Abstract
The generation of metameric body plans is a key process in development. In Drosophila segmentation, periodicity is established rapidly through the complex transcriptional regulation of the pair-rule genes. The ‘primary’ pair-rule genes generate their 7-stripe expression through stripe-specific cis-regulatory elements controlled by the preceding non-periodic maternal and gap gene patterns, whereas ‘secondary’ pair-rule genes are thought to rely on 7-stripe elements that read off the already periodic primary pair-rule patterns. Using a combination of computational and experimental approaches, we have conducted a comprehensive systems-level examination of the regulatory architecture underlying pair-rule stripe formation. We find that runt (run), fushi tarazu (ftz) and odd skipped (odd) establish most of their pattern through stripe-specific elements, arguing for a reclassification of ftz and odd as primary pair-rule genes. In the case of run, we observe long-range cis-regulation across multiple intervening genes. The 7-stripe elements of run, ftz and odd are active concurrently with the stripe-specific elements, indicating that maternal/gap-mediated control and pair-rule gene cross-regulation are closely integrated. Stripe-specific elements fall into three distinct classes based on their principal repressive gap factor input; stripe positions along the gap gradients correlate with the strength of predicted input. The prevalence of cis-elements that generate two stripes and their genomic organization suggest that single-stripe elements arose by splitting and subfunctionalization of ancestral dual-stripe elements. Overall, our study provides a greatly improved understanding of how periodic patterns are established in the Drosophila embryo.
Keywords: Segmentation, Pair-rule gene, Pattern formation, Transcription control, Cis-regulatory element, Regulatory network, Long-range regulation, Drosophila
INTRODUCTION
The Drosophila segmentation genes (Nusslein-Volhard et al., 1987; Nusslein-Volhard and Wieschaus, 1980) form a hierarchical regulatory network consisting of the maternal, gap, pair-rule and segment-polarity classes, which generate increasingly refined subdivisions along the anterior-posterior axis of the embryo (Akam, 1987). The maternal genes bicoid (bcd) and caudal (cad) establish long-range gradients that give basic polarity to the embryo. The gap genes hunchback (hb), giant (gt), Kruppel (Kr), knirps (kni), tailless (tll) and huckebein each form one or two shorter range gradients at different positions. The next step in the cascade is the crucial transition from these non-periodic to the periodic patterns of the pair-rule genes: with the exception of odd paired, all pair-rule genes, i.e. hairy (h), even skipped (eve), runt (run), fushi tarazu (ftz), odd skipped (odd), paired (prd) and sloppy paired (slp), are expressed in seven evenly spaced stripes, with patterns phase-shifted to produce well-defined overlaps (Nasiadka et al., 2002). The resulting positional code is read off in an inherently periodic manner by the segment-polarity genes, which generate sets of fourteen stripes that prefigure the fourteen segments of the larva.
The regulation within the segmentation network is almost entirely transcriptional, with the maternal factors functioning as activators, whereas the gap and pair-rule factors act largely as repressors. Expression patterns are generated through combinatorial binding of participating factors to modular cis-regulatory elements, which can be located upstream or downstream of the gene transcription start site; they typically range from 0.5 to 2 kb in length and contain multiple binding sites for both activators and repressors. In the case of the pair-rule genes, the regulatory task is particularly complex, as they need to translate the simpler patterns of the maternal and gap genes into a periodic series of seven stripes. Earlier work had argued that this takes place in a two-step process (Fig. 1A) (Ingham, 1988). First, a subset of genes, termed the ‘primary’ pair-rule genes, establish their patterns in a piecemeal fashion through stripe-specific cis-elements that drive expression in one or two individual stripes under the control of the maternal and gap factors. The ‘secondary’ pair-rule genes then establish their 7-stripe patterns wholesale through cis-elements that generate all seven stripes simultaneously in response to the already periodic positional cues provided by the primary pair-rule genes. Support for this subdivision came from genetic experiments showing that the 7-stripe patterns of primary pair-rule genes form correctly in mutants of the secondary pair-rule genes, but not vice versa (Ingham, 1988; Pankratz and Jackle, 1990). Based on such data and on experimental dissections of cis-regulatory regions, h, eve and run have been classified as primary pair-rule genes, with the cis-element that generates eve stripe 2 providing the classic example of a stripe-specific element (Arnosti et al., 1996a; Goto et al., 1989; Howard and Struhl, 1990; Klingler et al., 1996; Pankratz et al., 1990; Riddihough and Ish-Horowicz, 1991; Small et al., 1992; Stanojevic et al., 1991). The majority of pair-rule genes, namely ftz, odd, prd and slp, have been regarded as secondary pair-rule genes (Ingham, 1988; Nasiadka et al., 2002).
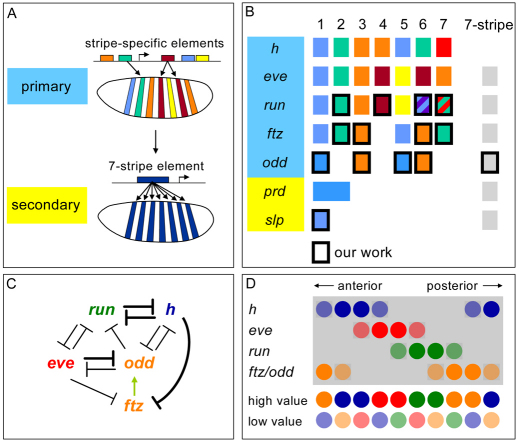
Fig. 1.
Regulation in the pair-rule gene network. (A) The mode of cis-regulatory control of primary versus secondary pair-rule genes according to the traditional model. (B) Overview showing the cis-elements and the patterns they produce for all pair-rule genes. Elements discovered in our work are framed in black. (C) Cross-regulatory relationships among the (expanded) group of primary pair-rule genes, based on ectopic expression studies conducted by Krause and colleagues (Manoukian and Krause, 1992; Manoukian and Krause, 1993; Nasiadka and Krause, 1999; Saulier-Le Drean et al., 1998), as well as loss-of-function analyses (Carroll and Vavra, 1989; Ingham and Gergen, 1988). The green arrow indicates activation and line thickness represents strength of regulation. (D) The positional code defined by phase-shifted pair-rule stripes within a two-segment unit, based on expression data from Myasnikova et al. (Myasnikova et al., 2001). Circles represent single nuclei.
Since the original studies that gave rise to this model, various inconsistencies have emerged: an extensive dissection of the run locus failed to identify a complete set of stripe-specific cis-elements; conversely, stripe-specific elements have been discovered in the regulatory regions of both ftz and odd (Berman et al., 2004; Calhoun and Levine, 2003; Schroeder et al., 2004), and there is evidence for early regulation of ‘primary’ by ‘secondary’ pair-rule genes (Andrioli et al., 2004; Saulier-Le Drean et al., 1998). These findings have led to calls for a reassessment (Nasiadka et al., 2002; Yu and Pick, 1995); however, a systematic reappraisal of the regulatory architecture underlying pair-rule stripe formation has not been carried out. In the present study, we investigate the temporal evolution of pair-rule gene patterns during the blastoderm, the organization of their cis-regulatory input, as well as the molecular epistasis among the pair-rule genes. We identify multiple novel stripe-specific cis-elements for ftz and run, as well as the missing 7-stripe element of odd. Our results indicate that ftz and odd should be placed among the primary pair-rule genes and point to a much closer integration of maternal/gap-mediated and pair-rule-mediated regulation than previously thought.
MATERIALS AND METHODS
cis-element prediction and binding site analysis
Stubb (Sinha et al., 2003) runs were performed as described previously for its predecessor Ahab (Schroeder et al., 2004). The strength of transcription factor input into a cis-element, represented by the integrated profile value, was assessed by single-factor Stubb runs without optimization (see Table S3 in the supplementary material). Position weight matrices (PWMs) were taken from Schroeder et al. (Schroeder et al., 2004), except in the cases of GT, KNI and TLL, for which we generated improved matrices by incorporating data from a recent bacterial one-hybrid study (Noyes et al., 2008b) (see Table S2 and Fig. S2 in the supplementary material), and of FTZ, EVE and ODD, for which we used the PWMs from Meng et al. (Meng et al., 2005) and Noyes et al. (Noyes et al., 2008a). cis-element expression patterns were taken from our previous work or measured at phase 3 as described (Schroeder et al., 2004). The likelihood that the cis-elements that generate stripes 1 and 5 co-occur by chance was computed by permuting the stripe labels for the different elements within each locus, determining the fraction of occasions that the two stripes are assigned to adjacent single-stripe elements or to the same dual-stripe element, and adjusting by the number of possible 2-stripe pairings.
