Abstract
The 69 insertion and Q151M mutations are multi-nucleoside/nucleotide resistance mutations (MNR). The prevalence among 4078 antiretroviral therapy (ART)-experienced individuals was <1.3%. Combined ART fully prevented MNR in subtype B infections. Case-control studies were performed to identify risk factors. Control subjects were patients with ≥3 thymidine-analogue mutations. The 69 insertion study (27 control subjects, 14 case patients) identified didanosine exposure as a risk (odds ratio, 5.0 per year; P = .019), whereas the Q151M study (which included 44 control subjects and 25 case patients) detected no associations. Following detection, individuals with Q151M tended to have lower suppression rates and higher mortality rates, relative to control subjects. Additional studies are needed to verify these findings in non-subtype B infections.
The introduction of highly-active antiretroviral therapy (HAART) reduced morbidity and mortality of human immunodeficiency virus (HIV) infected patients. However, drug resistance continues to emerge in association with treatment failure [1].
The 69 insertion and Q151M mutation are multi-nucleoside/nucleotide resistance (MNR) mutations on the reverse transcriptase that affect the activity of all approved nucleoside reverse transcriptase inhibitors (NRTIs). Although 69 insertion confers full resistance to all drugs, tenofovir retains some activity when Q151M is present [2–4]. MNR mutations occur rarely in European settings (prevalence, <1% to 3.6%). The prevalence in resource-limited countries has not been analyzed, but recent reports indicate that it has increased [5–8]. The possible increase in MNR HIV strains is of great concern because of the very limited options for salvage treatment in resource-limited settings and the general lack of understanding as to how to optimally treat patients who have MNR infections [9].
We aimed to identify predictors for the emergence of the 69 insertion and Q151M mutation in the Swiss HIV Cohort Study (SHCS) and studied outcomes of salvage regimens applied for treatment of MNR HIV infections.
METHODS
Data and Patient Selection
Our analysis included data from the SHCS and the SHCS drug resistance database up to February 2010 [10, 11]. The SHCS has been approved by the ethical committees of all participating institutions, and written informed consent has been obtained from participants.
Prevalence of Acquired and Transmitted MNR Mutations
The prevalence of the 69 insertion and the Q151M mutation was analyzed among treatment-experienced (at least 30 days exposure) and treatment-naive patients. Among treatment-naive patients, possible transmission clusters were identified through phylogenetic methods. These analyses were performed with PHYLIP 3.6 (distributed by J. Felsenstein), using the F84 nucleotide substitution model and the neighbor-joining tree algorithm with 1000 bootstraps. To avoid interference of treatment history, all major International AIDS Society–USA drug resistance–associated amino acid positions were deleted from the sequences prior to analysis [12].
Case-control Study to Determine Predictors for MNR
We compared patients with MNR detected with patients who carried viruses with ≥3 thymidine analogue mutations (TAMs), either from the TAM 1 (M41L, L210W, T215Y) or the TAM 2 pathway (D67N, K70R, T215F, K219K/E). The rationale was to establish a control group consisting of highly NRTI-experienced individuals with comparable characteristics, except for the occurrence of 69 insertion or Q151M. Separate matched case-control studies were performed for each of the 2 MNR mutations. Control patients were matched 2:1 on the basis of the first antiretroviral therapy (ART) received and the time between ART initiation and the detection of MNR mutations (for case patients) or ≥3 TAMs (for control subjects). Inclusion was restricted to individuals infected with HIV subtype B who started ART with single-class NRTI therapy.
Conditional logistic regression analyses were performed to identify risk factors for the emergence of 69 insertion and Q151M. Variables tested included the time spent on specific NRTIs and adjustments for the following potential confounders: sex, age, ethnicity, risk group, HIV-1 RNA level, and CD4+ cell count at time of detection of MNR mutations (for case patients) or ≥ 3 TAMs (for control subjects).
Factors Associated with Attaining Undetectable Viral Loads After Detection of MNR Mutations
Virological outcomes after detection of MNR mutations were analyzed for patients from the case-control studies with >1 follow-up HIV-1 RNA measurement. Characteristics and treatments were compared between patients who ever achieved 2 consecutive undetectable HIV-1 RNA levels <50 copies/mL and patients who did not. Fisher's exact test (categorical) and Wilcoxon rank-sum test (continuous variables) were used.
