Abstract
Recent lineage-tracing studies have produced conflicting results about whether the epicardium is a source of cardiac muscle cells during heart development. Here, we examined the developmental potential of epicardial tissue in zebrafish during both embryonic development and injury-induced heart regeneration. We found that upstream sequences of the transcription factor gene tcf21 activated robust, epicardium-specific expression throughout development and regeneration. Cre recombinase-based, genetic fate-mapping of larval or adult tcf21+ cells revealed contributions to perivascular cells, but not cardiomyocytes, during each form of cardiogenesis. Our findings indicate that natural epicardial fates are limited to non-myocardial cell types in zebrafish.
Keywords: Heart regeneration, Heart development, Cardiomyocyte, Epicardium, Zebrafish, Genetic fate-mapping
INTRODUCTION
Regenerative medicine has often been mentioned as a promising future treatment for heart disease. Yet, stimulation of new cardiac muscle regeneration for the injured mammalian heart by endogenous or exogenous cells remains a challenge. Current experimental strategies target manipulations of various cell types that could potentially supply new cardiac muscle, including the cardiomyocytes themselves, fibroblasts and various stem or progenitor cell populations (Bersell et al., 2009; Ieda et al., 2010; Mercola et al., 2011; Noseda et al., 2011; Passier et al., 2008).
The epicardium is a multipotent cardiac progenitor cell that has been investigated extensively for its natural developmental potential. During embryogenesis, the epicardium emerges from the proepicardial organ (PEO), a mesoderm-derived structure located at the atrioventricular junction, to cover the myocardium after the heart has completed looping. Lineage tracing studies in chick have shown that epicardial and epicardial-derived cells (EPDCs) within the subepicardial environment can differentiate into several cell types during heart development, including cardiac fibroblasts, smooth muscle and pericytes (Dettman et al., 1998; Gittenberger-de Groot et al., 1998; Manner, 1999; Mikawa and Gourdie, 1996). Some of these reports indicated additional capacity to form coronary vascular endothelial cells, although a recent examination identified sprouting from the sinus venosus as the major source of this cell type in mouse (Red-Horse et al., 2010).
In addition to being a source of non-myocardial cells, two independent lineage-tracing studies identified the epicardium as a significant source of cardiac muscle for the developing mouse heart. In these studies, Tbx18 and Wt1 regulatory sequences were used to direct permanent genetic labeling of epicardial tissue using Cre-loxP technology (Cai et al., 2008; Zhou et al., 2008). These particular findings, as well as additional data (Limana et al., 2007), positioned the epicardium as an attractive potential target for effecting cardiac muscle regeneration in situ after an injury like acute myocardial infarction. However, earlier genetic fate-mapping studies in mouse that did not report epicardial contributions to cardiomyocytes (Merki et al., 2005; Wilm et al., 2005), as well as a subsequent report indicating that Tbx18 is expressed by cardiomyocytes during murine development, raised additional questions regarding this lineage relationship (Christoffels et al., 2009). Thus, the developmental potential of epicardial cells remains unresolved.
Over the past 20 years, zebrafish have emerged as a key model system for embryonic heart development and function. More recently, it was shown that they also possess a striking natural capacity for adult myocardial regeneration (Poss et al., 2002). Notably, after surgical resection of the ventricular apex, epicardial cells proliferate intensely before incorporating into regenerating tissue in a fibroblast growth factor (Fgf)- and platelet-derived growth factor (Pdgf)-dependent manner (Kim et al., 2010; Lepilina et al., 2006). Although activation and proliferation of spared cardiomyocytes make a major contribution to regenerated cardiac muscle (Jopling et al., 2010; Kikuchi et al., 2010), whether epicardial cells also provide cardiomyocytes during regeneration has not been examined. Lineage tracing of epicardial cells in zebrafish thus provides the opportunity to define their contributions in the settings of embryonic heart development and adult heart regeneration.
Here, to explore the natural developmental potential of the epicardium, we screened several candidate genes for epicardial-specific expression as a prerequisite for genetic fate-mapping. We found that zebrafish tbx18 and wt1b regulatory sequences lacked epicardial specificity within the heart, showing additional activation in a subset of cardiomyocytes during development or regeneration. By contrast, a different epicardial marker, tcf21, displayed epicardial-specific expression throughout development and regeneration, and was at no point detected within cardiomyocytes. Using tcf21 regulatory sequences and inducible Cre recombinase technology, we examined the cellular contributions of the epicardium during development and regeneration. We found that larval cells lineage labeled by tcf21 expression gave rise to adult epicardial cells, subepicardial EPDCs and perivascular cells, including the smooth muscle of the outflow tract, but did not differentiate directly or indirectly into cardiomyocytes. Similarly, cells labeled for tcf21 expression either in larvae or adults contributed perivascular cells, but not cardiomyocytes, after cardiac injury and regeneration. Taken together, our results support the notion that epicardial tissue does not readily acquire a myocardial phenotype in vivo.
