT5C2 | Hypertension | 0.141 (0.044; 0.238) | 0.0043 |
| rs964184** | 11q23.3 | ZNF259, APOA5-A4-C3-A1 | HDL cholesterol | −1.926 mg/dl (−2.441; −1.411) | 2.28·10−13 |
| Total cholesterol | 4.578 mg/dl (3.191; 5.964) | 9.84·10−11 | |||
| LDL cholesterol | 1.699 mg/dl (0.417; 2.980) | 0.0094 |
Results from fixed effects meta-analysis based on beta-coefficients and standard errors from linear (total cholesterol, LDL, HDL) and logistic (hypertension) regression analysis of the single studies for which meta-analytic P<0.01.
estimated pooled regression coefficients with 95% confidence intervals. LDL = low-density lipoprotein cholesterol, HDL = high-density lipoprotein cholesterol.
Previous genome-wide studies have demonstrated strong association of rs964184 with triglycerides 21.
In silico interrogation revealed that the lead SNPs at four of the 13 novel loci were either non-synonymous coding variants or were in high LD with such SNPs. Specifically, the lead SNPs at 7q32.2 (rs11556924) and 15q25.1 (rs3825807) encoded changes in ZC3HC1 (R363H) and ADAMTS7 (S214P), respectively, while the lead SNP at 14q32.2 (rs2895811) is in strong LD (r2=0.82) with the V691A in the HHIPL1 gene. Lastly, the lead SNP at 17q21.32 (rs46522) is in strong LD (r2=0.94) with two potential functional variants in GIP: S103G (rs2291725) and a variant influencing the splice site of intron 3 (rs2291726) leading to a truncated transcript (Additional Table 3, see www.imbs-luebeck.de/imbs/sites/default/files/myfilemanager/500/AdditionalInformation.pdf) 7.
We next analyzed data from three genome-wide studies that also assessed gene expression in multiple tissues to assess potential effects of novel loci on the expression of regional genes (Supplementary Note)8–9. Three of the novel CAD risk variants showed convincing association with regional gene expression (cis effect) by either representing the most significant eSNP in the region or by being in high LD (r2≥0.85) with the strongest eSNP in the region: rs12190287 at 6q23.2 (TCF21), rs12936587 at 17p11.2 (RASD1, SMCR3 and PEMT) and rs46522 at 17q21.32 (UBE2Z) (Additional Table 4, see www.imbs-luebeck.de/imbs/sites/default/files/myfilemanager/500/AdditionalInformation.pdf). Subsequent interrogation of our novel loci in a genome-wide map of allelic expression imbalance provided further support for the eQTL findings at the 17q21.32 locus 10. This analysis also provided strong evidence of cis-effects for the 17p13.3 locus lead SNP (rs216172) on the expression of SMG6, (see Supplementary Note and Additional Table 5, see www.imbs-luebeck.de/imbs/sites/default/files/myfilemanager/500/AdditionalInformation.pdf)10.
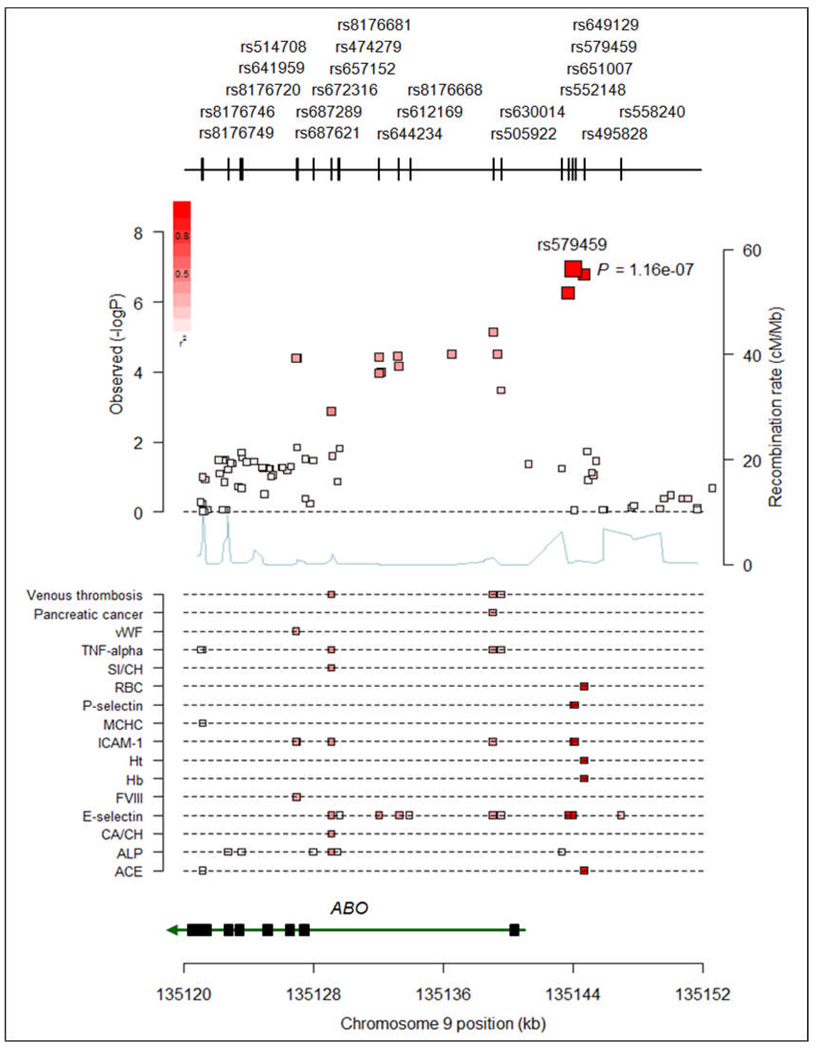
