Abstract
The chemical properties of zinc make it an ideal metal to study the role of coordination strain in enzymatic rate enhancement. The zinc ion and the protein residues that are bound directly to the zinc ion represent a functional charge/dipole complex, and polarization of this complex, which translates to coordination distortion, may tune electrophilicity, and hence, reactivity. Conserved protein residues outside of the charge/dipole complex, such as second-shell residues, may play a role in supporting the electronic strain produced as a consequence of functional polarization. To test the correlation between charge/dipole polarity and ligand binding affinity, structure-function studies were carried out on the di-zinc aminopeptidase from Vibrio proteolyticus. Alanine substitutions of S228 and M180 resulted in catalytically diminished enzymes whose crystal structures show very little change in the positions of the metal ions and the protein residues. However, more detailed inspections of the crystal structures show small positional changes that account for differences in the zinc ion coordination geometry. Measurements of the binding affinity of leucine phosphonic acid, a transition state analogue, and leucine, a product, show a correlation between coordination geometry and ligand binding affinity. These results suggest that the coordination number and polarity may tune the electrophilicity of zinc. This may have provided the evolving enzyme with the ability to discriminate between reaction coordinate species.
Metalloenzymes are some of the most powerful catalysts in the world. Understanding how the protein/metal partnership can give rise to dramatic rate enhancement will broaden the scope and understanding of enzyme evolution, protein engineering, and synthetic catalyst design. Two well known theories provide an evolutionary framework in the description of enzymatic rate enhancment: Arieh Warshel's preorganization theory (1–3) and the strain theory (4, 5) as first proposed by R.J.P Williams and B.L. Vallee.
Based on decades of chemical and computational research, Warshel's results indicate that enzymes provide preorganized networks, optimized by evolutionary pressure, paid for by folding energy, and aimed at enhancing the electrostatic force between charges. These preorganized features enhance the electrostatic effect, and lower the activation energy barrier, by orienting dipoles toward the stabilization of functional charges and the charged transition states (1–3).
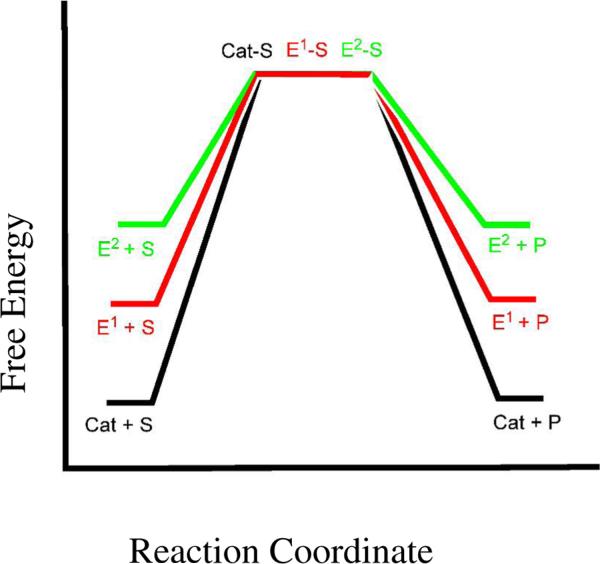
Enzymatic rate enhancement by the use of “strained” groups is an idea first postulated by R.J.P. Williams and B.L. Vallee. Based on the unusual absorption, EPR, magnetic, redox, and ligand binding properties of metalloenzymes, Vallee and Williams proposed that large molecules, like proteins, could hold functional groups (metals, cofactors, side-chains) in strained conformations as a strategy toward rate enhancement. They defined the entatic state as an energized state supported by the stable protein fold (4, 5). Figure 1 illustrates a simple example: a small molecule catalyst binds a substrate and forms product (black line). A larger, stronger molecule is able to hold the “catalytic group(s)” in a strained conformation, E1, a state that is closer in energy to the transition state, increasing the rate of the reaction (red line). A more rigid scaffold could maintain an even more energized active site, E2, and so forth. The strain theory predicts a tug-of-war relationship between the “group” and the “scaffold”; a rigid matrix may hold the “group” in an energized state, whereas a flexible (less rigid) matrix may become energized by the group (metals in structural sites). Unlike the preorganization theory, the entatic theory does not clearly define the meaning of strain (which can be steric or electronic) or how that strain relates to rate enhancement. Under certain circumstances, the entatic theory and the preorganization theory may be explaining the same effect – if the protein fold pays for the electronic strain that is produced as a consequence of functional polarization of the charge/dipole complex, then some portion of rate enhancement may be ascribed to the entatic enhancement of the electrostatic effect.
Figure 1. The Effect of Active Site Ground State Destabilization on the Activation Energy of a Unimolecular Reaction.
A small molecule catalyst binds substrate and produces product (black line). The evolution of the appropriate scaffold may hold the functional group(s) in a higher energy state, E1, and decrease the activation energy (red line). Functional groups held in a higher energy state, E2, may further increase the rate of the reaction (green line).
Zinc enzymes make an ideal system to test the entatic theory because the role played by zinc is strictly electrophilic, and the electrophilic properties are related to the charge/dipole interaction. Surveys of the Cambridge Structural Database (CSD) show zinc ion coordination number frequencies of 59% and 23% for 4 and 6 coordination numbers, respectively (6). In sites characterized by a lower dielectric than water, where zinc is in complex with ionized ligands, the difference between coordination number energies can be quite large. Calculations done using density functional and continuum dielectric theory compute that zinc complexes with one acidic or two or more neutral ligands prefer to be tetracoordinated in sites characterized by a lower dielectric than water, like in a protein (7, 8). Dudev and Lim compute that in a protein like environment, where zinc is sequestered from bulk water and in complex with heavy ligands, such as the ionized side-chains of cysteine, aspartic acid, and glutamic acid, the charge transfer from the first four coordinating atoms to the zinc ion is so great that the charge of zinc is virtually neutralized. Consequently, the addition of more ligands to the coordination sphere is accompanied by electron repulsion. This effect is unique to zinc and is a consequence of the difference in energy between the vacant and highest occupied molecular orbitals (7, 8). Dudev and Lim's calculations show that the magnesium ion, which prefers to be octahedrally coordinated in solvent exposed and buried sites, has a larger energy difference between the vacant and the highest occupied molecular orbitals, which translates into less charge transfer and less charge neutralization (8). These results are in agreement with previous studies done by Ryde on the zinc site of alcohol dehydrogenase, where differences in energy of 24–48 kcal/mol were calculated between tetracoordinated and pentacoordinated zinc species (9, 10).
Interestingly, the zinc ion coordination prevalence in protein sites depends on whether the zinc plays a structural or a catalytic role. In structural zinc sites (where the zinc ion acts to maintain the integrity of the fold) the occurrence rate for 4, 5, and 6 coordination numbers is 79%, 6%, and 12%, respectively; in catalytic zinc sites the occurrence rate for 4, 5, and 6 coordinations is 48%, 44%, and 6%, respectively (11). Five coordinate or geometrically strained zinc sites may represent energized sites ready for catalysis, whereas four coordinate, ideal tetrahedral zinc sites may represent stable sites primed for structural support.
Although surveys of protein zinc sites reveal that there is a 2:1 ratio of first-shell (metal coordinated) to second-shell (hydrogen bonded to first-shell) residues (12), in general, the role played by second-shell residues is not well understood. Studies done on carbonic anhydrase show that second-shell residues contribute to metal binding affinities and active site charge distribution (13, 14). Other studies suggest that second-shell residues anchor the direct metal ligands into a particular orientation that may be required for catalysis (15, 16). The subtle effects of second-shell residues on structure make them good candidates for structure-function studies.