Analysis of expression patterns and genetics
The generation of transgenic lines and RNA in situ hybridization were performed as described (Schroeder et al., 2004). For the h and odd basal elements, in-frame fusions were made between the N-terminal portion of the protein and lacZ. The sequences of all constructs were verified (see Table S1 in the supplementary material). The following strains were obtained from published sources: eve late element (M. Fujioka, Thomas Jefferson University); ftz LacC and zebra elements (L. Pick, University of Maryland, College Park); eve3, h25, run3, ftz13 (Bloomington Stock Center); prd4, Df(2L)ed1, which removes slp1 and slp2 (M. Fujioka); and drmp2 [J. Merriam (Green et al., 2002)], which removes odd as well as two adjacent paralogs (Hart et al., 1996) and leads to complete loss of all odd denticle belts. The ftz13 allele contains an amber mutation (Gln to stop) after 53 amino acids, as determined by sequencing genomic PCR products spanning the transcript from several individual flies with and without the mutation. The identity and phenotype of mutations were confirmed by cuticle preparations and, for transcript nulls, by RNA in situ hybridization. For genotyping, CyO or TM3 balancers carrying an hb-lacZ marker (S. Small, New York University) were used; run mutants were identified by their pattern defects, after establishing genotype and phenotype by double fluorescence in situ hybridization.
RESULTS
Temporal evolution of the 7-stripe pattern of the pair-rule genes
To gain a global overview of the expression dynamics of the pair-rule genes, we examined their RNA patterns in carefully staged blastoderm embryos (Fig. 2). We divided embryonic stage 5 into four phases based on the degree of cellularization, using nuclear morphology and the invagination of the plasma membrane as criteria (Fig. 2A) (Lecuit and Wieschaus, 2000). In h, eve, run, ftz and odd, despite subtle differences in the emergence of individual stripes, the overall spatiotemporal dynamic of stripe formation is very similar (Fig. 2B): striped patterns become visible in phase 1, all stripes except odd stripe 7 are expressed during phase 2, and their spacing and expression levels become largely uniform by phase 3. By contrast, prd and slp1 are initially expressed in a non-periodic gap-like pattern in the anterior, which begins to resolve into stripes in phase 2; however, the full 7-stripe pattern arises only during phase 3 (Fig. 2B). In addition, the stripes of h, run, eve, ftz and odd initially appear less clearly separated and more graded, whereas the 7-stripe patterns of prd and slp1 emerge fully refined, with sharper, more evenly spaced stripes. These observations suggest a natural grouping of h, run, eve, ftz and odd together, with prd and slp1 forming a separate class. The expression dynamics we observe at the transcript level correspond to the dynamics of protein expression reported by Reinitz and co-workers (Myasnikova et al., 2001; Surkova et al., 2008) (see Fig. S1 in the supplementary material).
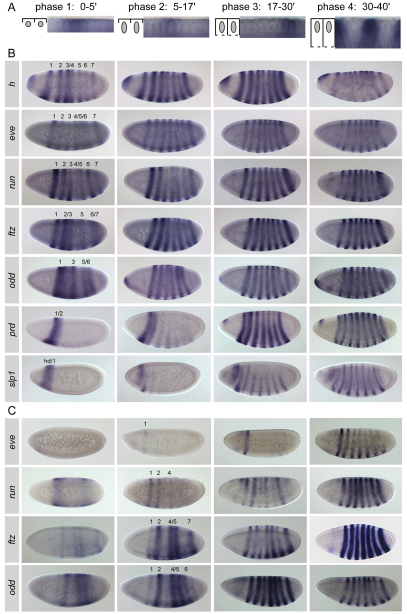
Fig. 2.
Expression dynamics of the pair-rule genes. (A) Schematics and high-magnification in situ hybridization images visualizing the four morphologically distinct phases of cellularization used to stage Drosophila embryos. Time is indicated in minutes. Initially small and spherical (phase 1), the nuclei elongate during phase 2; the plasma membrane then begins to ensheath the nuclei (phase 3), extending past them and closing to form cells during phase 4 (Lecuit and Wieschaus, 2000). Embryos are shown from the end of phases 1-3 and from the middle of phase 4. (B) RNA in situ hybridization of wild-type embryos using pair-rule gene antisense probes, for each of the four phases depicted in A. In these and all subsequent figures, lateral views of whole-mount embryos are shown, with anterior to the left and dorsal up. The stripes are labeled for phase 1, as the patterns are first forming. See also Fig. S1 in the supplementary material. (C) Expression of lacZ reporter genes for the primary pair-rule gene 7-stripe elements; shown are the eve late element (Fujioka et al., 1995), the run_(−3) element (see Fig. 3), the ftz LacC element (Hiromi et al., 1985) and odd_basal–1&–10 element (see Fig. 4B). Individual stripes expressed during phase 2 are labeled. The full 5 kb run 7-stripe element similarly shows early expression with complex dynamics for individual stripes (Klingler et al., 1996).
The early appearance of nearly all stripes of run, ftz and odd, as well as the temporal and spatial ‘irregularity’ of these early patterns, suggest that they are under the control of maternal and gap genes. Notably, stripe-specific cis-regulatory elements producing some of the early stripes in ftz and odd have been identified, as have maternal/gap-controlled cis-elements driving the early head expression of prd and slp1 (Calhoun and Levine, 2003; Ochoa-Espinosa et al., 2005; Schroeder et al., 2004); this suggests that early expression can serve as a heuristic to forecast the existence of maternal/gap-driven cis-elements. To systematically search for missing cis-elements, we examined the genomic regions surrounding the pair-rule genes using the computer algorithm Stubb (Sinha et al., 2003), which efficiently predicts cis-elements based on the local statistical over-representation of known factor binding motifs characterized by PWMs (Rajewsky et al., 2002; Schroeder et al., 2004; Sinha et al., 2004).
cis-regulation of the run locus
The expression timecourse of run shows that all stripes form early in the blastoderm (phase 1, Fig. 2B). However, stripe-specific cis-elements have only been identified for the odd-numbered stripes 1, 3 and 5, with the extended region encompassing these three elements also driving a weak stripe 7 (Klingler et al., 1996). The Stubb free energy profile, which provides a measure of the local density and strength of binding sites, showed several peaks with maternal/gap input in the run genomic region, which were tested for blastoderm expression using lacZ reporter gene constructs (Fig. 3). A particularly strong peak 17 kb upstream of the run transcription start site delineates a cis-element, run_(−17), that nicely drove reporter expression in stripe 4. A predicted element at −10 kb drove stripe 3 and represents a narrower delineation of the previously identified stripe 3 element. An element located 30 kb downstream, well outside the original dissection, drove expression in stripe 2 as well as a strong stripe 7, and an element at +19 kb drove expression in stripe 1 and in a broader domain encompassing stripes 6 and 7, although with diminished expression on the ventral side (Fig. 3). We tested additional regions with weaker free energy peaks immediately surrounding the run gene, but none drove striped expression.
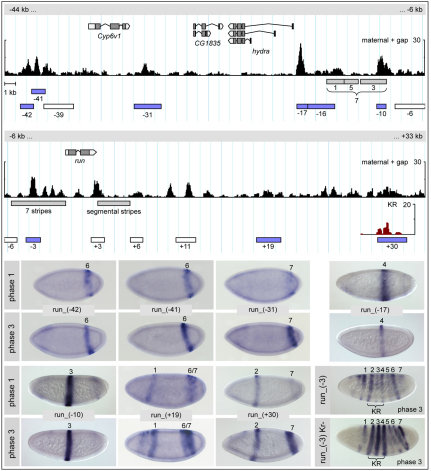
Fig. 3.
Dissection of the cis-regulatory region of run. (Top) The genomic region surrounding the Drosophila run locus, with annotated transcripts and Stubb free energy profiles as indicated. Boxes below the free energy profiles for maternal/gap input (black) depict the positions of previously known cis-elements (gray) (Klingler et al., 1996); cis-elements identified in this study that produce striped expression are shown in blue and those that do not in white. (Bottom) The RNA expression patterns of selected cis-element reporter constructs for phases 1 and 3. The run_(−16) element partially overlaps the known element for, and drives expression in, stripe 1. For the run_(+30) element, the inset in the top part of the figure shows the free energy profile for KR (brown); note the separate peaks discernible within the element. Curly brackets indicate the position of the KR expression domain.
Strikingly, however, we also found three regions with strong maternal/gap input near the Cyp6v1 gene, at 41, 42 and 31 kb upstream of run: the run_(−41) and run_(−42) elements partially overlap and drove expression in an identical pattern in the position of run stripe 6, while the run_(−31) element drove stripe 7 (Fig. 3). run is part of a gene cluster that is conserved in all Drosophilids and beyond; it encompasses two run paralogs located ~80 kb and ~160 kb upstream of run, as well as a variable number of unrelated intervening genes, with Cyp6v1 being the only constant among Drosophilids (Bao and Friedrich, 2008; Duncan et al., 2008). The remarkably deep conservation of microsynteny suggests the operation of functional constraints to keep the cluster intact. One possible mechanism is long-range cis-regulation, which to date has been observed mostly in vertebrates, but also in flies (Calhoun and Levine, 2003; Engstrom et al., 2006; Hong et al., 2008; Visel et al., 2009). In this case, the distal cis-elements might have arisen as part of the duplication of the ancestral run gene, and the available evidence suggests that they indeed regulate run: neither the run paralogs nor the intervening genes are expressed in the blastoderm, except for Cyp6v1, which shows weak non-striped expression (Duncan et al., 2008; Graveley et al., 2010; Tomancak et al., 2007).