MATERIALS AND METHODS
Zebrafish
Zebrafish at 4-5 months of age were used for ventricular resection surgeries as described previously (Poss et al., 2002). All transgenic strains were analyzed as hemizygotes; details of their construction are described below. Published transgenic strains used in this study were: wt1b:EGFP [Tg(wt1b:EGFP)li1] (Perner et al., 2007), cmlc2:EGFP [Tg(cmlc2:EGFP)f1] (Burns et al., 2005), cmlc2:DsRed2 [Tg(cmlc2:DsRed2)pd15] (Kikuchi et al., 2010), fli1:EGFP [Tg(fli1a:EGFP)y1] (Lawson and Weinstein, 2002), gata5:EGFP [Tg(gata5:EGFP)pd25] (Kikuchi et al., 2011), cmlc2:CreER [Tg(cmlc2:CreER)pd10] (Kikuchi et al., 2010), β-act2:RSG [Tg(bactin2:loxP-DsRed-STOP-loxP-EGFP)s928] (Kikuchi et al., 2010) and gata5:RnG [Tg(gata5:loxP-mCherry-STOP-loxP-nucEGFP)pd40] (Kikuchi et al., 2010). Animal density was maintained at four fish per liter in all experiments. For 4-hydroxytamoxifen (4-HT) labeling using tcf21:CreER, 3 days post-fertilization (dpf) embryos were placed in embryo medium with 4-HT added to a final concentration of 5 μM, from a 1 mM stock solution made in 100% ethanol. After 24 hours, embryos were placed in fresh 4-HT for an additional 24 hours. For cmlc2:CreER embryos, this protocol was similar except that embryos were incubated from 2 dpf to 4 dpf. To label adult zebrafish cells, double transgenic strains carrying either tcf21:CreER or cmlc2:CreER, with either gata5:RnG or β-act2:RSG reporter transgenes, were placed in a small beaker of aquarium water containing 5 μM 4-HT. Fish were maintained for 24 hours in this media, rinsed with fresh aquarium water and returned to a recirculating aquatic system.
Construction of transgenic animals
tbx18:DsRed2
The first exon of the tbx18 gene in the BAC clone CH211-197L9 was replaced with a DsRed2 cassette using Red/ET recombineering technology (GeneBridges). The recombined BAC was linearized by I-SceI digestion, and purified DNA was injected into one-cell zebrafish embryos. The full name of this transgenic line is Tg(tbx18:DsRed2)pd22.
tcf21:DsRed2
The first exon of the tcf21 gene in the BAC clone DKEYP-79F12 was replaced with a DsRed2 cassette using Red/ET recombineering technology. The recombined BAC was linearized by I-SceI digestion, and purified DNA was injected into one-cell zebrafish embryos. The full name of this transgenic line is Tg(tcf21:DsRed2)pd37.
tcf21:CreER
The first exon of the tcf21 gene in the BAC clone DKEYP-79F12 was replaced with a CreERT2 cassette using Red/ET recombineering technology. The same technology was used to replace an endogenous loxP site in the BAC vector with a cassette containing an I-SceI site, as well as a lens-specific promoter upstream of EGFP. The α-crystallin:EGFP cassette enables visual identification of transgenic animals by lens fluorescence (Waxman et al., 2008). The BAC construct was purified and co-injected with I-SceI into one-cell zebrafish embryos. The full name of this transgenic line is Tg(tcf21:CreER)pd42.
Immunofluorescence
Immunofluorescence was performed as described previously (Poss et al., 2002). Primary antibodies used in this study were as follows: anti-Mef2 (rabbit; Santa Cruz Biotechnology), anti-Myosin heavy chain (F59, mouse; Developmental Studies Hybridoma Bank), anti-zf Raldh2 (rabbit; Abmart), anti-MLCK (K36, mouse; Sigma), anti-GFP (rabbit; Invitrogen), anti-GFP (chicken; Abcam), anti-DsRed (rabbit; Invitrogen). Secondary antibodies (Invitrogen) used in this study were as follows: Alexa Fluor 594 goat anti-rabbit IgG (H+L) for anti-DsRed, Alexa Fluor 633 goat anti-rabbit IgG (H+L) for anti-Mef2 and anti-zf Raldh2, Alexa Fluor 633 goat anti-mouse IgG (H+L) for F59 and K36, and Alexa Fluor 488 goat anti-rabbit or chicken IgG (H+L) for anti-GFP. Fluorescent images were taken using a DM6000 microscope with a Retiga-EXi camera (Q-IMAGING), and confocal images were taken using a Leica SP2 or Zeiss LSM 710 confocal microscope.
Immunofluorescence detection of EGFP (enhanced green fluorescent protein) was used for all labeling experiments with the gata5:RnG line, allowing detection of both nuclear and cytosolic EGFP. The extent to which cytosolic EGFP was detectable depended on the cell type and animal stage. Larval epicardial cells had little detectable cytosolic EGFP compared with nuclear EGFP. Adult heart sections stained for EGFP fluorescence showed similar cytosolic and nuclear EGFP in epicardial cells, but predominantly nuclear EGFP in cardiomyocytes. These differences were likely to be based on different processing methods and effects of fixation between whole larvae and whole adult hearts, and the nuclear:cytosolic volume ratio differences between cell types. Both cytosolic (MHC) and nuclear (Mef2) markers of cardiomyocytes were thus employed in experiments that assessed potential cardiomyocyte co-labeling.