We identified five novel loci (9q34, 10q24, 11q23, 15q25, and 17p13) at which the CAD risk variant is fully or strongly correlated (r2>0.8) with variants that have previously been associated with other traits or diseases11. These traits include cerebral and abdominal aneurysm, aortic root size, celiac disease, lung adenocarcinoma, type 1 diabetes, venous thrombosis , LDL cholesterol, HDL cholesterol, triglycerides, smoking, and blood pressure, soluble levels of adhesion molecules, phytosterols (sitosterol and campesterol), angiotensin-converting enzyme (ACE) activity, coagulation factor VIII (FVIII), and von Willebrand factor (vWF) (at P<5×10−8, for references see Additional Table 6, see www.imbs-luebeck.de/imbs/sites/default/files/myfilemanager/500/AdditionalInformation.pdf). Thus, a substantial subset of the novel CAD risk loci appear to have pleiotropic effects. We illustrate a particularly striking example at the ABO locus in Figure 2.
Figure 2. Example of overlapping association signals for multiple traits at ABO gene region on chromosome 9q34.
In the upper panel the association signal for coronary disease at the ABO gene region in CARDIoGRAM and the positions and rs-numbers of SNPs in this region are shown. The size of boxes illustrates the number of individuals available for this respective SNP. In the lower panel all SNPs with P-values at genome-wide significance level of P<5×10−8 based on the NHGRI GWA study catalogue (http://www.genome.gov/gwastudies/; accessed on June 28th 2010) for all diseases and traits are shown. The degree of linkage disequilibrium (r2) between the lead SNPs for coronary disease and the other traits is reflected by the colour of the squares (upper panel) and the small bars (lower panel) (dark red (high LD) > faint red (low LD)). SI/CH = sitosterol normalized to cholesterol; CA/CH = campesterol normalized to cholesterol; ALP = alkaline phosphatase; ACE = angiotensin converting enzyme; FVIII = coagulation factor VIII; vWF = von Willebrand Factor.
The present genomic analysis of more than 135,000 individuals reveals three major findings. First, we more than doubled the number of loci with firm association to CAD. Specifically, our study yielded 13 novel and confirmed 10 previously reported loci. Second, we found that only a minority of the established and novel loci appear to act through traditional risk factors while the majority resides in gene regions that were not previously suspected in the pathogenesis of CAD. Third, a substantial proportion of the CAD risk variants are also strongly associated with various other human disease traits in GWA studies.
We anticipated that some of the genetic risk loci for CAD would act through established CAD risk factors, such as LDL cholesterol or blood pressure, which themselves have a significant genetic determination. Indeed, three of the novel risk loci (11q23.3, 9q34.2, 10q24.32) showed such associations. An association with higher LDL cholesterol or lipoprotein (a) concentration had also been found for four previously discovered risk variants including the PCSK9 locus that missed genome-wide significance level by a small margin in the present study (Table 1) 4,12–13. On the other hand, 17 out of the 23 confirmed loci appear to act through mechanisms that are independent of traditional risk factors. Elucidation of these mechanisms is critical for a more complete understanding of CAD and identification of novel therapeutic targets.