Aside from the free energy peaks described above, we also observed a narrow region with very strong maternal/gap input within the previously characterized, large 7-stripe element (Klingler et al., 1996). This was unexpected, given that 7-stripe elements are thought to be regulated in a periodic fashion by pair-rule genes. The cis-element defined by the peak, run_(−3), showed broad expression at phase 1, then developed a modulated striped pattern, with stronger expression of stripes 1 and 2 during phase 2 and of stripes 1, 2, 4 and 6 during phase 3, before resolving into a uniform 7-stripe pattern at phase 4 (Fig. 2C, Fig. 3). Aside from input by pair-rule factors (HAIRY, ODD), the element is predicted to receive input from maternal activators (BCD, CAD) and zygotic repressors (KR, GT), consistent with the early modulation of the pattern (see Table S3 in the supplementary material). In particular, the predicted KR input suggests that repression by this gap factor is responsible for the observed weakness of stripe 3. To test this, we mutated a central G, present in all footprinted KR sites, to T in the four strongest predicted sites within the run_(−3) element. The resulting construct showed greatly strengthened early expression of stripes 2, 3 and 4, which all fall within the KR expression domain, indicating that the gap input is indeed functional (Fig. 3, lower right panel).
In summary, we find that run has stripe-specific elements for all seven stripes, with partial redundancy for stripes 1, 6 and 7, as well as an early-acting 7-stripe element that integrates input from maternal, gap and pair-rule genes.
cis-regulation of the ftz locus
The expression timecourse of ftz shows most stripes present in phase 1, although stripe 4 arises and stripes 6 and 7 separate only later in phase 2 (Fig. 2B) (see Yu and Pick, 1995). The Stubb free energy profile of the locus revealed several regions with strong maternal/gap input, which we tested (Fig. 4A). The ftz_(+3) element drove expression of stripes 1 and 5, similar to a previously tested overlapping region (Calhoun and Levine, 2003). ftz_(−6) drove expression of stripes 2 and 7, an unexpected result given previous work: the smaller, overlapping ftz ps4 element drives expression only in stripe 2 and only when inserted in reverse orientation; therefore, it was thought to regulate the neighboring Antp gene, which is strongly expressed in parasegment 4 (Hiromi and Gehring, 1987; Pick et al., 1990). The ftz_(−7) element, which falls outside the originally dissected region, drove expression of stripes 3 and 6/7. These latter two elements jointly recapitulate the endogenous expression dynamics of ftz stripes 6 and 7: in phase 1, ftz_(−7) is expressed in a wider region covering stripes 6 and 7, which by phase 3 resolves into a strong stripe 6 and a diminishing stripe 7; concomitantly, ftz_(−6), which initially shows no expression in the posterior, gives rise to stripe 7. We have thus identified three stripe-specific elements that together account for all six early-arising ftz stripes.
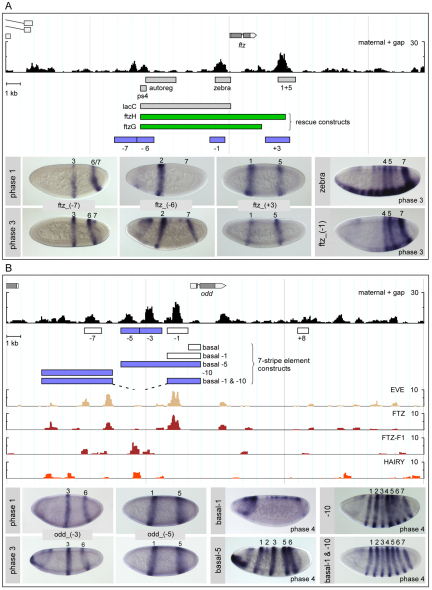
Fig. 4.
Dissection of the cis-regulatory regions of ftz and odd. (A) Dissection of Drosophila ftz. Constructs, including rescue constructs (green), are from Calhoun and Levine (Calhoun and Levine, 2003) and Hiromi et al. (Hiromi et al., 1985). (B) Dissection of odd, with constructs tested in search of the 7-stripe element as indicated. Note that the staining for the transgenic line containing the −10 element alone takes unusually long to develop, indicating that the element drives weak expression. Single-factor free energy profiles are shown for selected pair-rule genes. See Fig. 3 for further description of layout.
The last ftz stripe to become visible at phase 2 is stripe 4. How is this stripe generated? The Stubb free energy profile showed only one additional peak with strong maternal/gap input within the ftz genomic region; it overlaps the so-called zebra element (Hiromi et al., 1985), which forms part of the extended region necessary to drive a full ftz 7-stripe pattern (Yu and Pick, 1995) (LacC, Fig. 2C, Fig. 4A). A cis-element delineated to encompass this maternal/gap input while excluding much of the pair-rule input, ftz_(−1), drove modulated expression in the region of stripes 4-7, but with no clear separation of stripes (Fig. 4A). Given this somewhat artifactual expression, which resembles the zebra element and does not cleanly recapitulate the endogenous blastoderm pattern of ftz, we conclude that it is not possible to isolate a specific element for ftz stripe 4 and that, instead, this stripe is driven by an early-acting 7-stripe element that receives both maternal/gap and pair-rule input (see below).
cis-regulation of the odd locus
The expression timecourse of odd shows the early formation of stripes 1, 3, 5 and 6 (phase 1), whereas stripes 2 and 4 arise in phase 2 and stripe 7 becomes fully visible only in phase 3 (Fig. 2B). We previously identified two cis-elements with strong maternal/gap input (Schroeder et al., 2004): odd_(−3), which produces stripes 3 and 6, and odd_(−5), which produces stripes 1 and 5 (Fig. 4B). These two cis-elements thus account for all early odd stripes, and both elements are indeed active during phase 1. We tested other candidate elements within the odd genomic region that show reasonable maternal/gap input, but found no additional stripe-specific elements (Fig. 4B). This result supports the notion that maternal/gap-driven stripe-specific elements are predicted by phase 1 expression and suggests that the remaining odd stripes are established by a 7-stripe element.
Taken together, our computational analysis and redissection of pair-rule gene regulation reveal that stripe-specific cis-elements with strong maternal/gap input exist not only for the ‘classic’ primary pair-rule genes h and eve, but also for all run stripes, six ftz stripes and four odd stripes (see overview in Fig. 1B). All these stripes arise early in the blastoderm, during phase 1. The strong correlation between early expression and maternal/gap-driven stripe-specific cis-regulation suggests that the full repertoire of stripe-specific elements for the pair-rule genes has now been identified.
The role of 7-stripe elements in establishing the periodic pattern
7-stripe elements that drive the expression of all seven stripes simultaneously have been identified in the regulatory regions of all pair-rule genes except for odd and h. In the case of odd, our analysis strongly suggests that such an element has to exist. We used Stubb to predict binding sites for the pair-rule factors within the odd regulatory region, although the comparatively poor sampling of binding preferences for the pair-rule factors and reduced clustering compared with maternal/gap input make such predictions less reliable (see Discussion).
We tested several candidate elements and found that full 7-stripe expression requires a combined construct that encompasses both the sequence surrounding the odd basal promoter and an extended region upstream of the stripe-specific elements: the distal (−10) region alone gave rise to a weak 7-stripe pattern that never became entirely sharp or uniform (Fig. 4B); the odd basal (−1) region, which also includes the first intron, by itself mainly drove aberrant head expression; however, when joined together, the two elements drove a strong, properly delimited 7-stripe pattern. Notably, the ectopic head expression seen in the basal (−1) element was greatly reduced in the combined construct, suggesting non-additive interactions between the sub-elements. The (−1) region has very strong predicted input by EVE, which is thought to negatively regulate odd, and by FTZ, which acts as an activator, as well as substantial maternal and gap input. The (−10) region contains a co-clustering of weaker binding sites for FTZ and its co-factor FTZ-F1, an arrangement that has been observed in other FTZ targets (Florence et al., 1997; Hou et al., 2009; Yu et al., 1997).
The h genomic region has been dissected extensively, but no dedicated 7-stripe element has been found (Howard and Struhl, 1990; Pankratz et al., 1990; Riddihough and Ish-Horowicz, 1991). We tested regions with predicted pair-rule input, alone and in combination, but none of these constructs produced a 7-stripe pattern (see Fig. S3 in the supplementary material). The absence of a 7-stripe element is consistent with the fact that the h stripes fade soon after cellularization (Fig. 2B; not shown) (Hooper et al., 1989). By contrast, all other pair-rule genes retain their 7-stripe pattern through germ band extension, suggesting that an important role of 7-stripe elements is to maintain the pattern after cellularization (Fujioka et al., 1995; Fujioka et al., 1996; Goto et al., 1989; Klingler et al., 1996; Pick et al., 1990).