RESULTS
Regulatory sequences of tcf21, but not wt1b or tbx18, are epicardial-specific during cardiac development and regeneration
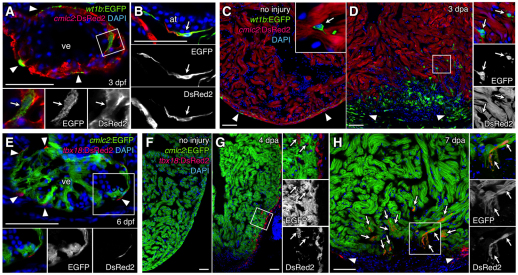
To facilitate lineage-tracing experiments, we first examined candidate gene regulatory sequences for their ability to drive expression specifically in larval and adult zebrafish epicardial cells. We obtained a wt1b:EGFP reporter strain (Perner et al., 2007) and generated a DsRed2 transgenic reporter strain with BAC sequences containing zebrafish tbx18, shown previously to be induced during heart regeneration (Lepilina et al., 2006). In 3 day post-fertilization (dpf) larvae, we detected wt1b-driven EGFP in most epicardial cells, in the process of fully covering the heart at this age, and also occasionally in ventricular and atrial cells expressing cmlc2-driven EGFP, a marker of cardiomyocytes (Fig. 1A,B). In adult hearts, we noticed that a low proportion of epicardial cells expressed wt1b:EGFP in the absence of injury, as did low numbers of small, intra-myocardial cells that are likely to be hematopoietic cells (Fig. 1C). After resection of the ventricular apex, wt1b:EGFP was induced in most epicardial cells at 3 days post-amputation (dpa), in addition to many intra-myocardial cells both distant from and within the wound site (Fig. 1D). tbx18-driven DsRed2 expression was weak in the larval heart, detectable only in epicardial cells at 3 and 6 dpf (Fig. 1E; data not shown). Few tbx18:DsRed2+ epicardial cells were detected surrounding the uninjured adult ventricle, but injury induced DsRed2 fluorescence in most epicardial cells (Fig. 1F,G). Interestingly, confocal analysis revealed that many cmlc2:EGFP+ cardiomyocytes adjacent to the injury site expressed tbx18:DsRed2 at 5-7 dpa (Fig. 1H), indicating that tbx18 regulatory sequences are activated in a subset of cardiomyocytes that participate in regeneration. Because tbx18 and wt1b regulatory sequences were not active in epicardial tissue at all life stages, and as they were activated in subpopulations of cardiomyocytes and other non-epicardial cells, they did not meet prerequisites for lineage tracing the epicardium.
Fig. 1.
Zebrafish wt1b and tbx18 regulatory sequences drive expression in epicardial and non-epicardial tissue including cardiomyocytes. (A,B) wt1b:EGFP (green) is detectable in epicardial cells (arrowheads), as well as in ventricular (A) and atrial (B) cardiac muscle (arrows), where it colocalizes with cmlc2:DsRed2 (red) at 3 days post-fertilization (dpf). (C,D) wt1b-driven EGFP marks a subset of epicardial cells (arrowheads) in the uninjured adult ventricle, as well as intra-myocardial cells (inset, arrow) that do not express cmlc2:DsRed2 (C). At 3 days post-amputation (dpa), many wt1b:EGFP+ cells lacking cmlc2:DsRed2 are evident in and around the injury site (D). A subset of these cells are epicardial (arrowheads), whereas many are intra-myocardial (arrows). Higher magnification of the boxed area is shown in each channel on the right. An antibody was used to detect EGFP in A-D. (E) tbx18:DsRed2 (red) labels epicardial cells (arrowheads) at 6 dpf, but does not colocalize with cmlc2:EGFP (green). (F,G) tbx18:DsRed2 has little or no detectable expression in the uninjured adult ventricle (F), but is detectable after injury in epicardial cells and EPDCs (G, arrows), where it does not colocalize with cmlc2:EGFP. (H) At 7 dpa, tbx18:DsRed2 colocalizes with cmlc2:EGFP in many cardiomyocytes at the injury site (arrows). Arrowheads indicate epicardial tbx18:DsRed2 fluorescence. An antibody was used to detect DsRed in E-H. at, atrium; ve, ventricle. Scale bars: 50 μm.
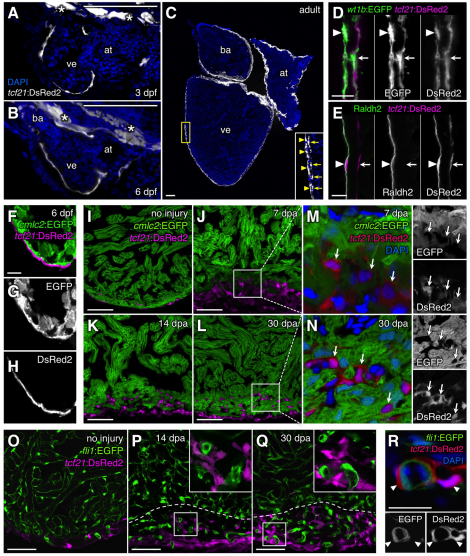
Tcf21 (also known as capsulin, Pod1 and epicardin) was identified as a basic helix-loop-helix transcription factor expressed in the PEO, epicardium and other mesoderm-derived tissues during embryonic mouse development (Hidai et al., 1998; Lu et al., 1998; Quaggin et al., 1999; Robb et al., 1998). tcf21 is also expressed in the epicardium of zebrafish larvae (Serluca, 2008). To visualize the activity of tcf21 regulatory sequences, we generated a BAC transgenic DsRed2 reporter line. DsRed2 fluorescence was detected in most epicardial cells at 3 dpf (Fig. 2A), as well as in other non-cardiac structures. By 6 dpf, through larval development and in mature adult animals, the entire heart was surrounded by tcf21:DsRed2+ epicardium (Fig. 2B,C). In adult ventricles, tcf21:DsRed2 was expressed throughout the outermost (epithelial) epicardial cell layer and subepicardial EPDCs (Fig. 2C,D). The epicardial (and endocardial) marker Raldh2 (Aldh1a2 – Zebrafish Information Network) was detected only in epithelial tcf21:DsRed2+wt1b:EGFP+ cells, consistent with findings indicating raldh2 downregulation after epicardial epithelial-mesenchymal transition in developing avian embryos (Perez-Pomares et al., 2002) (Fig. 2D,E). The tcf21:DsRed2+ epicardial domain, and often the subepicardial domain, appeared contiguous around the ventricle, suggesting representation of all ventricular epicardial cells. However, as it remains possible that there is more cell type diversity in the epicardium than is commonly appreciated, we cannot rule out the existence of occasional epicardial cells that do not activate tcf21 regulatory sequences. Thus, zebrafish tcf21 regulatory sequences drive robust, epicardial-specific expression within the larval and uninjured adult ventricle.