Association of All-cause Mortality with Detection of MNR Mutations
Cox proportional hazard models for matched case-control data were estimated to analyze the time to all-cause mortality after detection of MNR mutations (case patients) or ≥3 TAMs (control subjects). Models were stratified by years of detection of MNR mutations or ≥3 TAMs (1998, 1999–2003, after 2003). The proportional hazard assumption was verified by analyzing Schoenfeld residuals.
Statistical analyses were performed with Stata 11 SE (StataCorp), all confidence intervals (CI) are 95% CIs, and the level of significance was set at P = .05.
RESULTS
Prevalence of MNR Mutations in Treatment-experienced Patients
The SHCS included 19 (0.5%) of 4078 and 34 (0.8%) of 4078 treatment-experienced patients who carried viruses with the 69 insertion and Q151M mutation, respectively. Most patients in the 69 insertion and Q151M groups were treated with mono- or dual-NRTI therapy (14 [73.7%] of 19 and 30 [88.2%] of 34, respectively. MNR was never detected in patients who were exclusively treated with HAART (2 NRTIs and 1 boosted protease inhibitor [PI]/nonnucleoside reverse transcriptase inhibitor [NNRTI]). The median duration of ART until detection was 6.8 years for the 69 insertion group and 5.5 years for the Q151M group. The median years of detection were 2000.5 (range: 1995–2007) and 2000 (range: 1995–2006), respectively. Most individuals who carried viruses with MNR were infected with subtype B viruses (18 [94.7%] of 19 and 29 [85.3%] of 34, respectively). All additional analyses were restricted to this subtype.
Evidence for MNR Transmission among Treatment-naive Patients
We screened 5692 sequences from treatment-naive patients and detected the 69 insertion 3 times (0.05%) and Q151M once (0.02%). The phylogenetic analysis provided strong evidence of forward transmission of the 69 insertion from 1 index patient to 2 patients (100% bootstrap support). All of these patients were men who had sex with men and were from the same study center. For Q151M, no transmission cluster was detected.
Predictors for the Emergence of MNR in Patients Infected with HIV Subtype B
For the 69 insertion case-control study, matching criteria fitted for 14 case patients (69 insertion) and 27 control subjects (≥3 TAMs). Interestingly, years spent receiving didanosine were significantly associated with the emergence of 69 insertion in univariable (odds ratio [OR], 3.4; 95% CI, 1.2–9.6; P = .019) and multivariable models (OR, 5.0; 95% CI, 1.3–19.3; P = .019).
For the Q151M case-control study, 25 case patients and 44 control subjects (≥3 TAMs) were matched, but conditional logistic regressions failed to identify predictors for the emergence of Q151M.
Virological Response to Salvage Treatment upon Emergence of MNR
Virological outcomes for patients with >1 follow-up HIV-1 RNA measurement after MNR detection were analyzed. The probability of ever achieving viral suppression was comparable between the 69 insertion group (9 [69.2%] of 13; 95% CI, 38.6%–90.9%) and respective control subjects (15 [75.0%] of 20; 95% CI, 50.9%–91.3%). A large proportion of patients who received previously unseen drug classes achieved viral suppression (87.5% compared to 40.0% of other patients; P = .217). As shown in table 1, all patients detected without major NNRTI or PI mutations achieved viral suppression (patients 1, 4, and 8). Five of 7 patients with either NNRTI or PI mutations achieved viral suppression with the remaining active non-NRTI drug class (patients 2, 5, 6, 7, and 11).
Table 1.