The aminopeptidase from Vibrio proteolyticus (AAP) is a model enzyme for the study of the effect of extended residues on charge/dipole polarity and ligand binding affinity. AAP is a small, heat-stable protease that can remove most N-terminal residues from proteins and peptides, but is most efficient at removing bulky hydrophobic residues (17, 18). Studies on AAP have been carried out for decades. Two zinc ions in close proximity have been identified to form the metal component of the active site (Figure 2) (19, 20). Through structural and mutational studies, several protein residues have been shown to be critical to power the full potential of the enzyme. The E151 residue is a non-metal coordinated conserved residue whose mutant forms (alanine, histidine, and aspartate) render the protein inactive or nearly so (21–23). A recent quantum mechanical study on the mechanism of AAP implicates E151 as an acceptor of a proton from the amino-terminus of the incoming peptide, and subsequently, the donor of a proton to the newly formed amino-terminus - a step that is calculated as rate limiting (24).
Figure 2. Active Site Bond Lengths of AAP (Å).
Metal coordination and second-shell residue distances measured from the 0.95 Å AAP structure, PDB ID 1RTQ (26) (Figure made with Chemdraw 10.0).
The catalytic metal ions of AAP are situated in a cavity where they are held in distorted tetrahedral coordination in complex with aspartic acid, glutamic acid, and histidine residues (Figure 2). It is unknown if this distortion is important for high catalytic efficiency. Surveys done on zinc geometric coordination angles in small molecule structures show that the deviation of a tetrahedral complex from perfect geometry, calculated as the root mean square deviation, δ, has a mean value of 4.3° (with a 2.2° sample standard deviation) for non-chelated zinc complexes, and 11° (with a 5° sample standard deviation) for chelated complexes (25). Measurements of the zinc coordination bond angles of the 0.95 Å WT structure (PDB ID 1RTQ, (26)) show that the active site metals of AAP have r.m.s. deviations from ideal tetrahedral geometry of 24° and 16° for Zn1 and Zn2, respectively. Conserved residues that lie outside of the charge/dipole complex include S228, a second-shell residue; M180, which is located above the active site pocket; and D118, whose side chain forms hydrogen bonds with two loops behind the active site. The 0.95 Å structure of AAP (largest bond length errors of +/− 0.02 Å, PDB ID 1RTQ (26)) shows that S228 is in a short hydrogen bond (2.58 Å) with D179; a metal coordinated aspartate that displays bidentate-like coordination to Zn2 (Figure 2). A structural alignment of AAP with Streptomyces griseus aminopeptidase (SGAP) (1.58 Å, PDB ID 1CP7) (27), which has 25% sequence identity with AAP, reveals virtually superimposable active sites and other conserved features, such as a hydrogen bond network reaching out from Zn2-Asp179-Ser228 and ending at another short hydrogen bond (Figure 3).
Figure 3. Structural Features and Hydrogen Bond Network Shared by AAP and SGAP.
Structural features outside the active site that are common to AAP and SGAP include a series of beta sheets that house first-shell and extended residues. The conserved hydrogen bond network, Zn2-D179-S228-Q177-D229-S158, is virtually superimposable in a structural alignment (Figure made with PyMOL).
This paper presents a structure-function study carried out on the M180A, S228A, and D118N variants of AAP. Along with the determination of a series of high-resolution crystal structures, the binding affinity of leucine phosphonic acid (LPA), a transition state analogue, and leucine, a product, were measured. An empirical method was developed for the analysis of metal ion coordination geometries and use of this method shows a correlation between ligand binding affinity and zinc ion coordination geometry. A case for the entatic interpretation is presented and its implications are discussed.
METHODS AND MATERIALS
Mutagenesis, and Expression
All mutational forms of AAP (EC number 3.4.11.10) were constructed following the QuikChange™ Site Directed Mutagenesis protocol of Stratagene. AAP, in plasmid pet21b+ (Novagen), was kindly provided by Professor Rick Holz. AAP and variants of AAP were expressed using the Auto-Induction System (28). The plasmid encoding the variant form of AAP was transformed into the BL21 (DE3) strain of E. coli and a single colony was used to inoculate 30 mL of starter culture (28.08 mL ddH2O, 30 μL of 1 M MgSO4, 3 μL of 0.01 M ZnSO4, 375 μL of 40% glucose, 1.5 mL of 20 × NPS (0.5 M (NH4)2SO4, 1.0 M KH2PO4, 1.0 M Na2HPO4), & 60 μL of 50 mg/mL kanamycin) and grown overnight at 37 °C and 300 rpm. The −80 °C stocks were made by mixing 1.5 mL of starter culture with 150 μL of 80% glycerol. A 1–2 μL scrape of the frozen stock culture was used to inoculate 1 L of autoinduction media (928 mL of NZ-amine, 20 mL of 50 × 5052 (for 100 mL: 25 g glycerol, 73 mL H2O, 2.5 g glucose, 10 g alpha-lactose), 50 mL of 20 × NPS (aforementioned), and 2 mL of 50 mg/mL kanamycin). The culture was grown at 37 °C and 300 rpm for 18 hrs. The culture was spun down for 30 minutes at 8000 rpm at a temperature of 4 °C. The protein is secreted so the supernatant was removed and saved and the pellet was discarded.
Protein Purification
The secreted protein was precipitated with an ammonium sulfate cut as first reported by Prescott and coworkers with the modifications described below (29). After equilibrating the temperature of the supernatant to 4 °C, 364 g of (NH4)2SO4 were slowly added to every liter while stirring. The sample was stirred for two hours and left at 4 °C overnight. The next day the precipitant layer of protein at the bottom of the flask was resuspended by stirring and then spun down by centrifugation for 1 hr at 8000 rpm. The solution was carefully decanted and discarded. The protein precipitant on the side of the tube was gently resuspended into a total of 20 mL of Tricine buffer (50 mM Tricine, 1.0 mM ZnSO4, and pH 8.0) and dialyzed against 2 L the same solution (at 4 °C) to remove any excess (NH4)2SO4. Next, the protein was introduced into 1 L of Tricine buffer and re-cut with 364 g of ammonium sulfate as a step toward purification against an unidentified “brown pigment” that binds strongly to the protein but must be removed for crystallization. The sample was allowed to stir for 2 hours and then the precipitated protein was spun down, resuspended in Tricine buffer (10 mL), and dialyzed against 2 L of Tricine buffer to remove remaining ammonium sulfate. AAP is expressed with sequences that need to be proteolytically removed for full activity and crystallization. The protein was processed as previously reported (30). The mostly non-specific endopeptidase, proteinase K, was added to the AAP solution to give a final concentration of 20 nM proteinase K, and the sample was incubated at 37 °C with mild rocking. Samples of the protein were collected for an activity assay and an SDS gel every fifteen minutes. When the protein activity hit a plateau, the sample was transferred to a 65 °C water bath for 20 minutes to denature proteinase K. The sample was spun down and further purified from unprocessed protein, remaining brown pigment, and other contaminants. First, the protein was dialyzed against two exchanges of 2 L of acetate buffer (10 mM sodium acetate, 50 mM acetic acid, 0.2 mM ZnS04, pH 4.0) followed by dialysis against 2 L of Tricine buffer. The sample was spun down, filtered, and subject to anion exchange chromatography at pH 8.0 using a 300 mL linear gradient against elution buffer (Tricine buffer aforementioned with a 1 M NaCl) on a 20 mL Mono-Q column. Further cycles of acetate dialysis and chromatography were employed if brown color persisted.