Fig. 2.
Zebrafish tcf21 regulatory elements drive epicardial-specific expression. (A-C) Cardiac tcf21:DsRed2 expression (white) at 3 dpf (A), 6 dpf (B) and adult (C) stages. Asterisks indicate extracardiac expression in A and B. Inset in C shows enlargement of area in yellow box. (D,E) tcf21:DsRed2+ epicardial cells (magenta) within the adult ventricular wall, assessed for wt1b:EGFP (D) or antibody staining for Raldh2 (E; green) colocalization. Arrowheads and arrows in C-E indicate the outer (epithelial) epicardial cell layer and EPDCs, respectively. (F-I) tcf21:DsRed2 (magenta) and cmlc2:EGFP (green) expression in distinct cells of 6 dpf (F-H) and adult (I) ventricles. An antibody was used to detect DsRed in F-H; this antibody also failed to detect DsRed2+EGFP+ cells in adult tcf21:DsRed2; cmlc2:EGFP ventricles (data not shown). (J-N) tcf21:DsRed2 (magenta or red) and cmlc2:EGFP (green) expression also mark distinct cells during regeneration. Arrows indicate tcf21:DsRed2+ nuclei in M and N. (O-R) tcf21:DsRed2 (magenta or red) and fli1:EGFP (green) do not colocalize in uninjured or regenerating adult ventricles. Insets in P and Q are enlarged views of boxed areas. Dashed lines indicate approximate amputation planes. at, atrium; ba, bulbus arteriosus; ve, ventricle. Scale bars: 100 μm for A-C; 10 μm for D-H,R; 50 μm for I-Q.
We next assessed uninjured and injured tcf21:DsRed2 animals for potential expression in other cardiac cell types during development and regeneration. tcf21:DsRed2 did not colocalize with cmlc2-driven EGFP at 6 dpf and 4 months of age, indicating no expression in cardiomyocytes of the developing or mature ventricle (Fig. 2F-I). After resection of the apex of the adult ventricle, tcf21:DsRed2+ cells with mesenchymal morphology were observed in the injury site and regenerating tissues from 7 to 30 dpa (Fig. 2J-L). Although tcf21:DsRed2+ cells were closely associated with cardiomyocytes, confocal analysis confirmed the lack of cmlc2:EGFP expression in these cells (Fig. 2M,N). Similarly, tcf21:DsRed2 did not colocalize with fli1-driven EGFP, a marker of endocardial and vascular endothelial cells, in uninjured and regenerating cardiac tissue. Instead, fli1:EGFP-expressing vascular endothelium was closely associated with, and often surrounded by, tcf21:DsRed2+ cells, indicative of a perivascular cell phenotype (Fig. 2O-R). Together, these results indicate that zebrafish tcf21 regulatory sequences activate expression that is specific to the epicardium and absent from myocardial and endocardial/endothelial cells.
tcf21+ cells do not acquire myocardial fate during heart development
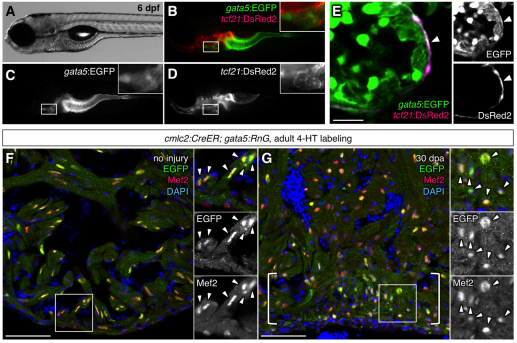
Studies that fate-mapped epicardial cells during murine development used the Wt1 or Tbx18 regulatory sequences to drive Cre recombinase expression, combined with a ubiquitous lineage-tracing reporter line (Cai et al., 2008; Zhou et al., 2008). To fate-map zebrafish epicardial cells, we generated a transgenic line with tcf21 regulatory sequences driving a tamoxifen-inducible Cre recombinase. We crossed this strain with an indicator line, gata5:RnG, to tag cells with a nuclear localization signal-tagged EGFP after excision of loxP-flanked stop sequences (Kikuchi et al., 2010). gata5 was expressed throughout the heart and endoderm-derived organs in larval and adult zebrafish (Fig. 3A-E), and gata5:RnG efficiently reported recombination-induced myocardial EGFP expression in combination with the cardiomyocyte-specific cmlc2:CreER line (Fig. 3F,G) (Kikuchi et al., 2010). Thus, through the overlapping expression domains of tcf21 and gata5, we expected precise labeling of epicardial cells (Fig. 3E). We incubated tcf21:CreER; gata5:RnG double transgenic larvae with 4-hydroxytamoxifen (4-HT) at 3-5 dpf, a time point at which PEO-derived epicardial cells have completely enveloped the heart during zebrafish development (Serluca, 2008). An antibody against EGFP was used to increase sensitivity of detecting labeled cells within tcf21:CreER; gata5:RnG tissue, and detected nuclear and occasionally cytosolic EGFP in labeled cells (see Materials and methods). Most cell nuclei with apparent epicardial morphology and position were labeled by this treatment, and no cells expressing cytosolic Myosin heavy chain (MHC) or the nuclear localized myocardial transcription factor Mef2 were labeled (Fig. 4A-D).
