Abstract
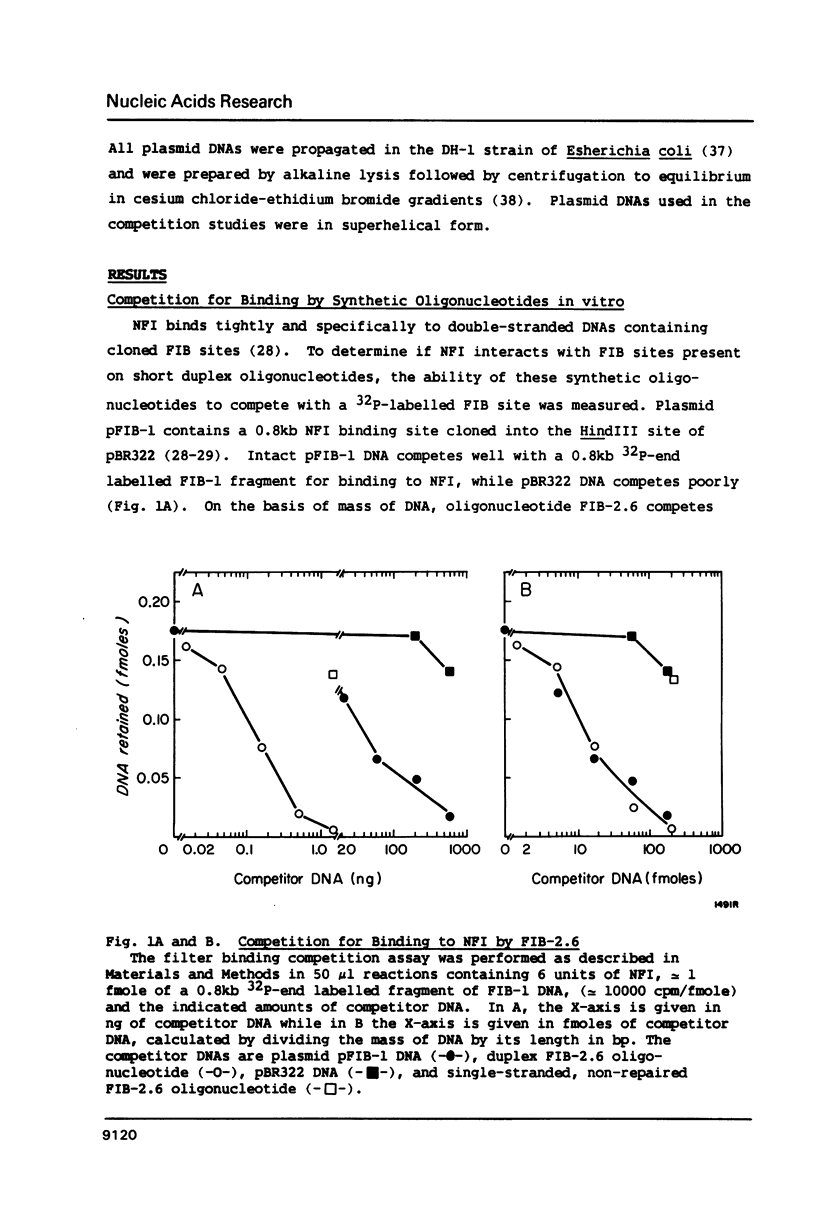
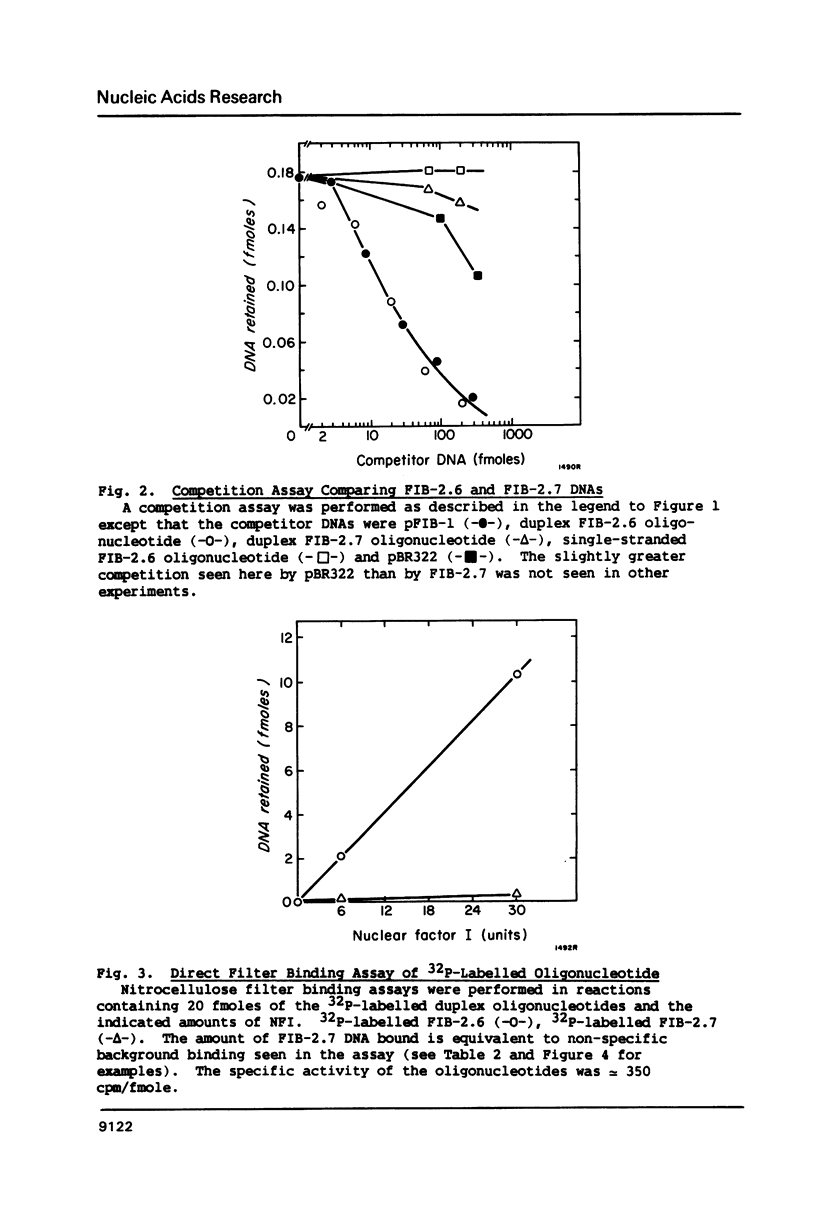
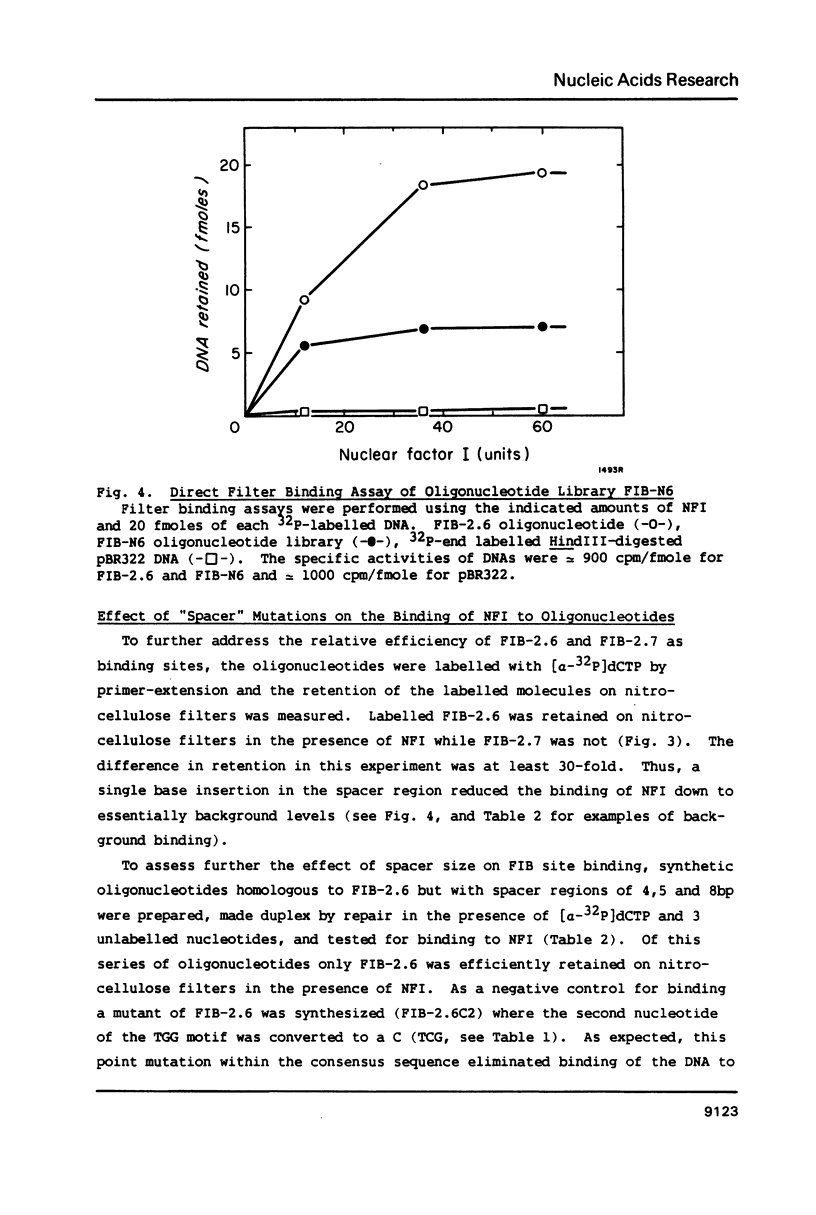
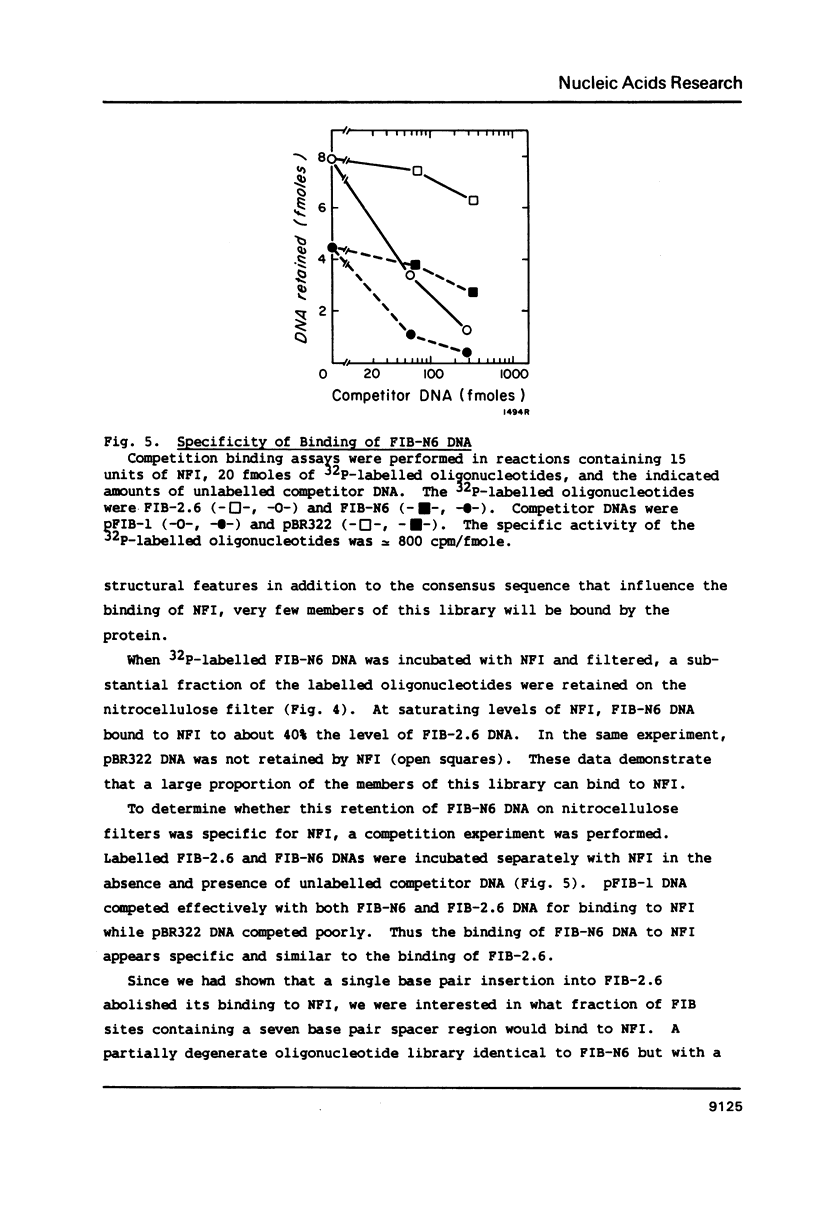
Nuclear factor I (NFI) binds tightly to DNA containing the consensus sequence TGG(N)6-7GCCAA. To study the role of the spacing between the TGG and GCCAA motifs, oligonucleotides homologous to the NFI binding site FIB-2 were synthesized and used for binding assays in vitro. The wild-type site (FIB-2.6) has a 6bp spacer region and binds tightly to NFI. When the size of this spacer was altered by +/- 1 or 2bp the binding to NFI was abolished. To further assess the role of the spacer and bases flanking the motifs, two oligonucleotide libraries were synthesized. Each member of these libraries had intact TGG and GCCAA motifs, but the sequence of the spacer and the 3bp next to each motif was degenerate. The library with a 6bp spacer bound to NFI to 40-50% the level of FIB-2.6. The library with a 7bp spacer bound to NFI to only 4% the level of FIB-2.6 and some of this binding was weaker than that of FIB-2.6 DNA. This novel use of degenerate DNA libraries has shown that: 1) the structural requirements for FIB sites with a 7bp spacer are more stringent than for sites with a 6bp spacer and 2) a limited number of DNA structural features can prevent the binding of NFI to sites with intact motifs and a 6bp spacer region.