Kinetic Studies
Initial rates of leucine p-nitroanalide (leu-PNA) hydrolysis were measured by monitoring absorbance at 405 nm (ε405 = 10,800 for p-nitroanaline). The formation of p-nitroanaline was followed at 25 °C in 50 mM Tricine, 1.0 mM ZnSO4, 0.2 M KCl, and pH 8.0 (31, 32). The activities of the enzymes (activity = μmoles of product/ min·mg) were measured at increasing zinc ion concentrations (0, 0.01, 0.1, 0.5, and 1.0 mM ZnSO4) to ensure kinetic assays were done under saturating metal conditions. To do this, the enzyme, which was stored in the aforementioned kinetics buffer, was added to kinetics buffer premixed with 1 mM substrate and the appropriate concentration of ZnSO4. The Vmax and Km were determined by following the reaction over a series of substrate concentrations ranging from 5 to 150 μM for WT, D118N, and the S228A enzymes and 100 to 2000 μM for the M180A variant. To dissolve the substrate to high concentrations, the substrate was first dissolved in ethanol and then introduced to the kinetics buffer, while stirring, to give a final ethanol concentration of 0.5%. An equivalent concentration of ethanol was added to all other kinetics solutions, and the activities of the enzymes were measured at 0.1, 0.5, and 1.0% ethanol to ensure that ethanol was not inhibiting. The Ki for LPA, leucine phosphonic acid ((1-amino-3-methylbutyl) phosphonic acid, Acros Organics), was determined as previously described for WT (33) by measuring the initial rate of leu-PNA hydrolysis with inhibitor concentrations ranging from 0–2 μM for WT, S228A, and D118N enzymes and 0–300 μM for the M180A mutant. Leucine inhibition was measured over a range of 0–5000 mM, and fine-tuned to 0–150 mM for the S228A variant.
Crystallization
WT and the variant forms of AAP crystallized by hanging drop method at 25°C from the same conditions reported for WT enzyme (20). For cocrystals, a 2 μL drop of 30 mg/mL protein in a solution of 10 mM HEPES, 10 mM KSCN, and 0.4 M NaCl at pH 8.0 was mixed by pipetting with a 2 μL drop of 1 mM leucine or 1 mM LPA in 10 mM HEPES, 10 mM KSCN, and 0.4 M NaCl at pH 8.0. The 4 μL mixture was suspended over a well containing 100 mM HEPES, 100 mM KSCN, and 4.0 M NaCl at pH 8.0. Crystals with dimensions of 0.2 × 0.2 × 0.3 mm appeared within 2–5 days and were isomorphous with WT, belonging to the hexagonal space group P6122 with one molecule per asymmetric unit.
Data Collection and Processing
The crystals were soaked in 50% sodium malonate solution at pH 6.7, mounted on a Hampton cryoloop, then frozen and stored in liquid nitrogen. The data were collected at 100 K at the Advanced Photon Source (APS) on the BioCARS Sector 14 BM-C fixed wavelength (λ = 0.90010), bending magnet beam line. To optimize the number of reflections collected for the S228A, D118N, and M180A crystals, two complete passes with differing exposure times were collected. The data were indexed and integrated using HKL2000 (34). Overlapping reflections were used to merge high and low resolution passes to yield complete data sets for the structure determination of the S228A, D118N, M180A and M180A/leucine crystals. Initial phases were obtained by molecular replacement using MOLREP (35) and the original crystal structure of AAP (PDB ID 1AMP) (all atom, no waters, no metals) as the search model (20). COOT (36) was used for rebuilding followed by isotropic B-factor restrained refinement with REFMAC5 (37) using the CCP4 (38) suite. Difference density for the mutations, ligands, and waters were fitted and refined. The S228A, D118N, M180A and M180A/leucine data were subject to anisotropic refinement. Table 1 lists the crystallographic data collection and refinement statistics.
Table 1. Crystallographic Collection and Refinement Statistics.
| STRUCTURE | D118N/Amino Acid | S228A/Leucine | S228A/Amino Acid | S228A/LPA | M180A | M180A/LPA | M180A/Leucine |
|---|---|---|---|---|---|---|---|
| PDB ID | 3B3T | 3B3W | 3B3V | 3B7I | 3B35 | 3B3C | 3B3S |
| Space group | P6122 | P6122 | P6122 | P6122 | P6122 | P6122 | P6122 |
| Unit cell parameters, Å |
a=b=109.3 c = 91.1 |
a=b=110.6 c = 91.6 |
a=b=109.3 c = 91.0 |
a=b=109.9 c = 91.1 |
a=b=109.2 c = 91.0 |
a=b=108.2 c = 97.2 |
a=b=109. c = 91.0 |
| Resolution | 42–1.17 | 15–1.75 | 22–1.22 | 20–1.75 | 41.0–1.15 | 30.6–1.46 | 17.2–1.18 |
| Number of Unique Reflections | 102,258 | 30,654 | 92,234 | 32,789 | 122,065 | 54,941 | 102,599 |
| Rmerge, %a | 7.5 (56.8) | 8.8 (66.2) | 9.3 (67.1) | 12.9 (65.9) | 5.9 (78.8) | 6.2 (50.2) | 8.7 (55.0) |
| Completeness % | 95.3 (91.3) | 92.8 (93.1) | 97.7 (81.4) | 98.6 (98.1) | 89.5 (95.1) | 99.7 (98.9) | 99.8 (98.4) |
| Cutoff I/σ(I) | 2.1 | 2.9 | 1.52 | 2.2 | 2.0 | 2.1 | 2.4 |
| B-factor model | anisotropic | isotropic | anisotropic | isotropic | anisotropic | isotropic | anisotropic |
| Rwork, % | 14.16 | 19.4 | 14.91 | 17.65 | 14.9 | 19.6 | 16.8 |
| Rfree, % | 16.09 | 23.4 | 17.05 | 20.87 | 16.6 | 22.6 | 19.6 |
| <B> | 11.3 | 27.4 | 19.6 | 14.1 | 14.6 | 23.6 | 15.5 |
| DPI, Åb (standard positional uncertainty, σ (x, Bave) | 0.03 | 0.12 | 0.04 | 0.10 | 0.03 | 0.07 | 0.04 |
| Estimated minimal error, Åb | 0.01 | 0.03 | 0.01 | 0.03 | 0.01 | 0.01 | 0.02 |
| No. of protein residues | 291 | 291 | 291 | 291 | 291 | 291 | 291 |
| No. of waters | 303 | 263 | 221 | 250 | 357 | 278 | 308 |
Values in highest resolution shell reported in parenthesis.
Values calculated by SFCHECK.