Fig. 3.
gata5 and tcf21 regulatory sequences drive expression that overlaps in the zebrafish epicardium. (A-E) A gata5:EGFP; tcf21:DsRed2 larva at 6 dpf. Insets are magnified images of the areas in white boxes in B-D. A single confocal slice visualizing gata5:EGFP; tcf21:DsRed2 ventricles shows colocalization limited to the epicardium (E, arrowhead). Each channel of the image is shown on the right (E). (F,G) The gata5:RnG indicator line efficiently reports induced Cre-mediated recombination in cardiomyocytes. cmlc2:CreER; gata5:RnG adults treated with 4-HT, showing EGFP labeling in Mef2+ cardiomyocytes nuclei of uninjured (F) and regenerated (G, brackets) cardiac tissue. Arrowheads in insets indicate EGFP immunofluorescence. Scale bars: 10 μm for E; 50 μm for F,G.
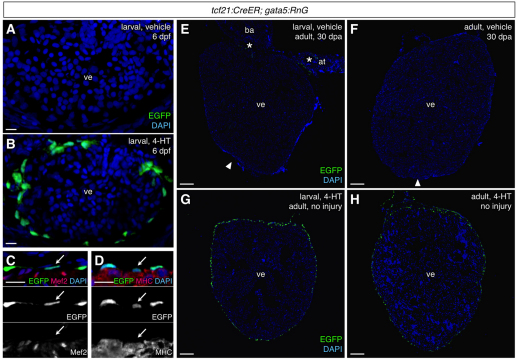
Fig. 4.
Efficient and specific induced epicardial labeling in tcf21:CreER; gata5:RnG zebrafish. (A,B) tcf21:CreER; gata5:RnG larvae incubated with vehicle (A) or 4-HT (B) at 3-5 days post-fertilization (dpf) and visualized at 6 dpf, indicating 4-HT-induced labeling of the larval epicardium (green). Confocal projections of 10 μm z-stacks are shown. (C,D) Confirmation of epicardial specificity at 6 dpf embryos, assessed by Myosin heavy chain (MHC) and Mef2 staining. Arrows indicate an epicardial cell nucleus in each image. (E,F) 30 days post-amputation (dpa) ventricles from adult tcf21:CreER; gata5:RnG animals treated with vehicle as larvae (E) or adults (F). Occasional epicardial labeling (E, asterisks) was observed in injured or uninjured hearts. Arrowheads indicate apical regenerate. (G,H) 4-HT treatment at larvae or adult stages specifically labeled the majority of adult epicardial cells. at, atrium; ba, bulbus arteriosus; ve, ventricle. An antibody was used to detect EGFP in these experiments. Scale bars: 10 μm for A-D; 100 μm for E-H.
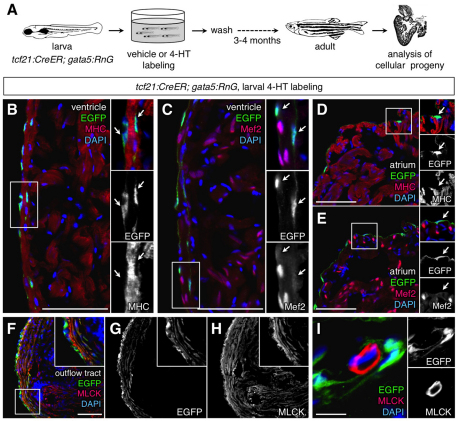
To assess the progeny of larval epicardial cells, we allowed labeled larvae to mature to adults and assessed EGFP immunofluorescence. EGFP+ cells covered the adult ventricular surface and subepicardial area, but never populated the inner trabecular myocardium (Fig. 4E,G; Fig. 5B,C). Similarly, atrial labeling was confined to the epicardial surface (Fig. 5D,E). These results suggest that larval epicardial cells give rise to a high percentage of adult epicardial cells and EPDCs. Immunofluorescence revealed that these EGFP+ cells expressed neither MHC nor Mef2 (Fig. 5B-E). We also used a second indicator line, β-act2:RSG, to detect potential myocardial contributions. β-actin2-driven expression in the heart is limited to myocardial cells, and the β-act2:RSG line was shown to label cardiomyocytes with particularly high efficiency in combination with the cmlc2:CreER line (Kikuchi et al., 2010). By contrast with hearts from cmlc2:CreER; β-act2:RSG animals treated with 4-HT as larvae, tcf21:CreER; β-act2:RSG hearts displayed no EGFP labeling (see Fig. S1A-D in the supplementary material). Thus, our data indicated no contributions by epicardium to cardiac muscle during zebrafish development.