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borgmeyer U., Nowock J., Sippel A. E. The TGGCA-binding protein: a eukaryotic nuclear protein recognizing a symmetrical sequence on double-stranded linear DNA. Nucleic Acids Res. 1984 May 25;12(10):4295–4311. doi: 10.1093/nar/12.10.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Challberg M. D., Desiderio S. V., Kelly T. J., Jr Adenovirus DNA replication in vitro: characterization of a protein covalently linked to nascent DNA strands. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5105–5109. doi: 10.1073/pnas.77.9.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J. Eukaryotic DNA replication: viral and plasmid model systems. Annu Rev Biochem. 1982;51:901–934. doi: 10.1146/annurev.bi.51.070182.004345. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Friefeld B. R., Lichy J. H., Field J., Gronostajski R. M., Guggenheimer R. A., Krevolin M. D., Nagata K., Hurwitz J., Horwitz M. S. The in vitro replication of adenovirus DNA. Curr Top Microbiol Immunol. 1984;110:221–255. doi: 10.1007/978-3-642-46494-2_8. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984 Oct;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Graves B. J., Johnson P. F., McKnight S. L. Homologous recognition of a promoter domain common to the MSV LTR and the HSV tk gene. Cell. 1986 Feb 28;44(4):565–576. doi: 10.1016/0092-8674(86)90266-7. [DOI] [PubMed] [Google Scholar]
- Greenbaum J. H., Marians K. J. The interaction of Escherichia coli replication factor Y with complementary strand origins of DNA replication. Contact points revealed by DNase footprinting and protection from methylation. J Biol Chem. 1984 Feb 25;259(4):2594–2601. [PubMed] [Google Scholar]
- Gronostajski R. M., Adhya S., Nagata K., Guggenheimer R. A., Hurwitz J. Site-specific DNA binding of nuclear factor I: analyses of cellular binding sites. Mol Cell Biol. 1985 May;5(5):964–971. doi: 10.1128/mcb.5.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski R. M., Nagata K., Hurwitz J. Isolation of human DNA sequences that bind to nuclear factor I, a host protein involved in adenovirus DNA replication. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4013–4017. doi: 10.1073/pnas.81.13.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski R. M., Sadowski P. D. Determination of DNA sequences essential for FLP-mediated recombination by a novel method. J Biol Chem. 1985 Oct 5;260(22):12320–12327. [PubMed] [Google Scholar]