Overall, the regulatory regions of eve, run, ftz and odd all contain multiple stripe-specific elements as well as 7-stripe elements. To assess the relative role of these two types of cis-elements in establishing periodic patterns, we examined the temporal dynamics of their expression and compared it with the expression timecourse of the endogenous genes. All the stripe-specific elements we tested are active in phase 1 and continue to drive expression through phase 3, i.e. at a time when the 7-stripe patterns of the endogenous genes are fully resolved (Figs 3 and 4). For the 7-stripe elements, we found interesting differences between the genes (Fig. 2C). The eve late element initiates at phase 3 with strong expression of stripe 1, whereas the remaining stripes emerge synchronously during phase 4. This timecourse mirrors the temporal dynamics of prd, the known activator of this element (Fujioka et al., 1996), with some delay, and suggests that the eve 7-stripe element is not required for the initial formation and refinement of stripes, but for their maintenance at later stages. By contrast, the 7-stripe elements of run, ftz and odd are active much earlier and show a more complex dynamic of stripe formation: all elements show expression at phase 1; during phase 2, they are expressed in a modulated striped pattern, which resolves into a uniform 7-stripe pattern by phase 3 (ftz, odd) or 4 (run). The modulated early expression suggests that in addition to (periodic) pair-rule factor input, these elements receive non-periodic input from the maternal and gap factors; this is supported by our computational analysis (Figs 3 and 4; see Table S3 in the supplementary material).
We thus find that both stripe-specific and 7-stripe elements are active during the establishment of the refined periodic pattern during phases 2 and 3. In run, this concurrent activity might be redundant. In the case of ftz and odd, however, it is clear that both types of cis-element are required: the stripes not produced by stripe-specific elements (ftz stripe 4, odd stripes 2, 4 and 7) appear in the 7-stripe element pattern by the time that they emerge in the endogenous transcript pattern, suggesting that they are indeed generated by these elements. Conversely, some stripes, such as stripe 3, are only weakly expressed from the 7-stripe elements at a time when they are already well established in the endogenous pattern (Fig. 2B,C), suggesting that they are primarily generated by the corresponding stripe-specific elements, which indeed drive strong and sharply defined stripes at this position (Fig. 4A,B). Overall, we observe a gradual increase in the role played by 7-stripe elements – from stripe formation entirely through stripe-specific elements without a 7-stripe element (h) to the de novo generation of a subset of stripes through an early-acting 7-stripe element (ftz, odd).
Pair-rule gene cross-regulation
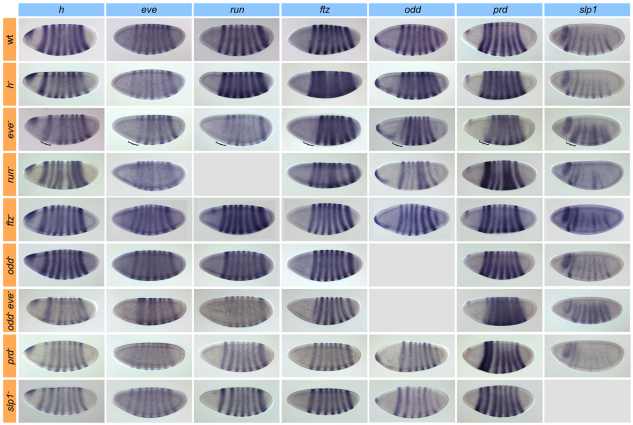
Given the extensive reliance on maternal and gap factor input in establishing the pair-rule gene expression patterns, what is the role of cross-regulation among the pair-rule genes? To address this question, we examined the effects of pair-rule gene mutants on pair-rule expression. Such experiments provided the initial motivation for the distinction between primary and secondary pair-rule genes (Ingham, 1988; Pankratz and Jackle, 1990), but most of the pertinent studies were carried out in the 1980s using now-outdated RNA detection methods, and they were often incomplete and heterogeneous in staging, making cross-comparison difficult (Carroll and Vavra, 1989; Frasch and Levine, 1987; Howard and Ingham, 1986; Ingham and Gergen, 1988). We therefore re-examined the issue in a comprehensive fashion by analyzing the entire matrix of transcript pattern versus mutant background for the seven patterned pair-rule genes under uniform conditions (Fig. 5). RNA transcript expression was evaluated at phase 3, i.e. at a time when 7-stripe patterns are fully resolved in the wild type. Null conditions were assayed for all genes; for h, eve, ftz and prd we used protein null alleles, permitting the visualization of transcript patterns; for odd and slp, we used small local deletions (see Materials and methods).
Fig. 5.
Pair-rule gene mutant analysis. Transcript patterns of all pair-rule genes in wild type (wt) and pair-rule gene mutants in Drosophila phase 3 blastoderm embryos. Genotypes are arrayed by row, transcript patterns by column. For eve mutant embryos, the position of the fused stripe 1/2 domain of slp1 is indicated by a curly bracket.
Overall, we observed irregularities in the strength (intensity and width) and spacing of pair-rule gene stripes in the mutants, but found that nearly all stripes are present (Fig. 5). The major exception was the (partial) loss of stripe 1 for most pair-rule genes in eve mutants, which is likely to be a secondary effect of the loss of proper separation of slp1 stripes (see below). In some other cases, we observed varying levels of stripe fusion or weakness, but an underlying 7-stripe pattern was always apparent. Thus, the regulatory input from individual pair-rule genes is not required for the generation of stripes per se, but rather for the refinement of the 7-stripe patterns, i.e. to achieve the proper intensity and spacing of stripes.
Looking in detail, we found that in eve and run mutants the regularity of both stripe spacing and intensity was impaired in all genes. In h mutants, we observed severe defects primarily in stripe intensity and width, whereas their spacing appeared regular. In odd mutants, we found a mild widening of the stripes of eve and run, showing that odd regulates these two genes at this stage (Saulier-Le Drean et al., 1998). Moreover, some of the defects observed in eve mutants were ameliorated in odd, eve double mutants, indicating that the effect of eve is partially mediated through odd (Fig. 5). By contrast, ftz mutants showed no serious defects, except for a weakening and loss of regularity of odd stripes. This is consistent with the role of ftz as an early local activator of odd, which is expressed in a highly similar overlapping pattern (Nasiadka et al., 2000; Nasiadka and Krause, 1999). Notably, we observed no obvious defects in the early transcript pattern of ftz itself in the ftz mutant (a protein null) indicating that, unlike previously thought, the establishment of the blastoderm pattern (and the activity of the responsible cis-elements) involves little contribution from autoactivation. However, ftz expression is indeed lost prematurely in ftz mutants during gastrulation (not shown), consistent with the notion of ftz autoactivation (Pick et al., 1990; Schier and Gehring, 1992; Schier and Gehring, 1993).
Interestingly, we also observed clear defects in pair-rule gene expression patterns in slp and prd mutants (Fig. 5, bottom rows). In slp, and to a lesser extent in prd mutants, stripe 1 of the other pair-rule genes was shifted anteriorly, leading to an abnormally wide spacing between stripes 1 and 2 (Andrioli et al., 2004). In prd mutants, moreover, expression of the first two stripes of h was substantially weakened (Fig. 5). The defects thus correspond to the early anterior expression domains of slp1 and prd, strongly arguing that the early maternal/gap-driven expression is functional in all pair-rule genes. Our data suggest that regulation of the other pair-rule genes by slp1 is also responsible for the main defect observed in eve mutants, namely the (partial) loss of stripe 1 in eve, run, ftz and odd, and of stripe 2 in h. In eve mutants, slp1 stripes 1 and 2 expand and fuse, filling the region normally occupied by, and thereby repressing, eve stripe 1 as well as the other pair-rule stripes (Fig. 5). Several lines of evidence support this interpretation: the missing stripes are initially present and only lost later (not shown), suggesting a secondary effect; the loss occurs from the ventral side, tracking the emergence of the fused slp1 stripes; strong SLP binding sites are predicted in or near the cis-elements that drive the repressed stripes (see Table S3 in the supplementary material); finally, misexpression of slp1 has been shown to lead to similar defects (Andrioli et al., 2004).
Altogether, our mutant analysis shows that among the five pair-rule genes active early, h, eve and run have the greatest impact on the establishment of expression patterns; however, odd has a role in regulating the other genes as well. More strikingly, even slp1 and prd are required for the proper establishment of primary pair-rule gene patterns, although spatially limited to the region where they show early expression. Functionally, the role of pair-rule gene cross-regulation lies in achieving the refinement and proper registration of the 7-stripe patterns. Given the temporal dynamics of the 7-stripe elements described above, our data imply that these cross-regulatory interactions are mediated either entirely (in the case of h and eve, which lack an early-acting 7-stripe element) or partially (in run, ftz and odd) through stripe-specific elements (see Discussion).