Fig. 5.
Adult progeny of labeled larval epicardial cells. (A) Schematic of the experimental design. (B,C) Portions of the ventricle from the lineage-traced heart, stained with an antibody for EGFP (green) and muscle markers (red). Lineage-labeled EGFP+ cells (arrows in inset) did not colocalize with cytosolic Myosin heavy chain (MHC; B) or nuclear Mef2 (C; n=10). The small EGFP+ region at the bottom of the inset of B is a thin strand of epicardial cytosol partially overlaying myocardial cytosol. A structure like this cannot be resolved by confocal imaging in processed tissue sections, but has neither the size nor morphology of a cardiomyocyte. Moreover, EGFP fluorescence was never observed in Mef2+ nuclei. (D,E) Portions of the atrium from the lineage-traced heart, stained with an antibody for EGFP (green) and muscle markers (red). Lineage-labeled EGFP+ cells (arrow in inset) did not colocalize with MHC (D) or Mef2 (E) (n=10). (F-I) EGFP+ cells in the outflow tract colocalize with Myosin light chain kinase (MLCK; red), a smooth muscle marker (F-H). This colocalization was not observed in coronary vascular smooth muscle (I). An antibody was used to detect EGFP in these experiments. Scale bars: 50 μm for B-H; 10 μm for I.
We also examined the contribution of tcf21-expressing larval cells to the adult bulbus arteriosus and coronary vessels. Interestingly, EGFP+ cells were found in subepicardial areas of the bulbus arteriosus, where the labeled cells had distinct morphology from epicardium and expressed the smooth muscle marker Myosin light chain kinase (MLCK) (Fig. 5F-H). These cells ostensibly arose from larval epicardium and not larval smooth muscle, as tcf21:DsRed2 was not detected in smooth muscle of the larval bulbus arteriosus (data not shown). Also, EGFP+ cells enveloped MLCK-positive smooth muscle cells within large vessels of the ventricle (Fig. 5I). Thus, larval tcf21+ epicardial cells contribute to adult epicardial tissue and perivascular cell types, but not cardiomyocytes.
tcf21+ epicardial cells contribute perivascular cells but not cardiomyocytes during heart regeneration
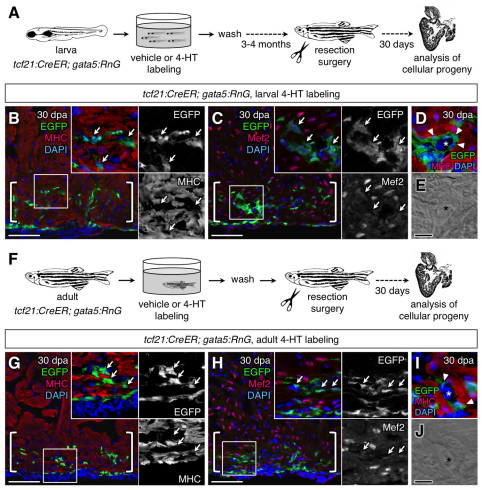
To examine epicardial contributions during cardiac regeneration, we grew 4-HT-labeled tcf21:CreER; gata5:RnG or tcf21:CreER; β-act2:RSG larvae to 4-5 months of age and resected ventricular apices (Fig. 6A). We found a substantial number of EGFP+ cells within 30 dpa tcf21:CreER; gata5:RnG regenerates; however, these cells did not express myocardial markers (Fig. 6B,C). Instead, confocal analysis identified EGFP-labeled cells with a mesenchymal appearance similar to DsRed2+ perivascular cells in regenerating tissue of the tcf21:DsRed2 reporter line (Fig. 6D,E). Furthermore, we detected no EGFP+ cardiomyocytes in tcf21:CreER; β-act2:RSG regenerates (see Fig. S1E,F in the supplementary material).
Fig. 6.
Epicardial cells contribute vascular support cells during heart regeneration. (A) Schematic of the experimental design. (B,C) Lineage-labeled EGFP+ cells (arrows, green) did not colocalize with Myosin heavy chain (MHC; B, red) or Mef2 (C, red) in 30 days post-amputation (dpa) regenerates of the larvally labeled animals (brackets; n=20). (D,E) EGFP+ cells (arrowheads) surrounds a vessel within the larvally labeled regenerate. Asterisk marks vascular lumen. (F) Schematic of the experimental design. (G,H) Lineage-labeled EGFP+ cells (arrows) did not colocalize with MHC (G) or Mef2 (H) in 30 dpa regenerates of the adult-labeled animals (brackets; n=6). (I,J) EGFP+ cells (arrowheads) surround a vessel within the adult-labeled regenerate. Asterisk marks vascular lumen. An antibody was used to detect EGFP in these experiments. Scale bars: 50 μm for B,C,G,H; 10 μm for D,E,I,J.