Characteristics of Patients who Achieved Viral Suppression and Characteristics of the First Successful Treatment following Detection of 69 Insertion or Q151M
| Mutation, patient | Year of detection | Log10 HIV-1 RNA level at detection, copies/mL | CD4+ cell count at detection, cells/μL | CDC stage | NRTI mutations at detection | Majora NNRTI mutations at detection | Majora PI mutations at detection | Year of first successful treatment | Successful treatment | Year of loss of follow-up |
| 69 insertion | ||||||||||
| 1 | 1995 | 4.4 | 50 | C | M41L, 69 insertion, L210W, T215Y | 2003 | TDF EFV ddI | 2009 | ||
| 2 | 1995 | 4.2 | 118 | B | M41L, 69 insertion, K70R, M184V, L210W, T215Y | Y188C | 1999 | NFV ZDV 3TC | 2009 | |
| 3 | 1997 | 5.9 | 32 | C | M41L, 69 insertion, L210W, T215Y | M46L, V82A, L90M | 2001b | |||
| 4 | 1997 | 3.3 | 472 | B | M41L, 69 insertion, M184I, T215Y | 1997 | SQV RTV NVP D4T | 2009 | ||
| 5 | 1997 | 4.3 | 315 | C | M41L, 69 insertion, T215Y | I84V | 2000 | LPV IDV D4T 3TC | 2009 | |
| 6 | 1999 | 3.1 | 180 | B | M41L, 69 insertion, L210W, T215Y | M46I, T74P, I84V, L90M | 2001 | LPV EFV ddI 3TC | 2009 | |
| 7 | 1999 | 5.1 | 139 | C | M41L, 69 insertion, M184I, T215Y | M46L, V82A, I84V | 2002 | EFV ddI ABC | 2009 | |
| 8 | 2001 | 4.2 | 415 | B | D67N, 69 insertion, K70R, K219Q | 2004 | RTV EFV ATV 3TC | 2009 | ||
| 9 | 2002 | 4.8 | 542 | B | M41L, 69 insertion, T215Y | Y181C | M46I, G48V, I84V, N88S, L90M | 2009 | ||
| 10 | 2003 | 5.7 | 13 | B | M41L, 69 insertion, L74V, L210W, T215Y | L100I, K103N | M46I, L76V, V82F, L90M | 2003b | ||
| 11 | 2004 | 4.8 | 198 | B | M41L, 69 insertion, M184V, L210W, T215Y | M46I, I84V, L90M | 2005 | LPV EFV ZDV 3TC | 2009 | |
| 12 | 2004 | 5.1 | 367 | C | M41L, 69 insertion, L210W, T215Y | M46L, V82A, L90M | 2009 | |||
| 13 | 2007 | 2.9 | 358 | B | M41L, 69 insertion, M184V, L210W, T215Y | K103N | I54L, I84V, L90M | 2008 | TDF RAL ETV ABC 3TC | 2009 |
| Q151M | ||||||||||
| 1 | 1995 | 5.4 | 160 | A | V75I, F77L, Y115F, F116Y, Q151M | Q58E | 2001b | |||
| 2 | 1996 | 5.7 | 50 | C | F77L, F116Y, Q151M | G48V, L90M | 1997b | |||
| 3 | 1996 | 4.2 | 100 | C | D67N, K70R, F77L, Q151M, T215F, K219E | V108I | 2000b | |||
| 4 | 1996 | 3.5 | 162 | B | F116Y, Q151M, | 1999 | NFV D4T 3TC | 2009 | ||
| 5 | 1996 | 4.4 | 133 | C | K65R, V75I, F77L, Y115F, F116Y, Q151M, K219Q | 2008 | TDF RTV RAL MVC ETV DRV ZDV 3TC | 2009 | ||
| 6 | 1997 | 5.2 | 75 | C | M41L, A62V, V75I, F77L, F116Y, Q151M, M184V | 1998b | ||||
| 7 | 1997 | 2.6 | 464 | A | D67N, Q151M, K219Q | 2001b | ||||
| 8 | 1997 | 3.6 | 61 | C | Q151M, M184V | 2001b | ||||
| 9 | 1997 | 3.8 | 476 | C | D67N, F116Y, Q151M | Q58E | 1997 | IDV D4T 3TC | 2009 | |
| 10 | 1998 | 5.1 | 15 | C | A62V, V75I, F77L, F116Y, Q151M, M184V | K103N | M46L, G48V, V82A | 2003b | ||
| 11 | 1998 | 5.0 | 8 | C | M41L, D67N, F77L, F116Y, Q151M, M184V, L210W, T215Y | M46I, V82A | 2000b | |||
| 12 | 1998 | 3.9 | 412 | B | A62V, K65R, V75I, F77L, Y115F, F116Y, Q151M, M184V | D30N | 1999 | SQV RTV EFV D4T ABC | 2007b | |
| 13 | 1999 | 5.8 | 5 | C | D67N, Q151M | V82A, I84V, L90M | 2000b | |||