Data Analysis
The coordination number and distortion of each metal ion was calculated by the empirical method described below. After the crystal structure was fully refined, each metal ion site was analyzed by measuring the distances from the zinc ion to oxygen and nitrogen atoms within a 2.5 Å radius. Distances greater than the average distance by 0.2–0.3 Å were omitted, and the remaining coordinations were fitted to perfect tetrahedral geometry (all angles 109.5°), if four coordinate; or perfect trigonal bipyramidal geometry (angles of 180°, 120°, and 90°), if five coordinate. The root mean square deviations were calculated using the equation:
Errors in Structure
The inversion of the least-square full matrix is needed to obtain the errors of a crystal structure. If this is not possible, methods have been developed that can give an approximate value of the parameter errors. The diffraction-component precision index (DPI) calculates an approximate value for the standard uncertainty in positions (for atoms with B = Bave) using R and an approximation to the least-square method,
where Ni = number of fully occupied sites, p = nobs-nparams, C = fractional completeness of structure factor data, R = Σ | ΔF | /Σ | F |, and dmin is the minimal d spacing (39). This method works best for resolutions greater than 2.0 Å. When tested, the DPI method has shown to yield positional uncertainties that are very similar to full matrix inversions (40). SFCHECK uses a modified DPI (41) method dependent on Rfree (42),
where nobs in the number of reflections used in refinement. The Rfree DPI method works for low and high-resolution data. Table 1 lists the standard uncertainties in atomic positions of the AAP structures using the DPI method.
RESULTS
Kinetic Parameters and Inhibition of Enzymes
All assays were done in an excess of zinc ion to ensure that the metal sites were saturated. Table 2 shows the average, normalized activity of each of the variants at increasing ZnSO4 concentrations. The maximum activity and the Km of the WT, D118N, S228A, and M180A enzymes were determined by fitting the experimental data to Michaelis-Menten curves using the SigmaPlot Kinetics Module. The data were subject to several goodness-of-fit tests, including AICc, R2, and sum of squares, to determine the best model of inhibition and the quality of that model. Leucine and LPA inhibition assays show that in all cases the data fitted best to a competitive inhibition model, and tests on that model show R2 value greater than 0.95. The kinetic parameters and the inhibitor constants, along with their standard errors, are presented in Table 3.
Table 2. Average, Normalized Activity of AAP Variants at Increasing ZnSO4 Concentrations.
To ensure that the kinetic assays were done at saturating zinc concentrations, the activity of each of the variants was measured in 50 mM Tricine buffer, 1 mM p-nitroanalide, 200 mM KCl, pH 8.0, 25 °C, and at increasing ZnSO4 concentrations.
| 0 μM | 10 μM | 100 μM | 500 μM | 1000 μM | |
|---|---|---|---|---|---|
| WT | 0.9 | 0.9 | 0.9 | 0.9 | 1.0 |
| D118N | 0.9 | 0.9 | 0.9 | 1.0 | 0.9 |
| S228A | 0.8 | 0.9 | 0.9 | 0.9 | 1.0 |
| M180A | 0.6 | 0.6 | 0.7 | 0.9 | 1.0 |
Table 3. Kinetic Parameters and Inhibitor Constants.
Kinetic parameters and inhibitor constants for WT, D118N, S228A, and M180A enzymes in 50mM Tricine buffer, 1.0mM ZnSO4, and 200mM KCl, at pH 8.0 and 25 °C.
| kcat (s−1) | Km (μM) | kcat/Km (s−1μM−1) | Kicompetitive Leucine (μM) | Kicompetitive LPA (μM) | |
|---|---|---|---|---|---|
| WT | 65 +/− 5 | 13 +/− 1.0 | 5 | 1100 +/− 150 | 1.9 +/− 0.3 |
| D118N | 60 +/− 3 | 10 +/− 0.4 | 6 | 870 +/− 100 | 1.3 +/− 0.1 |
| S228A | 7.0 +/− 0.7 | 25 +/− 2 | 0.3 | 65 +/− 8 | 1.5 +/− 0.2 |
| M180A | 0.7 +/− 0.1 | 340 +/− 30 | 0.002 | 2800 +/− 200 | 105 +/− 11 |
In a comparison with WT enzyme, at saturating substrate concentrations, the S228A enzyme is approximately ten-fold less active, M180A is approximately 100-fold less active, and the D118N enzyme has near the same activity. The Km of the M180A enzyme is approximately thirty-fold higher than WT, whereas the S228A and the D118N variants have Km values close to the WT Km. Compared to the other enzymes, S228A has a marked increase in leucine affinity, roughly twenty-fold higher than that of WT. In LPA affinity, M180A stands out with approximately fifty-fold lower affinity than WT.
X-ray Structure of the S228A Enzyme
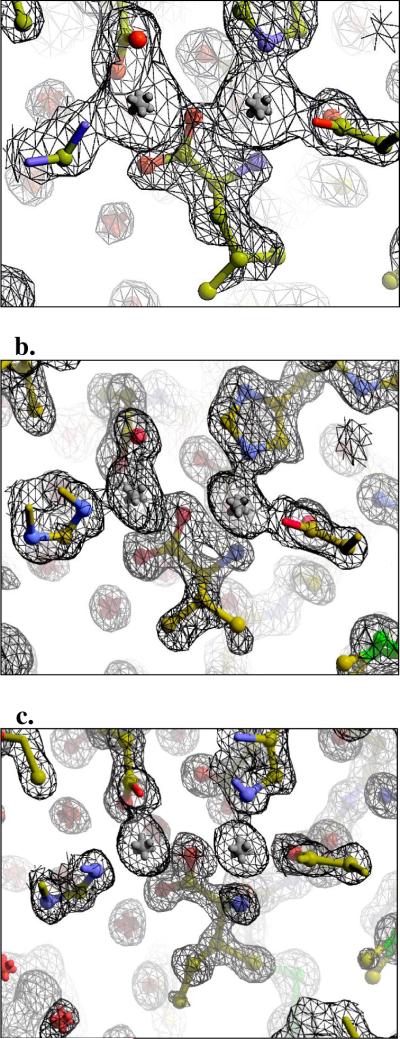
The recently solved high-resolution structure of WT AAP (0.95 Å) has allowed for precise measurements of the locations of the atoms in the active site (26). The conserved S228 residue forms a short hydrogen bond (2.58 Å) with the anti-orbital electrons of the D179 side chain, a residue bound directly to Zn2 (Figure 2). An alanine substitution at this position does not change the Km or the LPA Ki values much, but does decrease the activity by ten-fold and enhance the leucine affinity by twenty-fold (Table 3). S228A crystal structures have always revealed positive density in the active site. The 1.22 Å structure in Table 1 labeled S228A/Amino Acid is one such structure. The origin of this density is unknown, but given the extensive purification of AAP, the catalytic efficiency and loose specificity of AAP, and how well the density fitted to a fully occupied amino acid, most likely, the density comes from the amino acid products made as a consequence of AAP proteolyzing itself. Valine was modeled into the density, however, other amino acids, like leucine or isoleucine may have just as likely been used without much change to the quality of the difference density (Figure 4b). Regardless of side chain, the head group is always in the same position, at full occupancy.
Figure 4. Representative (2Fo−Fc) Active site density.
a.) S228A/Leucine structure contoured at 1.5 σ, b.) the S228A/Amino Acid structure contoured at 1.6 σ, and c.) the D118N/Amino Acid structure contoured 1.8 σ (Figure made with PyMOL). The gray spheres represent zinc 1 (on left) and zinc 2 (right).
An alignment of the high resolution WT structrue with the S228A structure shows little or no changes in the positions of the backbone atoms or the atoms in the active site, including the metal ions. One notable difference, however, is D179 (Figure 2), whose binding has gone from a bidentate binding mode to a more monodentate like binding mode. In the absence of S228, the oxygen of the side chain of D179 moves away from the metal by over 0.4 Å compared to its position in the WT enzyme (Figure 5a).