Although 4-HT treatment of tcf21:CreER; gata5:RnG larvae labeled the majority of adult epicardial cells, it remained possible that a population of epicardial cells with a distinct developmental potential arises in late larval or adult animals. To address this possibility, we incubated 3- to 4-month-old tcf21:CreER; gata5:RnG animals with 4-HT for one day and assessed progeny during regeneration (Fig. 6F). Although the majority of the adult epicardium appeared to be labeled in uninjured ventricles (Fig. 4F,H), and many EGFP+ EPDCs and perivascular cells were observed in 30 dpa regenerates, we detected no EGFP+ cells with myocardial marker expression (Fig. 6G-J). We performed similar experiments with tcf21:CreER; β-act2:RSG zebrafish that had been treated with 4-HT at adult stages, and did not detect EGFP+ cardiomyocytes in regenerated tissues (see Fig. S1G in the supplementary material). Our results indicate that the zebrafish epicardium is a source of perivascular support cells, but not cardiac muscle, during heart regeneration.
DISCUSSION
The extent to which epicardial cells act as a natural source of cardiac muscle during development or regeneration has been an unanswered question. Recent experiments in mice used genetic fate-mapping to indicate that epicardial cell have in vivo myogenic properties (Cai et al., 2008; Zhou et al., 2008), a result that was not reported in other epicardial lineage-tracing studies in mouse and chick (Dettman et al., 1998; Gittenberger-de Groot et al., 1998; Manner, 1999; Merki et al., 2005; Mikawa and Gourdie, 1996; Wilm et al., 2005). Here, to help clarify this issue, we investigated the developmental potential of epicardial cells in a third model system, zebrafish, shown recently to be amenable to Cre-based fate-mapping approaches. A key component of our study is the identification of a specific epicardial marker, tcf21. From our comparisons of zebrafish wt1b, tbx18 and tcf21 regulatory sequences, tcf21 emerged as the only marker with robust, epicardial expression that was never detected in cardiomyocytes themselves.
Whereas natural myocardial potential was attributed to embryonic murine Tbx18+ and Wt1+ epicardial cells, our genetic fate-mapping data exposed no similar property of the tcf21+ larval zebrafish epicardium. They instead suggest that epicardial cells, defined morphologically and not by marker expression, are unlikely to represent a source of cardiomyocytes during zebrafish heart development. These distinct results can be explained by the notion that developmental roles of epicardial cells have evolved differently in mammals versus teleosts. However, lineage-tracing experiments in avians also did not report epicardial-to-myocardial transitions. As many fundamental aspects of heart development are similar across species, the results might also be explained by activation of murine Tbx18+ and Wt1+ regulatory sequences in non-epicardial precursors of cardiomyocytes, or in cardiomyocytes themselves. Inducible, genetic fate-mapping of the murine counterpart of the tcf21+ population will be helpful to understand better the natural potential of epicardial cells during vertebrate development.
Our data indicate that epicardial cells give rise to vascular support cells in large vessels of the zebrafish ventricle, as well as smooth muscle in the bulbus arteriosus. These perivascular contributions are consistent with what has been reported in other species. Interestingly, the smooth muscle contributions of the larvally labeled epicardium to the bulbus arteriosus are limited to several layers situated under the epicardium. A large portion of smooth muscle in this structure was not labeled in tcf21 fate-mapping experiments, and thus appears to have a cellular origin distinct from the epicardium.
Regeneration of new cardiac muscle in zebrafish was reported to occur largely through the activation and proliferation of cardiomyocyte populations (Jopling et al., 2010; Kikuchi et al., 2010), a finding now supported by direct lineage-tracing of both the myocardium and the epicardium. Neovascularization occurs concomitant with new muscle creation after removal of the ventricular apex, and is important for continued regeneration. Our data reveal that epicardial cells contribute a large amount of perivascular cells during neovascularization of regenerating muscle, substantiating inferences from a previous study that did not involve lineage-tracing approaches (Lepilina et al., 2006). Although our findings indicate that epicardial cells are not a natural source of cardiomyocytes for regenerative therapies of the diseased heart, it remains possible that such a property can manifest from these cells through their experimental manipulation.