| 14 | 1999 | 5.3 | 23 | C | D67N, K70R, F116Y, Q151M, M184V, K219E | V82A, L90M | 2007 | RTV RAL ETV DRV 3TC | 2009 | |
| 15 | 2000 | 5.3 | 36 | C | M41L, D67N, K70R, Q151M, M184V, T215F, K219E | K103N, V108I | I54L, V82A, L90M | 2002b | ||
| 16 | 2000 | 4.9 | 481 | B | A62V, V75I, F77L, F116Y, Q151M | 2002 | LPV EFV ZDV ABC 3TC | 2009 | ||
| 17 | 2001 | 2.5 | 259 | C | A62V, F116Y, Q151M, M184V, | M46I | 2002 | EFV D4T 3TC | 2003b | |
| 18 | 2001 | 4.7 | 229 | A | A62V, V75I, F77L, F116Y, Q151M | I84V, L90M | 2003 | LPV EFV APV 3TC | 2009 | |
| 19 | 2001 | 5.0 | 253 | C | A62V, K65R, D67N, V75I, F77L, Y115F, F116Y, Q151M, M184V, K219E | D30N, M46I, I54M | 2002 | SQV NVP LPV | 2009 | |
| 20 | 2002 | 4.8 | 77 | B | K65R, K70R, V75I, F77L, Y115F, F116Y, Q151M, K219E | K101E, Y181C, G190A | M46I, V82A | 2004 | TDF RTV ETV ABC | 2009 |
| 21 | 2003 | 3.9 | 191 | C | D67N, K70R, F116Y, Q151M, M184V, K219Q | K103N | M46I, Q58E, V82F, L90M | 2006b | ||
| 22 | 2003 | 3.0 | 5 | B | A62V, D67N, K70R, V75I, F77L, F116Y, Q151M, M184V, K219E | K103N, V108I | I50V, V82A, L90M | 2004 | TPV TDF T20 LPV DDI ZDV APV 3TC | 2009 |
| 23 | 2004 | 4.5 | 84 | B | D67N, K70R, Y115F, F116Y, Q151M, M184V, K219Q | Y181C, Y188L | M46I, V82A | 2006 | TPV TDF RTV RAL ZDV 3TC | 2009 |
| 24 | 2004 | 3.9 | 461 | B | F116Y, Q151M, | M46I, L90M | 2007 | RTV DRV | 2008b | |
| 25 | 2006 | 4.8 | 773 | A | F116Y, Q151M | K103N, Y188L | 2006 | TDF RTV FTC ATV | 2009 |
NOTE. Viral suppression was defined as 2 consecutive viral loads <50 copies/mL. 3TC, lamivudine; ABC, abacavir; APV, amprenavir; CDC, Centers for Disease Control and Prevention; D4T, stavudine; ddI, didanosine; DRV, darunavir; EFV, efavirenz; ETV, etravirine; HIV-1, human immunodeficiency virus type 1; IDV, indinavir; LPV, lopinavir; MVC, maraviroc; NFV, nelfinavir; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NVP, nevirapine; PI, protease inhibitor; RAL, raltegravir; RTV, ritanovir; SQV, saquinavir; T20, enfuvirtide; TDF, tenofovir; TPV, tipranavir; ZDV, zidovudine.
Mutations printed in bold on the International AIDS Society-USA list [12].
Year of death.
Viral suppression rates between the Q151M group (14 [56.0%] of 25; 95% CI, 34.9%–75.6%) and the respective control subjects (27 [73.0%] of 37; 95% CI, 55.9%–86.2%) were similar. Generally, patients who achieved viral suppression had a higher median CD4+ cell count at detection (241 cells/μL vs 61 cells/μL; P = .030), a lower viral load (median log10 RNA level, 4.5 copies/mL vs 5.1 copies/mL; P = .089), a higher percentage of a previously unseen drug class (88.9% vs 37.5%; P = .033), and were detected later (median year, 1997 vs 2001; P = .027). As illustrated in Table 1, most patients with Q151M detected before the introduction of HAART never achieved viral suppression (patients 1–3 and 6–8). Four of 11 patients detected in the HAART era prior to the approval of enfuvirtide (T20) (1998–2002) never had a successful treatment (patients 10, 11, 13, and 15). All of these patients had low CD4+ cell counts (<50 cells/μL) and extensive PI and/or NNRTI mutations. Most of the other patients had PI mutations but achieved viral suppression with a regimen containing NNRTIs (patients 12, 14, and 16–20).