Figure 5. Active site coordination distances (Å).
a.) 1.22 Å S228A/amino acid structure, b.) 1.75 Å S228A/leucine structure, c.) 1.75 Å S228A/LPA structure, and d.) 1.17 Å D118N/amino acid structure. (Figure made with Chemdraw 10.0).
X-ray Structure of the S228A/Leucine Cocrystal
The S228A variant was crystallized in the presence of leucine and the crystal structure was solved to 1.75 Å resolution. The electron density maps show positive difference density in the active site that fitted the head group of an amino acid very well, and the side chain of leucine the best (Figure 4a). The coordinates of the active site residues, and the metal ions, are identical to their positions in the S228A structure; and the leucine is in the same location and orientation as the amino acid modeled in the S228A active site. Like the other S228 data, this structure shows that the oxygen of the D179 residue has moved back, leaving the D179-Zn2 coordination in a more monodentate-like mode than that seen in the WT structure (Figure 5b).
X-ray Structure of the S228A/LPA Cocrystal
The data collected from the crystal obtained from cocrystallization of S228A with LPA shows partial occupancy of the LPA molecule and what appears to be partial occupancy of an amino acid. LPA and leucine were modeled into the active site at half occupancy. The LPA fitted and refined into the same position and orientation seen in the WT enzyme in complex with LPA (2.20 Å, PDB ID 1FT7 (33)). Although the partial occupancy of the amino acid introduces errors into the analysis of the coordination bond distances and angles, the structure was included in the analysis (Figure 5c).
X-ray Structures of WT and D118N Enzymes
The aspartate in the 118 position is not conserved, as sequence alignments show that asparagine can occupy this position as well. The D118 residue is not in the first- or the second-shell, but it is located near the active site, within 5 Å of the nearest first-shell residue. The high-resolution structure of WT enzyme solved by Desmarias et al. (26) makes it possible to make bond length measurements with high precision. The r.m.s.d. from ideality was reported as +/− 0.2, making it possible to measure carboxylate bond lengths, which differ depending on the protonation state. A protonated D118 would shed light on why the sequence alignments show conservation of aspartate or asparagine at this position. The measurements of the two C-O bonds on the side chain of D118 are 1.24 and 1.32 Å, suggesting that D118 is protonated. To further study this, an asparagine mutant was made for structure/function studies. Catalytically, in terms of Km, kcat, and the Ki of leucine and LPA, the enzyme behaves much like WT (Table 3), and structurally, not much has changed. The 1.17 Å D118N structure aligned with WT shows that the nitrogen on the carbamide occupies the same space occupied by D118's carboxylate oxygen with the longer bond, supporting the hypothesis that D118 is protonated. Unlike WT protein, the N118 enzyme has difference density in the active site, which once again resembles a fully occupied amino acid. An isoleucine molecule was fitted and refined into the density (Figure 4c).
X-ray Structure of the M180A Variant
Methionine 180 is a conserved residue located above the active site (Figure 7a). The 1.15 Å structure of the M180A variant shows that an alanine substitution at position 180 does little to alter the positions of the atoms in the active site. The hole created by the missing methionine is filled by slight variations in packing. One notable change is the side chain of I255 rotates about its β-carbon to fill some of the space that would otherwise be occupied by M180 (Figure 7). The metal ions and the first-shell residues are in the same positions and orientations as they are in the WT structure, with the exception of D179. In the WT enzyme, D179 is coordinated to Zn2 and is a short hydrogen bond partner to S228 (Figure 2). In the M180A structure, D179 appears in two discrete positions, each fitted at half occupancy, and each position is over 3.2 Å away from S228: the 2.58 Å hydrogen bond seen in the WT structure is broken in the M180A variant (Figure 6a). Energetically, this is not the only curious happening. Compared to WT, the partially occupied D179 residue, D179b, depicted by red, hashed bonds in Figure 6a, exhibits a near 180° flip of the backbone carbonyl. The r.m.s.d. of both zinc complexes are strikingly lower than the r.m.s.d of WT, especially when calculated using D179b, the conformer that takes the backbone with it (Table 4).
Figure 7. Active site models.

a.) WT, b.) M180A, and the c) I255 in the WT/M180A alignment. Figure made with Molscript.
Figure 6. Active site coordination distances (Å).
a.) 1.15 Å M180A, b.) 1.46 Å M180A/LPA, c.) 1.18 Å M180A/leucine, and 2.2 Å WT/LPA (PDB ID 1FT7). Figure made with Chemdraw 10.0.
Table 4. Metal Coordination Numbers and R.M.S. Deviations.
Depending on the coordination number, the zinc ions were fitted to either perfect tetrahedral (all angles 109.5°) or perfect trigonal bipyramidal (angles of 180°, 120°, and 90°) coordination geometry, and the root mean square deviations were calculated (25). After the structure was fully refined, each metal ion site was analyzed by measuring the distances from the zinc ions to oxygen and nitrogen atoms within a 2.5 Å radius. Distances greater than the average distance by 0.2–0.3 Å were omitted, and the remaining coordinations were fitted to perfect tetrahedral geometry or perfect trigonal bipyramidal geometry.
| Structure | Resolution | Zn1 coordination number | Zn1 r.m.s. deviation | Zn2 coordination number | Zn2 r.m.s. deviation |
|---|---|---|---|---|---|
| AAPc | 0.95 Å | 4 | 24° | 4 | 16° |
| AAP/LPAd | 2.20 Å | 4 | 11° | 5 | 17° |
| D118N/amino acid | 1.17 Å | 4 | 15° | 5 | 18° |
| S228A/amino acid | 1.22 Å | 4 | 17° | 5 | 14° |
| S228A/leucine | 1.75 Å | 4 | 17° | 5 | 13° |
| S228A/LPA | 1.75 Å | 4 | 20° | 5 | 17° |
| M180A | 1.15 Å | 4 | 20° | 4 | 14°a/11°b |
| M180A/LPA | 1.46 Å | 4 | 15° | 5 | 15° |
| M180A/leucine | 1.18 Å | 4 | 19° | 4 | 14°a/11°b |
PDB ID 1RTQ,
PDB ID 1FT7.
X-ray Structure of the M180A/LPA Crystal
The crystal structure of M180A in the presence of LPA was solved to see how the molecule binds to the mutant active site. Comparisons of the 1.46 Å M180A/LPA crystal structure with the previously solved WT/LPA structure (PDB ID 1FT7) show that the LPA molecule is bound in the same position and orientation in both enzymes (Figure 6b, Figure 6d). Unlike the M180A structure, the cocrystal M180A/LPA structure shows D179 in one position, at full occupancy, hydrogen bonded to S228. Although the kinetics show a fifty-fold decrease in LPA affinity, the structure shows no change in LPA binding. The bonding interactions that LPA makes with M180A are the same interactions that LPA makes with WT.
X-ray Structure of the M180A/Leucine Crystal
Leucine has a three-fold lower affinity for the active site of M180A than it has for the active site of WT. The 1.18 Å structure of M180A in the presence of leucine shows that the hydrophobic leucine moiety is nestled inside of the hydrophobic pocket, thus blocking the active site; however, the head group is pointed away from the zinc ions. Other than the leucine molecule, the structure is identical to M180A without cocrystallization: the zinc complexes are tetrahedral with the same low r.m.s. deviations (Table 4), D179 is in two conformations, the D179 conformation with lower r.m.s.d. exhibits a backbone flip, and the D179-S228 hydrogen bond is broken (Figure 6c).