Supplementary Material
Acknowledgements
We thank C. Englert, R. Anderson, D. Stainier and ZIRC for transgenic animals; J. Burris, A. Eastes, and P. Williams for zebrafish care; X. Meng (Abmart) and Developmental Studies Hybridoma Bank for antibodies; and Poss lab members for comments on the manuscript. K.K. (AHA and JSPS), J.W. (AHA) and Y.F. (AHA) acknowledge postdoctoral fellowship support. V.G. was supported by an NHLBI Medical Scientist Training Program supplement. This work was supported by grants from NHLBI, AHA and Pew Charitable Trusts to K.D.P. K.D.P. is an Early Career Scientist of the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.067041/-/DC1
References
- Bersell K., Arab S., Haring B., Kuhn B. (2009). Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138, 257-270 [DOI] [PubMed] [Google Scholar]
- Burns C. G., Milan D. J., Grande E. J., Rottbauer W., MacRae C. A., Fishman M. C. (2005). High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol. 1, 263-264 [DOI] [PubMed] [Google Scholar]
- Cai C. L., Martin J. C., Sun Y., Cui L., Wang L., Ouyang K., Yang L., Bu L., Liang X., Zhang X., et al. (2008). A myocardial lineage derives from Tbx18 epicardial cells. Nature 454, 104-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels V. M., Grieskamp T., Norden J., Mommersteeg M. T., Rudat C., Kispert A. (2009). Tbx18 and the fate of epicardial progenitors. Nature 458, E8-E9; discussion E9-E10 [DOI] [PubMed] [Google Scholar]
- Dettman R. W., Denetclaw W., Jr, Ordahl C. P., Bristow J. (1998). Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev. Biol. 193, 169-181 [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot A. C., Vrancken Peeters M. P., Mentink M. M., Gourdie R. G., Poelmann R. E. (1998). Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ. Res. 82, 1043-1052 [DOI] [PubMed] [Google Scholar]
- Hidai H., Bardales R., Goodwin R., Quertermous T., Quertermous E. E. (1998). Cloning of capsulin, a basic helix-loop-helix factor expressed in progenitor cells of the pericardium and the coronary arteries. Mech. Dev. 73, 33-43 [DOI] [PubMed] [Google Scholar]
- Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., Srivastava D. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Marti M., Raya A., Belmonte J. C. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606-609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Werdich A. A., Anderson R. M., Fang Y., Egnaczyk G. F., Evans T., Macrae C. A., Stainier D. Y., Poss K. D. (2010). Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464, 601-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Major R. J., Blum N., Dahn R. D., Begemann G., Poss K. D. (2011). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Wu Q., Zhang Y., Wiens K. M., Huang Y., Rubin N., Shimada H., Handin R. I., Chao M. Y., Tuan T. L., et al. (2010). PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc. Natl. Acad. Sci. USA 107, 17206-17210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N. D., Weinstein B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307-318 [DOI] [PubMed] [Google Scholar]
- Lepilina A., Coon A. N., Kikuchi K., Holdway J. E., Roberts R. W., Burns C. G., Poss K. D. (2006). A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607-619 [DOI] [PubMed] [Google Scholar]
- Limana F., Zacheo A., Mocini D., Mangoni A., Borsellino G., Diamantini A., De Mori R., Battistini L., Vigna E., Santini M., et al. (2007). Identification of myocardial and vascular precursor cells in human and mouse epicardium. Circ. Res. 101, 1255-1265 [DOI] [PubMed] [Google Scholar]
- Lu J., Richardson J. A., Olson E. N. (1998). Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mech. Dev. 73, 23-32 [DOI] [PubMed] [Google Scholar]
- Manner J. (1999). Does the subepicardial mesenchyme contribute myocardioblasts to the myocardium of the chick embryo heart? A quail-chick chimera study tracing the fate of the epicardial primordium. Anat. Rec. 255, 212-226 [DOI] [PubMed] [Google Scholar]
- Mercola M., Ruiz-Lozano P., Schneider M. D. (2011). Cardiac muscle regeneration: lessons from development. Genes Dev. 25, 299-309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merki E., Zamora M., Raya A., Kawakami Y., Wang J., Zhang X., Burch J., Kubalak S. W., Kaliman P., Belmonte J. C., et al. (2005). Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc. Natl. Acad. Sci. USA 102, 18455-18460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T., Gourdie R. G. (1996). Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev. Biol. 174, 221-232 [DOI] [PubMed] [Google Scholar]
- Noseda M., Peterkin T., Simoes F. C., Patient R., Schneider M. D. (2011). Cardiopoietic factors: extracellular signals for cardiac lineage commitment. Circ. Res. 108, 129-152 [DOI] [PubMed] [Google Scholar]
- Passier R., van Laake L. W., Mummery C. L. (2008). Stem-cell-based therapy and lessons from the heart. Nature 453, 322-329 [DOI] [PubMed] [Google Scholar]
- Perez-Pomares J. M., Phelps A., Sedmerova M., Carmona R., Gonzalez-Iriarte M., Munoz-Chapuli R., Wessels A. (2002). Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs). Dev. Biol. 247, 307-326 [DOI] [PubMed] [Google Scholar]
- Perner B., Englert C., Bollig F. (2007). The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev. Biol. 309, 87-96 [DOI] [PubMed] [Google Scholar]
- Poss K. D., Wilson L. G., Keating M. T. (2002). Heart regeneration in zebrafish. Science 298, 2188-2190 [DOI] [PubMed] [Google Scholar]
- Quaggin S. E., Schwartz L., Cui S., Igarashi P., Deimling J., Post M., Rossant J. (1999). The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development 126, 5771-5783 [DOI] [PubMed] [Google Scholar]
- Red-Horse K., Ueno H., Weissman I. L., Krasnow M. A. (2010). Coronary arteries form by developmental reprogramming of venous cells. Nature 464, 549-553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb L., Mifsud L., Hartley L., Biben C., Copeland N. G., Gilbert D. J., Jenkins N. A., Harvey R. P. (1998). epicardin: A novel basic helix-loop-helix transcription factor gene expressed in epicardium, branchial arch myoblasts, and mesenchyme of developing lung, gut, kidney, and gonads. Dev. Dyn. 213, 105-113 [DOI] [PubMed] [Google Scholar]
- Serluca F. C. (2008). Development of the proepicardial organ in the zebrafish. Dev. Biol. 315, 18-27 [DOI] [PubMed] [Google Scholar]
- Waxman J. S., Keegan B. R., Roberts R. W., Poss K. D., Yelon D. (2008). Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev. Cell 15, 923-934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm B., Ipenberg A., Hastie N. D., Burch J. B., Bader D. M. (2005). The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development 132, 5317-5328 [DOI] [PubMed] [Google Scholar]
- Zhou B., Ma Q., Rajagopal S., Wu S. M., Domian I., Rivera-Feliciano J., Jiang D., von Gise A., Ikeda S., Chien K. R., et al. (2008). Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.