Three major classes of stripe-specific elements
As a result of previous studies and our work described here, we now have a set of 22 cis-elements that drive expression of either one or two pair-rule gene stripes in response to regulatory input primarily by maternal and gap factors. Given this extensive and, we believe, complete catalog of elements, we sought to identify common features and understand how their binding site composition relates to their expression. To maximize the reliability of our computational binding site predictions, we improved the PWMs for KNI, TLL and GT, which had previously been problematic (Schroeder et al., 2004), and ran Stubb in a mode that aids the cross-comparison of binding site content between different elements (see Materials and methods). We focused on the repressive gap factors, which provide much of the precise positional information (Ochoa-Espinosa et al., 2005; Segal et al., 2008).
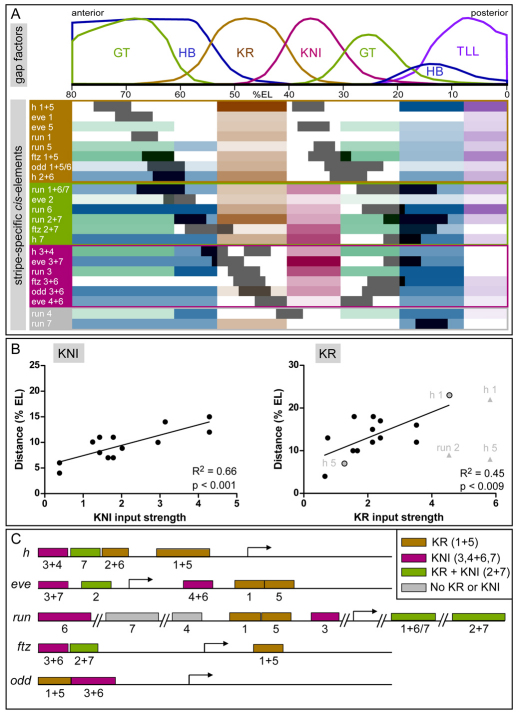
A remarkable feature of stripe-specific cis-regulation is the prevalence of dual-stripe elements: 12 of the 22 elements generate two stripes simultaneously. These elements naturally fall into three classes, based on their relationship to the expression domains of the three gap factors KR, KNI and GT, which form partially overlapping, bilaterally symmetric gradients in the trunk region of the embryo (Fig. 6A). The first class consists of the stripe 1+5 elements of h, odd, ftz and the stripe 2+6 element of h: all these elements receive strong repressive input from KR and produce two stripes flanking the KR domain. The elements in the second class, comprising the 3+7 and 4+6 stripe elements of eve (Clyde et al., 2003) and the 3+6 stripe elements of ftz and odd, receive strong repressive input from KNI and produce two stripes flanking the KNI domain. The elements in the third class (run_2+7, run_1+6/7 and ftz_2+7) drive stripes that flank the KR domain on one side and the KNI or posterior GT domain on the other, and receive input from the interjacent repressors KR, KNI and, sometimes, GT. With the exception of the elements driving run stripes 4 and 7, all single-stripe elements can also be placed into one of the three classes, based on the position of the stripes and their primary repressive inputs (Fig. 6A).
Fig. 6.
Binding site composition and genomic organization of stripe-specific cis-elements. (A) The spatial correlation of cis-element expression domains (dark gray) with the repressive gap factor input they receive. HB, blue; KR, brown; KNI, magenta; GT, green; TLL, purple; the strength of input is represented by the color intensity. Elements are grouped according to the central gap factors that provide the dominant repressive input. Expression patterns of gap factors are shown above [FRDWT 10% strip, time class 14A 4, data from Myasnikova et al. (Myasnikova et al., 2001)]. (B) Scatter plots showing the relationship between strength of predicted input (x, Stubb integrated profile value) and distance from the center of the gap factor domain to the proximal stripe border (y; % EL, percentage egg length). The results of linear regression analysis are indicated. The data points for the full h 1+5 element and the run 2+7 element are shown in gray, and for the separable h stripe 1 and 5 elements in gray with a black border. (C) Genomic organization of stripe-specific cis-elements within the pair-rule gene loci, with color-coding based on the classification shown in A. See also Fig. S2 and Tables S1, S2 and S3 in the supplementary material.
Within each class, the cis-elements drive expression at slightly different positions relative to the KR or KNI gradients. If these factors determine stripe boundaries, we would expect stripes driven by cis-elements with stronger factor input to be positioned further away from the respective gradient. To test this, we measured the distance between the center of the expression domain and the proximal stripe borders, and correlated it with the strength of predicted factor input into the cis-element (Fig. 6B); we restricted the analysis to those cis-elements for which the factor provides the most proximal predicted repressive input. We found a strong correlation for KNI (R2=0.66, P<0.001), but not for KR (R2=0.02, not significant). This appears to be due to the fact that the two stripes generated by some of the KR-regulated dual-stripe elements are positioned at different distances from the center of the KR domain, whereas in the case of KNI they are positioned more symmetrically on both sides of the gradient. The most dramatic case is the h 1+5 element, which has a very high predicted KR input, although stripe 5 lies close to the center of the KR domain (Fig. 6A). Since the same level of KR input cannot position two stripes at different locations on the gradient, there must be either additional inputs or a mechanism that allows differential regulation of the two stripes. Interestingly, the h 1+5 element, which at 3 kb is particularly long, is separable into two elements that drive the individual stripes, although only the combined element correctly specifies the posterior border of stripe 5 (Howard and Struhl, 1990; Langeland et al., 1994; Riddihough and Ish-Horowicz, 1991). Stubb predicts two distinct clusters of KR binding sites (see Fig. S3 in the supplementary material) within the element, suggesting that the KR inputs for the two stripes have been separated; the stronger cluster falls within the sub-element driving stripe 1, which is farther from the KR domain. A similar situation might obtain in the case of the run_(+30) element, which generates stripes 2 and 7 and also contains two distinct KR binding site clusters (Fig. 3). If we exclude this element and use the split h 1 and h 5 elements, we obtain a strong correlation between predicted KR input and positioning of the stripe borders (R2=0.45, P<0.009).
Taken together, our data strongly support the notion that the gap factors KR and KNI act as repressive morphogens in specifying pair-rule stripe positions. The prevalence of dual-stripe elements that drive two stripes flanking their expression domains further suggests that evolution has favored the efficiency offered by symmetrical reuse of similar inputs. However, the need to be optimally positioned along gap factor gradients may also provide the driving force for the separation of dual-stripe elements into single-stripe elements. Strong evidence for this comes from the genomic organization of pair-rule gene cis-elements: in all genes examined, stripes 1 and 5 are either generated by a dual-stripe element (ftz, odd), by two adjacent but separable elements (eve, run), or by a partially separable element (h) (Fig. 6C). Such an arrangement is unlikely to occur by chance (P<0.02). In both eve and run, unlike in ftz and odd, the distances of stripes 1 and 5 to the center of the KR domain differ substantially, as do their positions relative to the flanking GT domains (Fig. 6A). It thus appears likely that the single-stripe elements arose by splitting of an ancestral stripe 1+5 element, driven by the need for different levels of gap factor input.
DISCUSSION
The transition from non-periodic to periodic gene expression patterns is a key step in the establishment of the segmented body plan of the Drosophila embryo. The pair-rule genes that lie at the heart of this process have been the subject of much investigation, but important questions, in particular regarding the organization of cis-regulation, have remained unresolved. We have revisited the issue in a comprehensive fashion by combining computational and experimental cis-dissection with mutant analysis and a detailed characterization of expression dynamics, both of endogenous genes and of cis-regulatory elements. This systems-level analysis under uniform conditions reveals important insights and gives rise to a refined and in some respects substantially revised view of how periodic patterns are generated.
ftz and odd as primary pair-rule genes
Our most important finding is that ftz and odd, which had been regarded as secondary pair-rule genes, closely resemble eve, h and run in nearly all respects and should thus be co-classified with them as primary pair-rule genes. Expression dynamics clearly subdivide the pair-rule genes into two distinct groups, with eve, h, run, ftz and odd all showing an early and non-synchronous appearance of most stripes, whereas the 7-stripe patterns of prd and slp1 arise late and synchronously. The early expression of stripes is associated with the existence of stripe-specific cis-elements that respond to positional cues provided by the maternal and gap genes. Unlike previously thought, maternal and gap input is used pervasively in the initial patterning not only of eve, h and run, but also of ftz and odd (Fig. 1B). Aided by computational predictions, we were able to identify 8 new stripe-specific cis-elements for ftz, odd, and run. Although molecular epistasis experiments reveal a stronger role for eve, h and run in pair-rule gene cross-regulation, odd clearly affects the blastoderm patterning of other primary factors as well (Saulier-Le Drean et al., 1998) and ftz affects the patterning of odd (Nasiadka et al., 2000; Nasiadka and Krause, 1999).