We explored several molecular mechanisms by which the novel loci could affect CAD risk. We show that some lead SNPs - or linked variants - affect the primary structure of the protein product in which the variant is located, while in other instances the risk variant is associated with expression of a specific gene or genes in one or more tissues. A more detailed discussion of the genes in each locus is provided in the Supplementary Note. While these data help to prioritize genes for follow-up functional studies, it should be emphasized that substantial work is necessary to define the mechanisms involved for each of the novel loci, as exemplified recently for the chromosome 1p13 locus14–15.
We also observed that eight of 23 CAD loci (five of the 13 novel loci: 9q34, 10q24, 11q23, 15q25, and 17p13 and three of the 10 established loci: 1p13, 9p21.3, and 12q24) not only affect the risk of CAD but also associate with multiple other diseases and traits (Additional Table 6, see www.imbs-luebeck.de/imbs/sites/default/files/myfilemanager/500/AdditionalInformation.pdf). Each of these findings requires further analysis to determine whether co-localization of SNPs for CAD and other traits points to intermediate phenotypes, and thus mechanistic links in a joint etiology, results from pleiotropic effects of a single allele affecting multiple phenotypes, or identifies chromosomal regions harbouring multiple genes and alleles that participate in the regulation of multiple independent traits via diverse mechanisms.
By design, our study focused on common risk variants. Assuming a heritability of 40% for CAD1, the lead SNPs of previously established loci combined with the novel loci discovered in this study explain approximately 10 % of the additive genetic variance of CAD. Our inability to explain a greater fraction of CAD heritability even after a large meta-analysis and replication effort is in line with results of most other complex traits examined by current GWA methods16. These results suggest that many other common susceptibility variants of similar or lower effects and/or rare variants contribute to risk of CAD.
The clinical utility of CAD risk alleles for the prediction of risk may be best determined in samples that are independent from this discovery study. In order to provide a framework for future research we explored a weighted score based on the 23 CAD risk variants validated in this investigation. We observed a greater than three-fold difference in CAD risk between top and bottom 10% of the risk scores although this may be a slight overestimation since risk scores were extracted from a subset of the discovery sample (Supplementary Figure 3). Nonetheless, this increase in risk is at least comparable to that of several other traditional risk factors for CAD including hypertension, diabetes and smoking13. Whether risk allele information may improve the performance of current risk profiling strategies for CAD prediction17–18 and whether such an approach is cost-effective requires further evaluation in prospective studies. Our findings provide a firm framework for such research.
In summary, our large-scale GWA meta-analysis discovered the association with CAD of 13 novel chromosomal loci. We observed only limited association between these CAD SNPs and traditional risk factors, suggesting that most SNPs act though novel pathways. Elucidation of the mechanisms by which these loci affect CAD risk carries the potential for better prevention and treatment of this common disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank the participants and staff in each of the studies who contributed to the present article. The sources of funding are listed in the supplementary materials.
Footnotes
AUTHORS CONTRIBUTIONS
Manuscript writing: H.S., I.R.K., S.K., M.P.R., T.L.A., H.H., A.F.R.S., P.D., R.R., R.M., J.E., N.J.S.
GWA meta-analysis samples, genotyping and analysis: H.S., I.R.K., S.K., M.P.R., T.L.A., H.H., M.P., A.F.R.S., M.B., C.G., D.Absher, D.A., K.A., S.G.B., A.J.B., J.C.B., E.B., P.S.B., M.S.B., L.C., A.D., S.D., J.D., A.Doering, N.E.E.M., R.E., S.E., M.F., A.R.F, S.G., J.R.G., E.H., A.H., T.I., C.I., M.A.K., J.W.K., A.K., R.L., M.L., W.L., P.L.-N., C.L., C.M., T.M., O.M., V.M., K.M., T.M., J.N., C.P.N., A.P., L.Q., D.J.R., V.S., A.S., A.Schillert, S.S., J.S., S.M.S., D.S.S., K.S., G.Thorgeirsson, G.T., M.T., A.G.U., B.F.V., G.A.W., H.E.W., C.W., P.W., J.C.M.W., B.J.W.,T.Z., A.Z., F.C., L.A.C., T.Q., W.M., C.H., S.B., A.S.H., P.D., U.T., R.R., J.R.T., C.J.O´D., R.M., J.E., N.J.S.