DISCUSSION
Enzymes are large, complex molecules; a theory of the origin of enzymatic rate enhancment must account for this complexity. Both the preorganization and the entatic theory provide an evolutionary framework in their description of enzymatic rate enhancement: the theories relate complexity to function. They share the viewpoint that strain (whether it be electronic or steric) is a consequence of features that enhance catalysis, and that the rest of the enzyme has evolved stabilizing interactions to pay for this strain. This viewpoint is supported by several studies: surveys of the PDB show localization of strained residues in functional areas of proteins (43–45), many independent studies show stability for function trade-offs (46–48), and fold and function are poorly correlated (49, 50).
For the purpose of testability, it is important to define what is meant by strain. The entatic hypothesis, as first reported, is unclear in the meaning of strain, which can be steric (mechanical) or electronic. In the case of metalloenzymes, Williams and Vallee use the term “strain” to describe the distortion of metal coordination geometry from ideal, but “coordination strain” does not mean anything unless the energetic contribution to such an effect can be described. Furthermore, Warshel's work has focused mainly on the use of mechanical strain in catalytic rate enhancement. The adaptation of coordination geometry distortion may be to enhance the electrostatic effect. In the preorganization of protein dipoles around a charge, the reactivity of the charged center may be enhanced by a polarity in the charge/dipole complex (Figure 8). For the sake of reactivity, it may be important to keep one side of the charge-shell exposed - a polarized beacon for the incoming charge; not only do the orientations of protein dipoles matter, but their positions as well. Thus, the cost of preorganization will include the repulsion energy of localizing charge, and in the case of metals, this may translate to breaking symmetry. If the protein matrix absorbs the electronic strain produced by functional geometric distortion of metal ion coordination geometry, then some portion of rate enhancement can be ascribed to the entatic enhancement of the electrostatic effect.
Figure 8. The Effect of Symmetry on the Polarity of the Charge/Dipole Complex.
The orientation and the positions of dipoles around a charge will influence the overall polarity of the charge/dipole complex. Localization of the dipoles around a charge will result in greater polarity of the charge/dipole complex, b., compared to a more symmetric orientation of the dipoles around a charge, a.
Zinc/protein complexes in biological systems provide a unique framework to study the effect of charge/dipole polarization on function because in protein sites the number and the nature of the coordinating ligands may tune the electrophilicity of zinc. Certain considerations should be taken into account before entatic studies on zinc sites are undertaken. First, it is important to choose residues that have subtle effects on the charge/dipole interaction. For instance, choosing residues that are part of secondary structure may destroy the protein fold altogether. Second-shell residues make a good starting point for mutational studies because previous studies show that these residues often have subtle effects, both in terms of structure and catalytic function. Second, the zinc site under inspection must be at least partially sequestered from bulk water and in complex with at least one ionized residue. Zinc enzymes whose active site ions are solvent exposed and not in complex with ionized residues may have zinc ions whose charge is not quenched by the coordinating ligands and may not need as much entatic activation. This is not to say that strain is not a factor in these sites - the coordinating dipoles must still preorient to form the site, and optimization of this orientation may cause both electronic and steric strain.
If zinc sites can be activated without the need for geometric distortion, what evolutionary advantage do coordinately strained sites have? These sites may provide a discrimination function, a built in specificity toward a particular subset of species along the reaction coordinate. By keeping the charge of zinc close to neutral, the enzyme may tune the electrophilicity of zinc along the reaction coordinate - providing electrostatic stabilization to some species and electrostatic destabilization for others - which may be crucial under conditions where the concentrations of disfavored ligands, like products, are high. AAP has a 1000-fold lower affinity for leucine than for LPA. It is easy to imagine why, AAP is a protease and product builds up quickly. Thus, the adaptation of a discrimination function would give the enzyme a selective advantage.
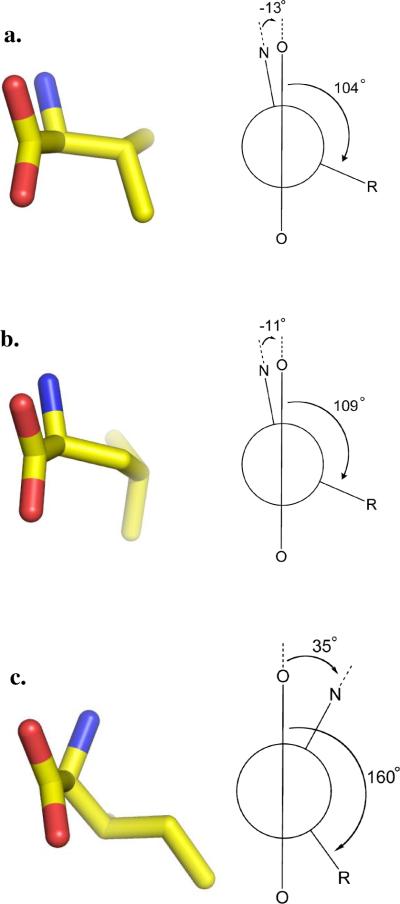
An alanine substitution at the S228 position changes the active site affinity for leucine by twenty-fold without changing the affinity for LPA. A comparison of the amino acids in the active sites of S228A and D118N, a low affinity site, reveals a curious finding. Measurements made of the torsion angle between the carbonyl carbon and the alpha carbon show that the amino acid with near eclipse conformation is in the higher affinity site (Figure 9). In this respect, the amino acid in the higher affinity S228A active site is more strained than the amino acid in the low affinity, D118N active site. This means that the S228A enzyme binds the amino acid tightly while also paying the cost of mechanically straining it. Inspection of the active site shows that in the absence of S228, the D179 oxygen moves back toward A228. This leaves Zn2 looking trigonal bipyramidal in coordination, compared to octahedral in the D118N, low affinity site. Based on these observations, an empirical method was developed to analyze the coordination number and distortion. Use of this method shows that in complex with leucine, both D118N and S228A have a tetrahedral Zn1 and a trigonal bipyramidal Zn2. However, the calculation of the r.m.s. deviation of the complexes from ideality shows that for D118N, the Zn2 is significantly more distorted in trigonal bipyramidal coordination geometry (Table 4) and resembles a six coordinate, octahedral complex.
Figure 9. Amino acid models and conformation diagrams.
a.) valine in the S228A/Amino Acid structure, b.) leucine in the S228A/Leucine structure, and c.) isoleucine in the D118N/Amino Acid structure (Figure made with PyMOL).
Compared to WT, the ground state structure of the M180A enzyme shows that each of the metal ions has significantly lower distorted tetrahedral coordination (Table 4). The M180A structure also reveals some energetically peculiar changes. For one, the S228-D179 hydrogen bond is gone and D179 has moved into a new position. D179 can actually be found in two conformations, and each of the conformers broke the S228 hydrogen bond to be where they are. The second peculiarity is that one of the D179 conformers has a flipped backbone, a feat that can be very energetically costly. Supported by the aforementioned calculations, surveys, and studies done on zinc sites; the entatic interpretation describes this force.
In a low leucine affinity site, such as WT or D118N, the hybridization of the product molecule, in conjunction with the constraining nature of the active site (the short S228-D179 hydrogen bond, bridging metals, and the hydrophobic cavity) forces Zn2 into octahedral-like coordination geometry, causing electron repulsion. The detailed inspection of the S228A structure shows that in the absence of the short S228A-D179 hydrogen bond, the D179 oxygen is free to move back in the product complex - allowing Zn2 to form trigonal bipyramidal coordination, which may alleviate some of the electron repulsion. S228, and the army of conserved residues behind it, may function as a buttress, keeping the D179 oxygen near the constrained product. Perhaps one of the advantages of short hydrogen bonds in or near active sites is their incompressibility, which may act to stabilize repulsive interactions.