The revised grouping brings the classification of pair-rule genes in line with their role in transmitting positional information to the subsequent tiers in the segmentation hierarchy. By the end of cellularization, the expression patterns of h, eve, run and ftz/odd are offset against each other to produce neatly tiled overlaps of four-nuclei-wide stripes (Fig. 1D). In combination, these patterns define a unique expression code for each nucleus within the two-segment unit that specifies both position and polarity in the segment and is read off by the segment-polarity genes (Baumgartner and Noll, 1990; Jaynes and Fujioka, 2004; Nasiadka et al., 2002). ftz (as an activator of odd) and odd together represent the fourth essential component of this positional code and are thus functionally equivalent to h, eve and run (Hughes and Krause, 2001). Placing all components of this code under the same direct control of the preceding tier of regulators presumably increases both the speed and the robustness of the process.
Although our results support a subdivision between primary and secondary pair-rule genes, they also reveal surprising complexities in the regulatory architecture that are not captured by any rigid dichotomy. The five primary pair-rule genes all have different cis-regulatory repertoires: stripe formation in h appears to rely solely on stripe-specific cis-elements, whereas eve employs a handoff from stripe-specific elements to a late-acting 7-stripe element. In run, ftz and odd, by contrast, the emergence of the full 7-stripe pattern is the result of joint action by stripe-specific elements and early-acting 7-stripe elements, with ftz and odd requiring the 7-stripe element to generate a subset of stripes. This difference in the importance of 7-stripe elements in stripe formation is supported by the results of rescue experiments: whereas the 7-stripe elements of both run and ftz provide partial rescue of the blastoderm expression pattern when the stripe-specific elements are missing (Butler et al., 1992; Hiromi et al., 1985), this is not the case for eve (Fujioka et al., 2002). Interestingly, run appears to occupy a unique and particularly crucial position among the five genes: it has both a full complement of stripe-specific elements and an early-acting 7-stripe element, and thus serves as an early integration point of maternal/gap input and pair-rule cross-regulation. Its regulatory region is particularly large and complex, with cis-elements acting over long distances across intervening genes and with partial redundancy between elements. Moreover, the removal of run has the most severe effects on pair-rule stripe positioning among the five genes. Another complexity lies in the fact that the early anterior expression of slp1 and prd, which otherwise exhibit all the characteristics of secondary pair-rule genes, has a clear role in patterning the anteriormost stripes of the primary pair-rule genes. In fact, we provide evidence that the most severe defect observed in the primary pair-rule gene mutants, namely the loss of pair-rule gene stripes 1 or 2 under eve loss-of-function conditions, is an indirect effect attributable to the regulation of these stripes by slp1 (Andrioli et al., 2004).
Integration of regulatory input from maternal/gap and pair-rule genes
Our investigation provides important new insight regarding the relative roles of maternal/gap input and pair-rule gene cross-regulation in stripe formation. As described, nearly all stripes of the primary pair-rule genes are initially formed through stripe-specific cis-elements. Cross-regulatory interactions between the pair-rule genes then refine these patterns by ensuring the proper spacing, width and intensity of stripes, as revealed by mutant analysis. However, these two aspects or layers of regulation are not as clearly separated as has often been thought. For example, 7-stripe elements have been regarded primarily as conduits of pair-rule cross-regulation; by contrast, we find that the 7-stripe elements of run, ftz and odd are active from phase 1 onwards and contain significant input from the maternal and gap genes, resulting in modulated or partial 7-stripe expression early. In the case of the KR binding sites predicted within the run 7-stripe element, we have shown by mutational analysis that this input is indeed functional. Conversely, stripe-specific cis-elements receive significant regulatory input from pair-rule genes. This is particularly clear in the case of eve and h: expression dynamics indicate that their 7-stripe patterns become properly resolved without the involvement of 7-stripe elements, and the defects in the h and eve expression patterns that we observe in pair-rule gene mutants occur at a time when only stripe-specific elements are active. In a few cases, pair-rule input into stripe-specific elements has been demonstrated directly (Hartmann et al., 1994; Riddihough and Ish-Horowicz, 1991; Tsai and Gergen, 1994), but our comparative analysis of expression dynamics underscores that it is indeed a pervasive feature of pair-rule cis-regulation.
An important question arising from these findings is how the different types of regulatory input are integrated, both at the level of individual cis-elements and at the level of the locus as a whole. Binding sites for maternal and gap factors typically form tight clusters, supporting a modular organization of cis-regulation. Such modularity is crucial for the ‘regional’ expression of individual stripes, which can be achieved only if repressive gap inputs can be properly separated between cis-elements. By contrast, binding sites for the pair-rule factors appear to be more dispersed across the cis-regulatory regions and not tightly co-clustered with the maternal and gap inputs. Unlike the gap factors, which are thought to act as short-range repressors, with a range of roughly 100 bp (Arnosti et al., 1996b; Hewitt et al., 1999; Nibu et al., 1998), most pair-rule genes act as long-range repressors, with an effective range of at least 2 kb (Aronson et al., 1997; Barolo and Levine, 1997; Courey and Jia, 2001; Goldstein et al., 2005; Kobayashi et al., 2001). Pair-rule factor inputs may therefore influence expression outcomes at greater distances along the DNA, which would allow a single cluster of binding sites to affect multiple cis-elements. Remarkably, the 7-stripe elements of ftz, run, eve, prd and odd all combine inputs from promoter-proximal and more distal sequence over distances of at least 5 kb (Goto et al., 1989; Gutjahr et al., 1994; Klingler et al., 1996; Pick et al., 1990), and, as we have seen in the case of odd, the combination can involve non-additive effects. Thus, the refinement of expression patterns through pair-rule cross-regulation might rely on interactions over greater distances, consistent with the ‘global’ role of pair-rule genes as regulators of the entire 7-stripe pattern. How such interactions are realized at the molecular level and how the inputs from stripe-specific and 7-stripe elements are combined to produce a defined transcriptional outcome is currently unknown.
Principles of stripe formation
Taken together with previous studies, our data support a coherent and conceptually straightforward model of how stripes are made in the Drosophila blastoderm. In the trunk region of the embryo, the maternal activators BCD and CAD form two overlapping but anti-correlated gradients; they coarsely specify the expression domains of region-specific gap genes, which become refined by cross-repressive interactions among the gap factors. The result is a tiled array of overlapping gap factor gradients, centered around the bilaterally symmetric domains of KR and KNI, which are flanked on either side by the bimodal domains of GT and HB. The same basic principles are used again in the next step: following the initial positioning of stripes by maternal and gap factor input, cross-repressive interactions among the primary pair-rule genes serve to refine the pattern and ensure the uniform spacing and width of expression domains. The significant correlations that we observe between the strength of KR/KNI input and the position of the resulting stripes relative to the respective gradients supports the notion that these factors function as repressive morphogens in defining the proximal borders of the nascent pair-rule stripes. Importantly, owing to the symmetry of the KR and KNI gradients, combined with the largely symmetric positioning of the flanking GT and HB gradients, the same regulatory input can be used to specify two distinct positions, one on either slope of the gradient; this is exploited in a systematic fashion by dual-stripe cis-elements, which account for the majority of stripe-generating elements. Thus, the key to stripe formation is the translation of transcription factor gradients into an array of narrower, partially overlapping expression domains that are stabilized by cross-repressive interactions; the iteration of this process, combined with the duplication of position through bilateral gradient symmetry, creates a periodic array of stripes that is sufficient to impart a unique identity to each nucleus in the trunk region of the embryo.
With the exception of the anteriormost pair-rule gene stripes, which are subject to more complex regulation (as evidenced by the crucial role of slp1), this model accounts for most of the stripe formation process. Particularly striking is the similarity between the central gap factors and the primary pair-rule factors with respect to regulatory interactions and expression domain positions. The offset arrangement of two pairs of mutually exclusive gap domains, KNI/HB and KR/GT (Fig. 6A), is mirrored at the pair-rule gene level, with the anti-correlated and mutually repressive stripes of HAIRY and RUN phase-shifted against the similarly anti-correlated expression patterns of EVE and ODD (Fig. 1C,D). The parallel suggests that this regulatory geometry is particularly suited to robustly specify a multiplicity of positions. However, such a circuitry of cross-repressive relationships is compatible with a range of potentially stable expression states and thus is insufficient by itself to uniquely define position along the anterior-posterior axis (Albert and Othmer, 2003; Jaeger and Reinitz, 2006; Tyson et al., 2003). Therefore, the initial priming of position by the preceding tier of the regulatory hierarchy is crucial. At the level of the pair-rule genes, the extensive repertoire of maternal/gap-driven cis-elements that initiate stripe expression for all four components of the array is thus necessary to ensure that the cross-regulatory dynamics will drive the correct overall pattern. Finally, to achieve a smooth transition between the tasks of transmitting spatial information from the preceding tier of the hierarchy and refinement/stabilization of the pattern, the two layers of regulation need to be closely integrated. This is likely to be facilitated by the fact that in each of the mutually repressive pairs, one gene generates stripes purely through stripe-specific elements (h, eve), whereas the other has an early-acting 7-stripe element that mediates pair-rule cross-repression and acts concurrently with stripe-specific elements (run, odd).