Survival after Detection of MNR
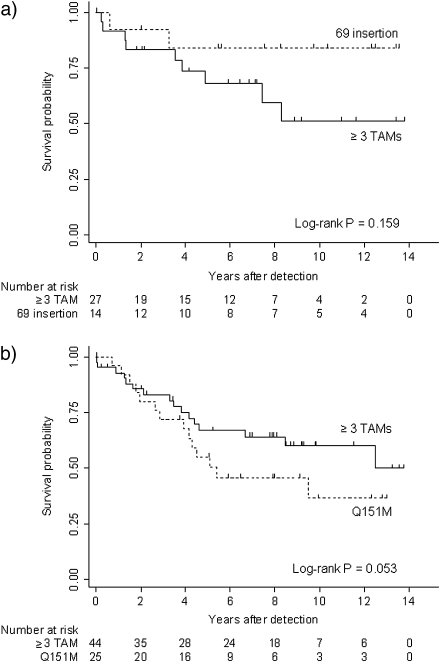
The crude incidence of mortality after detection of the 69 insertion was 1.9 deaths (95% CI, 0.2–6.9) per 100 person-years of follow-up, compared with 6.3 deaths (95% CI, 2.9–12.0) per 100 person-years of follow-up among control subjects (≥3 TAMs) [Figure 1]. Of the 2 deaths noted in the 69 insertion group, 1 death was HIV-related, and the cause of the other death was unknown. The risk of mortality was not significantly different between patients with the 69 insertion and those with ≥3 TAMs (univariable hazard ratio [HR], 0.3 [95% CI, 0.1–1.6]; P = .178). The small number of events did not allow stratified or multivariable models.
Figure 1.
Kaplan-Meier curves showing survival after detection of 69 insertion (a) or Q151M (b). Patients detected with ≥3 thymidine analogue mutations (TAMs) were matched (2:1) for comparison. Log-rank test was stratified for matched pairs.
Patients with Q151M detected tended to have a higher crude incidence of 9.8 deaths per 100 person-years (95% CI, 5.3–16.4), compared with 5.8 death per 100 person-years (95% CI, 3.3–9.4) in control subjects (with ≥3 TAMs) [Figure 1]. HIV infection was the cause of death for 6 (42.9%) of 14 patients from the Q151M group and 7 (43.8%) of 16 control subjects. Additional causes of death reported in the Q151M group were neoplasm (14.3%), cardio-vascular diseases (14.3%), chronic hepatitis C (7.1%), suicide (7.1%), or unknown (14.3%). The detection of Q151M was associated with increased mortality but was of marginal statistical significance in the univariable (HR, 2.7 [95% CI, 0.9–8.0]; P = .075) and multivariable model, adjusted for sex, ethnicity, risk group, CD4+ cell count, and age (HR, 7.5 [95% CI, 0.9–64.6]; P = .068).
Sensitivity analyses including only those deaths associated with HIV infection (univariable HR, 2.6 [95% CI, 0.6–10.4]; P = .189) or by additionally matching case patients and control subjects by CD4+ cell count at time of detection of Q151M or ≥3 TAMs showed similar results (univariable HR, 3.2 [95% CI, 1.0–10.9]; P = .058; and multivariable HR, 6.2 [95% CI, 0.7–56.0]; P = .105).
DISCUSSION
Because the prevalence of MNR is increasing in resource-limited countries, and because 69 insertion and Q151M affect the activity of an entire drug class, it is of great importance to identify risk factors and to optimize treatment strategies.
Our study currently represents the largest longitudinal dataset with full treatment history available. We found high evidence of forward transmission of 69 insertions. This finding is of relevance, because the presence of 69 insertion results in a substantial reduction of treatment options, which can be devastating in settings with limited access to potent salvage therapies.