The M180A mutant exhibits approximately fifty-fold lower affinity for LPA; however, inspection of the M180A variant in complex with LPA reveals that the ligand binds the mutant active site much like it binds the active site of WT enzyme. In the ground state structure, the charge/dipole of the M180A mutant is less polarized than the charge/dipole complex of WT. The M180A variant only exhibits a three-fold decrease for the leucine molecule. The structure of the M180A variant in complex with leucine shows that the leucine molecule is not in contact with the metals. In fact, the metals are in the same coordination, and have the same distortion, as the M180A structure with no ligands bound. Because the kinetics measure inhibition and not metal binding, this structure makes it easy to understand why the leucine affinity did not change as much as the LPA affinity.
Under certain circumstances, rate enhancment may take place by the entatic enhancment of the electrostatic effect. In particluar, tetrahedral zinc sites that are sequestered from bulk water and in complex with one or more ionized residues may need polarization of the charge/dipole complex for electrophilic function. What these sites gain is a discrimination function. By keeping the charge on zinc close to neutral, the enzyme can tune the electrophilicity of zinc along the reaction coordinate. Enzymes work by preorganizing features that enhance the electrostatic attraction between the enzyme and the charged transition states, but some also work by preorganizing features that enhance the electrostatic repulsion between enzyme and products.
ACKNOWLEDGMENTS
Thanks to the Biochemistry Department at Brandeis University and all the members of the Petsko/Ringe lab. Thanks to Dr. Aaron Moulin and Dr. Edwin Pozharski for helpful discussions about data processing and crystallization.
Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, and Office of Science, under Contract No W-31-109-Eng-38. Use of the BioCARS Sector 14 was supported by the National Institute of Health, National Center for Research Resources, under grant number RR007707.
Abbreviations
- AAP
aminopeptidase from Vibrio proteolyticus
- LPA
Leucine phosphonic acid
- CSD
Cambridge Structural Database
- PDB
Protein Database
- SGAP
Streptomyces griseus aminopeptidase
REFERENCES
- 1.Warshel A. Energetics of Enzyme Catalysis. Proc Natl Acad Sci USA. 1978;75:5250–5254. doi: 10.1073/pnas.75.11.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warshel A, Sussman F, Hwang JK. Evaluation of Catalytic Free Energies in Genetically Modified Proteins. J. Mol. Biol. 1988;201:139–159. doi: 10.1016/0022-2836(88)90445-7. [DOI] [PubMed] [Google Scholar]
- 3.Warshel A. Electrostatic Origin of the Catalytic Power of Preorganized Active Sites. J. Biol. Chem. 1998;273:27035–27038. doi: 10.1074/jbc.273.42.27035. [DOI] [PubMed] [Google Scholar]
- 4.Vallee BL, Williams RJP. Metalloenzymes: The Entatic Nature of Their Active Sites. Proc Natl Acad Sci USA. 1968;59:498–505. doi: 10.1073/pnas.59.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams RJP. Energized (Entatic) States of Groups and of Secondary Structures in Proteins and Metalloproteins. Eur. J. Biochem. 1995;234:363–381. doi: 10.1111/j.1432-1033.1995.363_b.x. [DOI] [PubMed] [Google Scholar]
- 6.Dudev M, Wang J, Dudev T, Lim C. Factors Governing the Metal Coordination Number in Metal Complexes From Cambridge Structural Database Analyses. J. Phys. Chem. 2006;110:1889–1895. doi: 10.1021/jp054975n. [DOI] [PubMed] [Google Scholar]
- 7.Dudev T, Lim C. Tetrahedral vs. Octahedral Zinc Complexes With Ligands of Biological Interest: A DFT/CDM Study. J. Am. Chem. Soc. 2000;122:11146–11153. [Google Scholar]
- 8.Dudev T, Lim C. Metal Binding and Selectivity in Zinc Proteins. Journal of the Chinese Chemical Society. 2003;50:1093–1102. [Google Scholar]
- 9.Ryde U. Molecular Dynamics Simulations of Alcohol Dehydrogenase With a Four- or Five- Coordinate Zinc Ion. Proteins. 1995;21:40–56. doi: 10.1002/prot.340210106. [DOI] [PubMed] [Google Scholar]
- 10.Ryde U. The Coordination of Catalytic Zinc in Alcohol Dehydrogenase by Combined Quantum-Chemical and Molecular Mechanical Calculations. J. Comput. Aided Mol. Des. 1996;10:153–164. doi: 10.1007/BF00402823. [DOI] [PubMed] [Google Scholar]
- 11.Alberts I, Nadassy K, Wodak SJ. Analysis of Zinc Binding Sites in Protein Crystal Structures. Protein Science. 1998;7:1700–1716. doi: 10.1002/pro.5560070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudev T, Yen-lin L, Dudev M, Carmey L. First-Second Shell Interactions in Metal Binding Sites in Proteins: a PDB Survey and DFT/CDM Calculations. J. Am. Chem. Soc. 2003;125:3168–3180. doi: 10.1021/ja0209722. [DOI] [PubMed] [Google Scholar]
- 13.Huang C, Lesburg C, Kiefer L, Fierke C, Christianson D. Reversal of the Hydrogen Bond to Zinc Ligand Histidine-119 Dramatically Diminishes Catalysis and Enhances Metal Equilibration Kinetics in Carbonic Anhydrase II. Biochemistry. 1996;35:3439–3446. doi: 10.1021/bi9526692. [DOI] [PubMed] [Google Scholar]
- 14.DiTusa C, McCall K, Christensen T, Mahapatro M, Fierke CA, Toone EJ. Thermodynamics of Metal Ion Binding. 2. Metal Ion Binding by Carbonic Anhydrase Variants. Biochemistry. 2001;40:5345–5351. doi: 10.1021/bi0017327. [DOI] [PubMed] [Google Scholar]
- 15.Mertz P, Yu L, Sikkink R, Rusnak F. Kinetic and Spectroscopic Analysis of a Conserved Histidine in the Metallophophatases Calcineurin and Lambda Protein Phosphatase. J. Biol. Chem. 1997;272:21296–21302. doi: 10.1074/jbc.272.34.21296. [DOI] [PubMed] [Google Scholar]
- 16.Marino SF, Regan L. Secondary Ligands Enhance Affinity at a Designed Metal-Binding Site. Chem. Biol. 1999;6:649–655. doi: 10.1016/s1074-5521(99)80116-1. [DOI] [PubMed] [Google Scholar]
- 17.Prescott JM, Wilkes SH. Aeromonas Aminopeptidase. Methods Enzymol. 1976;45:530–543. doi: 10.1016/s0076-6879(76)45047-4. [DOI] [PubMed] [Google Scholar]
- 18.Wagner FW, Wilkes SH, Prescott JM. Specificity of Aeromonas Aminopeptidase Toward Amino Acid Amides and Dipeptides. J. Biol. Chem. 1972;247:1208–1210. [PubMed] [Google Scholar]
- 19.Griffin TB, Prescott JM. Some Physical Characteristics of the Proteinase from Aeromonas proteolytica. J. Biol. Chem. 1970;245:1348–1356. [PubMed] [Google Scholar]