Stripe positioning is not always symmetric around the central gap factor gradients. In such cases, the conflicting needs for appropriate regulatory input into the relevant cis-elements appear to have driven the separation of ancestral dual-stripe elements into more specialized elements optimized for generating a single stripe. The co-localization of the cis-elements that generate stripes 1 and 5 within all pair-rule gene regulatory regions, be it in the form of dual-stripe elements or of adjacent but separable single-stripe elements, provides strong evidence that this is indeed a key mechanism underlying the emergence of single-stripe cis-elements. Given the evolutionary plasticity of regulatory sequence (Carroll, 2008; Ludwig et al., 2000), it is not difficult to envision such a separation and subfunctionalization of cis-elements. How the crucial transition from a non-periodic to periodic pattern is achieved in other insects is a fascinating question (Cerny et al., 2008; Peel et al., 2005; Rosenberg et al., 2009; Tautz, 2004). Intriguingly, most of the molecular players and general features of their expression patterns are well conserved beyond Diptera, and it is tempting to speculate that dual-stripe cis-regulation might have arisen through co-option as the blastoderm fate map shifted to include more posterior positions. It will be interesting to see to what extent the mechanisms and principles of stripe formation that we have outlined here apply in other species.
Supplementary Material
Acknowledgements
We thank the Bloomington Stock Center, J. Jaynes, M. Fujioka, P. Gergen, M. Klingler, J. Merriam, S. Small and L. Pick for sharing materials; U. Unnerstall for assistance in preparing the manuscript; and C. Desplan for valuable input and discussion. This work was supported in part by National Institutes of Health grant GM066434, by an Alexander von Humboldt Professorship and by the Center for Integrated Protein Science Munich (U.G.) and by a Rockefeller University Graduate Fellowship (M.D.S.). Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.062141/-/DC1
References
- Akam M. (1987). The molecular basis for metameric pattern in the Drosophila embryo. Development 101, 1-22 [PubMed] [Google Scholar]
- Albert R., Othmer H. G. (2003). The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster. J. Theor. Biol. 223, 1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrioli L. P., Oberstein A. L., Corado M. S., Yu D., Small S. (2004). Groucho-dependent repression by sloppy-paired 1 differentially positions anterior pair-rule stripes in the Drosophila embryo. Dev. Biol. 276, 541-551 [DOI] [PubMed] [Google Scholar]
- Arnosti D. N., Barolo S., Levine M., Small S. (1996a). The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122, 205-214 [DOI] [PubMed] [Google Scholar]
- Arnosti D. N., Gray S., Barolo S., Zhou J., Levine M. (1996b). The gap protein knirps mediates both quenching and direct repression in the Drosophila embryo. EMBO J. 15, 3659-3666 [PMC free article] [PubMed] [Google Scholar]
- Aronson B. D., Fisher A. L., Blechman K., Caudy M., Gergen J. P. (1997). Groucho-dependent and -independent repression activities of Runt domain proteins. Mol. Cell. Biol. 17, 5581-5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao R., Friedrich M. (2008). Conserved cluster organization of insect Runx genes. Dev. Genes Evol. 218, 567-574 [DOI] [PubMed] [Google Scholar]
- Barolo S., Levine M. (1997). hairy mediates dominant repression in the Drosophila embryo. EMBO J. 16, 2883-2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner S., Noll M. (1990). Network of interactions among pair-rule genes regulating paired expression during primordial segmentation of Drosophila. Mech. Dev. 33, 1-18 [DOI] [PubMed] [Google Scholar]
- Berman B. P., Pfeiffer B. D., Laverty T. R., Salzberg S. L., Rubin G. M., Eisen M. B., Celniker S. E. (2004). Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 5, R61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler B. A., Soong J., Gergen J. P. (1992). The Drosophila segmentation gene runt has an extended cis-regulatory region that is required for vital expression at other stages of development. Mech. Dev. 39, 17-28 [DOI] [PubMed] [Google Scholar]
- Calhoun V. C., Levine M. (2003). Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. USA 100, 9878-9883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. B. (2008). Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25-36 [DOI] [PubMed] [Google Scholar]
- Carroll S. B., Vavra S. H. (1989). The zygotic control of Drosophila pair-rule gene expression. II. Spatial repression by gap and pair-rule gene products. Development 107, 673-683 [DOI] [PubMed] [Google Scholar]
- Cerny A. C., Grossmann D., Bucher G., Klingler M. (2008). The Tribolium ortholog of knirps and knirps-related is crucial for head segmentation but plays a minor role during abdominal patterning. Dev. Biol. 32, 284-294 [DOI] [PubMed] [Google Scholar]
- Clyde D. E., Corado M. S., Wu X., Pare A., Papatsenko D., Small S. (2003). A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature 426, 849-853 [DOI] [PubMed] [Google Scholar]
- Courey A. J., Jia S. (2001). Transcriptional repression: the long and the short of it. Genes Dev. 15, 2786-2796 [DOI] [PubMed] [Google Scholar]
- Duncan E. J., Wilson M. J., Smith J. M., Dearden P. K. (2008). Evolutionary origin and genomic organisation of runt-domain containing genes in arthropods. BMC Genomics 9, 558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom P. G., Suzuki H., Ninomiya N., Akalin A., Sessa L., Lavorgna G., Brozzi A., Luzi L., Tan S. L., Yang L., et al. (2006). Complex Loci in human and mouse genomes. PLoS Genet. 2, e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florence B., Guichet A., Ephrussi A., Laughon A. (1997). Ftz-F1 is a cofactor in Ftz activation of the Drosophila engrailed gene. Development 124, 839-847 [DOI] [PubMed] [Google Scholar]
- Frasch M., Levine M. (1987). Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1, 981-995 [DOI] [PubMed] [Google Scholar]
- Fujioka M., Jaynes J. B., Goto T. (1995). Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development 121, 4371-4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Miskiewicz P., Raj L., Gulledge A. A., Weir M., Goto T. (1996). Drosophila Paired regulates late even-skipped expression through a composite binding site for the paired domain and the homeodomain. Development 122, 2697-2707 [DOI] [PubMed] [Google Scholar]
- Fujioka M., Yusibova G. L., Patel N. H., Brown S. J., Jaynes J. B. (2002). The repressor activity of Even-skipped is highly conserved, and is sufficient to activate engrailed and to regulate both the spacing and stability of parasegment boundaries. Development 129, 4411-4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein R. E., Cook O., Dinur T., Pisante A., Karandikar U. C., Bidwai A., Paroush Z. (2005). An eh1-like motif in odd-skipped mediates recruitment of Groucho and repression in vivo. Mol. Cell. Biol. 25, 10711-10720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T., Macdonald P., Maniatis T. (1989). Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell 57, 413-422 [DOI] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., Yang L., Artieri C. G., van Baren M. J., Boley N., Booth B. W., et al. (2010). The developmental transcriptome of Drosophila melanogaster. Nature 471, 473-479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. B., Hatini V., Johansen K. A., Liu X. J., Lengyel J. A. (2002). Drumstick is a zinc finger protein that antagonizes Lines to control patterning and morphogenesis of the Drosophila hindgut. Development 129, 3645-3656 [DOI] [PubMed] [Google Scholar]
- Gutjahr T., Vanario-Alonso C. E., Pick L., Noll M. (1994). Multiple regulatory elements direct the complex expression pattern of the Drosophila segmentation gene paired. Mech. Dev. 48, 119-128 [DOI] [PubMed] [Google Scholar]
- Hart M. C., Wang L., Coulter D. E. (1996). Comparison of the structure and expression of odd-skipped and two related genes that encode a new family of zinc finger proteins in Drosophila. Genetics 144, 171-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C., Taubert H., Jackle H., Pankratz M. J. (1994). A two-step mode of stripe formation in the Drosophila blastoderm requires interactions among primary pair rule genes. Mech. Dev. 45, 3-13 [DOI] [PubMed] [Google Scholar]
- Hewitt G. F., Strunk B. S., Margulies C., Priputin T., Wang X. D., Amey R., Pabst B. A., Kosman D., Reinitz J., Arnosti D. N. (1999). Transcriptional repression by the Drosophila giant protein: cis element positioning provides an alternative means of interpreting an effector gradient. Development 126, 1201-1210 [DOI] [PubMed] [Google Scholar]
- Hiromi Y., Gehring W. J. (1987). Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell 50, 963-974 [DOI] [PubMed] [Google Scholar]
- Hiromi Y., Kuroiwa A., Gehring W. J. (1985). Control elements of the Drosophila segmentation gene fushi tarazu. Cell 43, 603-613 [DOI] [PubMed] [Google Scholar]
- Hong J. W., Hendrix D. A., Levine M. S. (2008). Shadow enhancers as a source of evolutionary novelty. Science 321, 1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper K. L., Parkhurst S. M., Ish-Horowicz D. (1989). Spatial control of hairy protein expression during embryogenesis. Development 107, 489-504 [DOI] [PubMed] [Google Scholar]