Replication phase samples, genotyping and analysis: H.S., I.R.K., S.K., M.P.R., H.H., M.P., H.A., S.A., K.A., J.L.A., D.Ardissino, T.A.B., L.C.B, D.M.B., K.B., S.M.B., M.J.B., I.B., J.F.C., R.W.D., G.D., R.D., S.G.E., J.C.E., U.d.F., B.G., D.G., V.G., N.H., S.L.H., B.D.H., G.T.J., J.W.J., L.M.K., J.J.P.K., K.T.K., G.K., D.L., K.L., P.L.-N., A.J.L., P.M.M., N.M., P.P.M., P.A.M., T.Morgan, T.W.M., J.B.M., S.C., M.M.N., O.O., F.P., R.S.P., C.C.P., A.A.Q., L.S.R., F.R.R., D.R., M.L.S., M.S.S., M.S., S.Sivapalaratnam, A.V.S., T.B.S., J.D.S., N.S., J.A.S., K.Stark, K.S., M.Stoll, W.H.W.T., A.M.v.R., N.J.W., S.Y., P.D., U.T., R.R., R.M., J.E., N.J.S.
Analysis group: I.R.K., M.P., D.Absher, L.C., E.H., M.L., K.M., A.Schillert, G.T., B.F.V., G.A.W., L.A.C., J.R.T.
Biological analyses: H.S., T.L.A., H.H., M.B., C.G., Z.A., P.S.B., V.C., J.F., S.G., P.L.-N., G.L., S.M., C.R., E.S., M.T., F.C., A.H.G., T.Q., C.H., W.H.O., P.D., U.T., J.E., N.J.S.
CARDIoGRAM consortium executive group: H.S., S.K., M.P.R., J.E., N.J.S.
CARDIoGRAM consortium steering group: H.S., I.R.K., S.K., M.P.R., T.L.A., E.B., R.L., A.Z., C.H., A.S.H., U.T., J.R.T., R.M., J.E., N.J.S.
DISCLOSURES AND COMPETING FINANCIAL INTERESTS
Genotyping of PennCATH and Medstar was supported by GlaxoSmithKline. Dawn M. Waterworth, Max C. Walker and Vincent Mooser are employees of GlaxoSmithKline. Hilma Holm, Solveig Gretarsdottir, Jeffrey R. Gulcher, Augustine Kong, Kari Stefansson, Gudmar Thorleifsson, Gudmundur Thorgeirsson, and Unnur Thorsteinsdottir are employees of and/or own stock/stock options in deCODE genetics. There are no other disclosures.
References
- 1.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. The New England journal of medicine. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 2.Schunkert H, Erdmann J, Samani NJ. Genetics of myocardial infarction: a progress report. Eur Heart J. 2010;31:918–925. doi: 10.1093/eurheartj/ehq038. [DOI] [PubMed] [Google Scholar]
- 3.Preuss M, et al. Design of the Coronary Artery Disease Genome-Wide Replication and Meta-Analysis (CARDIoGRAM) Study -- A Genome-Wide Association Meta-Analysis Involving More than 22,000 Cases and 60,000 Controls. Circ Cardiovasc Genet. 2010 doi: 10.1161/CIRCGENETICS.109.899443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke R, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. The New England Journal of Medicine. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 5.Tregouet DA, et al. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nature Genetics. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 6.Psaty BM, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nitz I, et al. Association analyses of GIP and GIPR polymorphisms with traits of the metabolic syndrome. Mol Nutr Food Res. 2007;51:1046–1052. doi: 10.1002/mnfr.200700048. [DOI] [PubMed] [Google Scholar]
- 8.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 9.Zhong H, et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6:e1000932. doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge B, et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nature Genetics. 2009;41:1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 11.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown MS, Goldstein JL. Expression of the familial hypercholesterolemia gene in heterozygotes: mechanism for a dominant disorder in man. Science. 1974;185:61–63. doi: 10.1126/science.185.4145.61. [DOI] [PubMed] [Google Scholar]
- 13.Linsel-Nitschke P, et al. Lifelong reduction of LDL-cholesterol related to a common variant in the LDL-receptor gene decreases the risk of coronary artery disease--a Mendelian Randomisation study. PLoS One. 2008;3:e2986. doi: 10.1371/journal.pone.0002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linsel-Nitschke P, Samani NJ, Schunkert H. Sorting out cholesterol and coronary artery disease. The New England Journal of Medicine. 2010;363:2462–2463. doi: 10.1056/NEJMcibr1010765. [DOI] [PubMed] [Google Scholar]
- 16.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson PW, et al. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 18.Ripatti S, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samani NJ, et al. Genomewide association analysis of coronary artery disease. The New England Journal of Medicine. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudbjartsson DF, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nature Genetics. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 21.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nature Genetics. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.