Moreover, our study identified a significant association of ddI exposure with the emergence of the 69 insertion [13]. No specific NRTI was associated with the emergence of Q151M. Our study did not confirm a previously reported negative association of lamivudine with Q151M [2, 14], nor was d4T exposure correlated with an increased risk for Q151M (data not shown).
Of note, MNR mutations were never detected in patients who were exclusively treated with HAART. This is in contrast to the high prevalence of MNR mutations observed in resource-limited settings [7, 8]. Free access to potent antiretroviral drugs in Switzerland and close monitoring are the most likely explanations for this difference.
Moreover, this study widened the very limited knowledge for treatment strategies of patients detected with MNR. These patients can be successfully treated if potent drugs, such as boosted PI, raltegravir, or T20 are available. The descriptive analysis of therapy success further suggests that extensive resistance to NNRTIs and PIs, as well as a low CD4+ cell count at the time of detection of MNR mutations, were prognostically unfavorable [15].
Patients with viruses possessing Q151M tended to have an increased mortality risk, compared with patients with ≥3 TAMs. Although this finding was robust throughout several sensitivity analyses, conclusions regarding causality between Q151M and death should be drawn with care.
This study has some limitations. Even though our sample is the largest study to date to address MNR, it is still limited in power [11]. Our study was restricted to subtype B–infected individuals, and results may therefore not be readily transferable to other subtypes.
Taken together, our data indicate that modern antiretroviral therapies in combination with adequate viral monitoring are able to prevent the emergence of MNR mutations in developed settings. In Switzerland, detected MNR mutations are mainly a relic of the mono- or dual-NRTI therapy era, although 2 cases of transmitted MNR were observed. This analysis further demonstrates that salvage treatment can be successful even when MNR mutations are present if at least 1 previously unused drug class is available. Today, the development of MNR seems to be becoming an emerging problem in resource-limited settings, where most patients are infected with non–subtype B strains. Thus, additional studies are needed to investigate whether our findings are also true for non–subtype B infections.
MEMBERS OF THE SHCS
M. Battegay, E. Bernasconi, J. Böni, H. C. Bucher, P. Bürgisser, A. Calmy, S. Cattacin, M. Cavassini, R. Dubs, M. Egger, L. Elzi, M. Fischer, M. Flepp, A. Fontana, P. Francioli (President of the SHCS), H. Furrer (Chairman of the Clinical and Laboratory Committee), C. A. Fux, M. Gorgievski, H. Günthard (Chairman of the Scientific Board), H. H. Hirsch, B. Hirschel, I. Hösli, C. Kahlert, L. Kaiser, U. Karrer, C. Kind, T. Klimkait, B. Ledergerber, G. Martinetti, N. Müller, D. Nadal, F. Paccaud, G. Pantaleo, A. Rauch, S. Regenass, M. Rickenbach (Head of Data Center), C. Rudin (Chairman of the Mother & Child Substudy), P. Schmid, D. Schultze, J. Schüpbach, R. Speck, B. M. de Tejada, P. Taffé, A. Telenti, A. Trkola, P. Vernazza, R. Weber, S. Yerly.
Funding
Swiss National Science Foundation (SNF grant 3345-062041 to the Swiss HIV Cohort Study and SNF grant 3247B0-112594 to H.F.G., S.Y., and B.L.), Swiss HIV Cohort Study (SHCS) projects #470, 528, and 569, the SHCS Research Foundation, the European Community's Seventh Framework Programme (grant FP7/2007–2013), under the Collaborative HIV and Anti-HIV Drug Resistance Network (grant 223131), the Union Bank of Switzerland (to H.F.G.), Tibotec, and the Novartis Foundation, formerly Ciba-Geigy Jubilee Foundation (to V.V.W.).
Acknowledgments
We thank the patients who participate in the Swis HIV Cohort Study; the physicians and study nurses, for excellent patient care; the resistance laboratories, for high-quality genotypic drug resistance testing; SmartGene (Zug, Switzerland), for technical support; Brigitte Remy, Martin Rickenbach, F. Schöni-Affolter, and Yannick Vallet from the SHCS Data Center in Lausanne, Switzerland, for the data management; and Marie-Christine Francioli, for administrative assistance.