- Hou H. Y., Heffer A., Anderson W. R., Liu J., Bowler T., Pick L. (2009). Stripy Ftz target genes are coordinately regulated by Ftz-F1. Dev. Biol. 335, 442-453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K., Ingham P. (1986). Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell 44, 949-957 [DOI] [PubMed] [Google Scholar]
- Howard K. R., Struhl G. (1990). Decoding positional information: regulation of the pair-rule gene hairy. Development 110, 1223-1231 [DOI] [PubMed] [Google Scholar]
- Hughes S. C., Krause H. M. (2001). Establishment and maintenance of parasegmental compartments. Development 128, 1109-1118 [DOI] [PubMed] [Google Scholar]
- Ingham P., Gergen P. (1988). Interactions between the pair-rule genes runt, hairy, even-skipped and fushi tarazu and the establishment of periodic pattern in the Drosophila embryo. Development 104, 51-60 3075544 [Google Scholar]
- Ingham P. W. (1988). The molecular genetics of embryonic pattern formation in Drosophila. Nature 335, 25-34 [DOI] [PubMed] [Google Scholar]
- Jaeger J., Reinitz J. (2006). On the dynamic nature of positional information. BioEssays 28, 1102-1111 [DOI] [PubMed] [Google Scholar]
- Jaynes J. B., Fujioka M. (2004). Drawing lines in the sand: even skipped et al. and parasegment boundaries. Dev. Biol. 269, 609-622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler M., Soong J., Butler B., Gergen J. P. (1996). Disperse versus compact elements for the regulation of runt stripes in Drosophila. Dev. Biol. 177, 73-84 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Goldstein R. E., Fujioka M., Paroush Z., Jaynes J. B. (2001). Groucho augments the repression of multiple Even skipped target genes in establishing parasegment boundaries. Development 128, 1805-1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeland J. A., Attai S. F., Vorwerk K., Carroll S. B. (1994). Positioning adjacent pair-rule stripes in the posterior Drosophila embryo. Development 120, 2945-2955 [DOI] [PubMed] [Google Scholar]
- Lecuit T., Wieschaus E. (2000). Polarized insertion of new membrane from a cytoplasmic reservoir during cleavage of the Drosophila embryo. J. Cell Biol. 150, 849-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M. Z., Bergman C., Patel N. H., Kreitman M. (2000). Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403, 564-567 [DOI] [PubMed] [Google Scholar]
- Manoukian A. S., Krause H. M. (1992). Concentration-dependent activities of the even-skipped protein in Drosophila embryos. Genes Dev. 6, 1740-1751 [DOI] [PubMed] [Google Scholar]
- Manoukian A. S., Krause H. M. (1993). Control of segmental asymmetry in Drosophila embryos. Development 118, 785-796 [DOI] [PubMed] [Google Scholar]
- Meng X., Brodsky M. H., Wolfe S. A. (2005). A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol. 23, 988-994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myasnikova E., Samsonova A., Kozlov K., Samsonova M., Reinitz J. (2001). Registration of the expression patterns of Drosophila segmentation genes by two independent methods. Bioinformatics 17, 3-12 [DOI] [PubMed] [Google Scholar]
- Nasiadka A., Krause H. M. (1999). Kinetic analysis of segmentation gene interactions in Drosophila embryos. Development 126, 1515-1526 [DOI] [PubMed] [Google Scholar]
- Nasiadka A., Grill A., Krause H. M. (2000). Mechanisms regulating target gene selection by the homeodomain-containing protein Fushi tarazu. Development 127, 2965-2976 [DOI] [PubMed] [Google Scholar]
- Nasiadka A., Dietrich B. H., Krause H. M. (2002). Anterior-posterior patterning in the Drosophila embryo. Adv. Dev. Biol. Biochem. 12, 155-204 [Google Scholar]
- Nibu Y., Zhang H., Bajor E., Barolo S., Small S., Levine M. (1998). dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 17, 7009-7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes M. B., Christensen R. G., Wakabayashi A., Stormo G. D., Brodsky M. H., Wolfe S. A. (2008a). Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133, 1277-1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes M. B., Meng X., Wakabayashi A., Sinha S., Brodsky M. H., Wolfe S. A. (2008b). A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 36, 2547-2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Wieschaus E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795-801 [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Frohnhofer H. G., Lehmann R. (1987). Determination of anteroposterior polarity in Drosophila. Science 238, 1675-1681 [DOI] [PubMed] [Google Scholar]
- Ochoa-Espinosa A., Yucel G., Kaplan L., Pare A., Pura N., Oberstein A., Papatsenko D., Small S. (2005). The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila. Proc. Natl. Acad. Sci. USA 102, 4960-4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz M. J., Jackle H. (1990). Making stripes in the Drosophila embryo. Trends Genet. 6, 287-292 [DOI] [PubMed] [Google Scholar]
- Pankratz M. J., Seifert E., Gerwin N., Billi B., Nauber U., Jackle H. (1990). Gradients of Kruppel and knirps gene products direct pair-rule gene stripe patterning in the posterior region of the Drosophila embryo. Cell 61, 309-317 [DOI] [PubMed] [Google Scholar]
- Peel A. D., Chipman A. D., Akam M. (2005). Arthropod segmentation: beyond the Drosophila paradigm. Nat. Rev. Genet. 6, 905-916 [DOI] [PubMed] [Google Scholar]
- Pick L., Schier A., Affolter M., Schmidt-Glenewinkel T., Gehring W. J. (1990). Analysis of the ftz upstream element: germ layer-specific enhancers are independently autoregulated. Genes Dev. 4, 1224-1239 [DOI] [PubMed] [Google Scholar]
- Rajewsky N., Vergassola M., Gaul U., Siggia E. D. (2002). Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo. BMC Bioinformatics 3, 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddihough G., Ish-Horowicz D. (1991). Individual stripe regulatory elements in the Drosophila hairy promoter respond to maternal, gap, and pair-rule genes. Genes Dev. 5, 840-854 [DOI] [PubMed] [Google Scholar]
- Rosenberg M. I., Lynch J. A., Desplan C. (2009). Heads and tails: evolution of antero-posterior patterning in insects. Biochim. Biophys. Acta 1789, 333-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulier-Le Drean B., Nasiadka A., Dong J., Krause H. M. (1998). Dynamic changes in the functions of Odd-skipped during early Drosophila embryogenesis. Development 125, 4851-4861 [DOI] [PubMed] [Google Scholar]
- Schier A. F., Gehring W. J. (1992). Direct homeodomain-DNA interaction in the autoregulation of the fushi tarazu gene. Nature 356, 804-807 [DOI] [PubMed] [Google Scholar]
- Schier A. F., Gehring W. J. (1993). Analysis of a fushi tarazu autoregulatory element: multiple sequence elements contribute to enhancer activity. EMBO J. 12, 1111-1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M. D., Pearce M., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E. D., Gaul U. (2004). Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2, E271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., Raveh-Sadka T., Schroeder M., Unnerstall U., Gaul U. (2008). Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451, 535-540 [DOI] [PubMed] [Google Scholar]
- Sinha S., van Nimwegen E., Siggia E. D. (2003). A probabilistic method to detect regulatory modules. Bioinformatics 19, i292-i301 [DOI] [PubMed] [Google Scholar]
- Sinha S., Schroeder M. D., Unnerstall U., Gaul U., Siggia E. D. (2004). Cross-species comparison significantly improves genome-wide prediction of cis-regulatory modules in Drosophila. BMC Bioinformatics 5, 129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S., Blair A., Levine M. (1992). Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 11, 4047-4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic D., Small S., Levine M. (1991). Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science 254, 1385-1387 [DOI] [PubMed] [Google Scholar]
- Surkova S., Kosman D., Kozlov K., Manu Myasnikova E., Samsonova A. A., Spirov A., Vanario-Alonso C. E., Samsonova M., Reinitz J. (2008). Characterization of the Drosophila segment determination morphome. Dev. Biol. 313, 844-862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. (2004). Segmentation. Dev. Cell 7, 301-312 [DOI] [PubMed] [Google Scholar]
- Tomancak P., Berman B. P., Beaton A., Weiszmann R., Kwan E., Hartenstein V., Celniker S. E., Rubin G. M. (2007). Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8, R145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C., Gergen J. P. (1994). Gap gene properties of the pair-rule gene runt during Drosophila segmentation. Development 120, 1671-1683 [DOI] [PubMed] [Google Scholar]
- Tyson J. J., Chen K. C., Novak B. (2003). Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 15, 221-231 [DOI] [PubMed] [Google Scholar]
- Visel A., Rubin E. M., Pennacchio L. A. (2009). Genomic views of distant-acting enhancers. Nature 461, 199-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Pick L. (1995). Non-periodic cues generate seven ftz stripes in the Drosophila embryo. Mech. Dev. 50, 163-175 [DOI] [PubMed] [Google Scholar]
- Yu Y., Li W., Su K., Yussa M., Han W., Perrimon N., Pick L. (1997). The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature 385, 552-555 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.