- 20.Chevrier B, Schalk C, D'Orchymont H, Rondeau JM, Moras D, Tarnus C. Crystal Structure of Aeromonas proteolytica Aminopeptidase: A Prototypical Member of the Co-Catalytic Zinc Enzyme Family. Structure. 1994;2:283–291. doi: 10.1016/s0969-2126(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 21.Bzymek KP, Holz RC. The Catalytic Role of Glutamate 151 in the Leucine Aminopeptidase From Aeromonas proteolytica. J. Biol. Chem. 2004;279:31018–31025. doi: 10.1074/jbc.M404035200. [DOI] [PubMed] [Google Scholar]
- 22.Bzymek KP, Swierczek SI, Bennett B, Holz RC. Spectroscopic and Thermodynamic Characterization of the E151D and E151A Altered Leucine Aminopeptidase From Aeromonas proteolytica. Inorg. Chem. 2005;44:8574–8580. doi: 10.1021/ic051034g. [DOI] [PubMed] [Google Scholar]
- 23.Bzymek KP, Moulin A, Swierczek SI, Ringe D, Petsko GA, Bennett B, Holz RC. Kinetic, Spectroscopic, and X-ray Crystallographic Characterization of the Functional E151H Aminopeptidase From Aeromonas proteolytica. Biochemistry. 2005;44:12030–12040. doi: 10.1021/bi0505823. [DOI] [PubMed] [Google Scholar]
- 24.Schurer G, Lanig H, Clark T. Aeromonas proteolytica Aminopeptidase: an Investigation of the Mode of Action Using a Quantum Mechanical/Molecular Mechanical Approach. Biochemistry. 2004;43:5414–5427. doi: 10.1021/bi0340191. [DOI] [PubMed] [Google Scholar]
- 25.Harding MM. The Geometry of Metal-Ligand Interactions Relevant to Proteins. II. Angles at the Metal Atom, Additional Weak Metal-Donor Interactions. Acta Cryst. 2000;56:857–867. doi: 10.1107/s0907444900005849. [DOI] [PubMed] [Google Scholar]
- 26.Desmarais W, Bienvenue DL, Bzymek KP, Petsko GA, Ringe D, Holz RC. The High-Resolution Structures of the Neutral and the Low pH Crystals of Aminopeptidase from Aeromonas proteolytica. J. Biol. Inorg. Chem. 2006;11:398–408. doi: 10.1007/s00775-006-0093-x. [DOI] [PubMed] [Google Scholar]
- 27.Gliboa R, Greenblatt HM, Perach M, Spungin-Bialik A, Lessel U, Wohlfahrt G, Schomburg D, Blumberg S, Shoham G. Interactions of Streptomyces griseus Aminopeptidase with Methionine Product Analogue: A Structural Study at 1.53 Å Resolution. Acta. Cryst. 2000;56:551–558. doi: 10.1107/s0907444900002420. [DOI] [PubMed] [Google Scholar]
- 28.Studier FW. Protein Production by Auto-Induction in High Density Shaking Cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Prescott JM, Wilkes SH, Wagner FW, Wilson KJ. Aeromonas Aminopeptidase. Improved Isolation and Some Physical Properties. J. Biol. Chem. 1971;25:1756–1764. [PubMed] [Google Scholar]
- 30.Bzymek KP, D'Souza VM, Chen G, Campbell H, Mitchell A, Holz RC. Function of the Signal Peptide and the N- and C-terminal Propeptides in the Leucine Aminopeptidase from Aeromonas proteolytica. Protein Expr. Purif. 2004;37:294–305. doi: 10.1016/j.pep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Prescott JM, Wilkes SH. Aeromonas Aminopeptidase: Purification and Some General Properties. Arch. Biochem. Biophys. 1966;117:328–336. doi: 10.1016/0003-9861(66)90420-6. [DOI] [PubMed] [Google Scholar]
- 32.Baker JO, Wilkes SH, Bayliss ME, Prescott JM. Hydroxamates and Aliphatic Boronic Acids: Marker Inhibitors for Aminopeptidase. Biochemistry. 1983;26:2098–2103. doi: 10.1021/bi00278a009. [DOI] [PubMed] [Google Scholar]
- 33.Stamper C, Bennett B, Edwards T, Holz RC, Ringe D, Petsko G. Inhibition of the Aminopeptidase from Aeromonas proteolytica by l-Leucinephosphonic Acid. Spectroscopic and Crystallographic Characterization of the Transition State of Peptide Hydrolysis. Biochemistry. 2001;40:7035–7046. doi: 10.1021/bi0100891. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Vagin A, Teplyakov A. MOLREP: An Automated Program for Molecular Replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 36.Emsley P, Cowtan K. COOT: Model-Building Tools for Molecular Graphics. Acta Cryst. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 Dictionary: Organization of Prior Chemical Knowledge and Guidelines For Its Use. Acta Cryst. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project, Number 4 The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Cruickshank DWJ. Macromolecular Refinement. In: Dodson E, Moore M, Ralph A, Bailey S, editors. Proceedings of the CCP4 Weekend. Daresbury Laboratory; Warrington: 1996. pp. 11–22. [Google Scholar]
- 40.Cruickshank DWJ. Remarks About Protein Structure Precision. Acta Cryst. 1999;55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, Dodson EJ. Newslett. Protein Crystallogr. 1997;33:25–30. [Google Scholar]
- 42.Brunger AT. Assessment of Phase Accuracy By Cross Validation: the Free R Value. Methods and Applications. Acta Crystallogr. 1993;49:24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- 43.Herzberg O, Moult J. Analysis of the Steric Strain in the Polypeptide Backbone of Protein Molecules. Proteins: Struct. Funct. Genet. 1991;11:223–229. doi: 10.1002/prot.340110307. [DOI] [PubMed] [Google Scholar]
- 44.Karplus PA. Experimentally Observed Conformation-Dependent Geometry and Hidden Strain in Proteins. Protein Science. 1996;5:1406–1420. doi: 10.1002/pro.5560050719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petock JM, Torshin IY, Weber IT, Harrison RW. Analysis of Protein Structures Reveals Regions of Rare Backbone Conformations at Functional Sites. Proteins: Struct. Func. Genet. 2003;53:872–879. doi: 10.1002/prot.10484. [DOI] [PubMed] [Google Scholar]
- 46.Meiering EM, Serrano L, Fersht AR. Effect of active site residues in barnase on activity and stability. J. Mol. Biol. 1992;225:585–589. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- 47.Schreiber G, Buckle AM, Fersht AR. Stability and function: two constraints in the evolution of barstar and other proteins. Structure. 1994;2:945–951. doi: 10.1016/s0969-2126(94)00096-4. [DOI] [PubMed] [Google Scholar]
- 48.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. USA. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin ACR, Orengo CA, Hutchinson EG, Jones S, Karmirantzou M, Laskowski RA, Mitchel JBO, Taroni C, Thornton JM. Protein Folds and Functions. Structure. 1998;6:875–884. doi: 10.1016/s0969-2126(98)00089-6. [DOI] [PubMed] [Google Scholar]
- 50.Hegyi H, Gerstein M. The Relationship Between Protein Structure and Function: A Comprehensive Survey with Applications to the Yeast Genome. J. Mol. Biol. 1999;288:147–164. doi: 10.1006/jmbi.1999.2661. [DOI] [PubMed] [Google Scholar]