References
- 1.Lodwick R, Costagliola D, Reiss P. Triple-class virologic failure in HIV-infected patients undergoing antiretroviral therapy for up to 10 years. Arch Intern Med. 170:410–419. doi: 10.1001/archinternmed.2009.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Vaerenbergh K, Van Laethem K, Albert J, et al. Prevalence and characteristics of multinucleoside-resistant human immunodeficiency virus type 1 among European patients receiving combinations of nucleoside analogues. Antimicrob Agents Chemother. 2000;44:2109–17. doi: 10.1128/aac.44.8.2109-2117.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shafer RW, Schapiro JM. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 2008;10:67–84. [PMC free article] [PubMed] [Google Scholar]
- 4.Miller MD, Margot N, Lu B, et al. Genotypic and phenotypic predictors of the magnitude of response to tenofovir disoproxil fumarate treatment in antiretroviral-experienced patients. J Infect Dis. 2004;189:837–46. doi: 10.1086/381784. [DOI] [PubMed] [Google Scholar]
- 5.Re MC, Borderi M, Monari P, et al. Prevalence of multiple dideoxynucleoside analogue resistance (MddNR) in a cohort of Italian HIV-1 seropositive patients extensively treated with antiretroviral drugs. Int J Antimicrob Agents. 2001;18:519–23. doi: 10.1016/s0924-8579(01)00440-x. [DOI] [PubMed] [Google Scholar]
- 6.Schmit JC, Van Laethem K, Ruiz L, et al. Multiple dideoxynucleoside analogue-resistant (MddNR) HIV-1 strains isolated from patients from different European countries. AIDS. 1998;12:2007–15. doi: 10.1097/00002030-199815000-00012. [DOI] [PubMed] [Google Scholar]
- 7.Sungkanuparph S, Manosuthi W, Kiertiburanakul S, Piyavong B, Chumpathat N, Chantratita W. Options for a second-line antiretroviral regimen for HIV type 1-infected patients whose initial regimen of a fixed-dose combination of stavudine, lamivudine, and nevirapine fails. Clin Infect Dis. 2007;44:447–52. doi: 10.1086/510745. [DOI] [PubMed] [Google Scholar]
- 8.Hosseinipour MC, van Oosterhout JJ, Weigel R, et al. The public health approach to identify antiretroviral therapy failure: high-level nucleoside reverse transcriptase inhibitor resistance among Malawians failing first-line antiretroviral therapy. AIDS. 2009;23:1127–34. doi: 10.1097/QAD.0b013e32832ac34e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallego O, d Mendoza C, Labarga P, et al. Long-term outcome of HIV-infected patients with multinucleoside-resistant genotypes. HIV Clin Trials. 2003;4:372–81. doi: 10.1310/X618-KWKJ-WCTQ-LQ2L. [DOI] [PubMed] [Google Scholar]
- 10.von Wyl V, Yerly S, Boni J, et al. Emergence of HIV-1 drug resistance in previously untreated patients initiating combination antiretroviral treatment: a comparison of different regimen types. Arch Intern Med. 2007;167:1782–90. doi: 10.1001/archinte.167.16.1782. [DOI] [PubMed] [Google Scholar]
- 11.Schoeni-Affolter F, Ledergerber B, Rickenbach M, et al. Cohort profile: the Swiss HIV Cohort Study. Int J Epidemiol. 2010;39(5):1179–89. doi: 10.1093/ije/dyp321. [DOI] [PubMed] [Google Scholar]
- 12.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1: December 2009. Top HIV Med. 2009;17:138–45. [PubMed] [Google Scholar]
- 13.Winters MA, Coolley KL, Girard YA, et al. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Invest. 1998;102:1769–75. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaccarelli M, Perno CF, Forbici F, et al. Q151M-mediated multinucleoside resistance: prevalence, risk factors, and response to salvage therapy. Clin Infect Dis. 2004;38:433–437. doi: 10.1086/381097. [DOI] [PubMed] [Google Scholar]
- 15.Gupta RK, Hill A, Sawyer AW, et al. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. Lancet Infect Dis. 2009;9:409–17. doi: 10.1016/S1473-3099(09)70136-7. [DOI] [PubMed] [Google Scholar]