- Guggenheimer R. A., Stillman B. W., Nagata K., Tamanoi F., Hurwitz J. DNA sequences required for the in vitro replication of adenovirus DNA. Proc Natl Acad Sci U S A. 1984 May;81(10):3069–3073. doi: 10.1073/pnas.81.10.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hay R. T. The origin of adenovirus DNA replication: minimal DNA sequence requirement in vivo. EMBO J. 1985 Feb;4(2):421–426. doi: 10.1002/j.1460-2075.1985.tb03645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L., Fleckenstein B. Nuclear factor 1 interacts with five DNA elements in the promoter region of the human cytomegalovirus major immediate early gene. EMBO J. 1986 Jun;5(6):1367–1371. doi: 10.1002/j.1460-2075.1986.tb04368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L., Siebenlist U., Danner D., Leder P., Rawlins D., Rosenfeld P., Kelly T., Jr High-affinity binding site for a specific nuclear protein in the human IgM gene. Nature. 1985 Mar 21;314(6008):289–292. doi: 10.1038/314289a0. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol. 1985 Feb 5;181(3):351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Leegwater P. A., van Driel W., van der Vliet P. C. Recognition site of nuclear factor I, a sequence-specific DNA-binding protein from HeLa cells that stimulates adenovirus DNA replication. EMBO J. 1985 Jun;4(6):1515–1521. doi: 10.1002/j.1460-2075.1985.tb03811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger L. F. D1 protein of Drosophila melanogaster. Purification and AT-DNA binding properties. J Biol Chem. 1985 Nov 15;260(26):14311–14318. [PubMed] [Google Scholar]
- Lichy J. H., Field J., Horwitz M. S., Hurwitz J. Separation of the adenovirus terminal protein precursor from its associated DNA polymerase: role of both proteins in the initiation of adenovirus DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5225–5229. doi: 10.1073/pnas.79.17.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichy J. H., Horwitz M. S., Hurwitz J. Formation of a covalent complex between the 80,000-dalton adenovirus terminal protein and 5'-dCMP in vitro. Proc Natl Acad Sci U S A. 1981 May;78(5):2678–2682. doi: 10.1073/pnas.78.5.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miyamoto N. G., Moncollin V., Egly J. M., Chambon P. Specific interaction between a transcription factor and the upstream element of the adenovirus-2 major late promoter. EMBO J. 1985 Dec 16;4(13A):3563–3570. doi: 10.1002/j.1460-2075.1985.tb04118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Enomoto T., Lichy J. H., Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Hurwitz J. Adenovirus DNA replication in vitro: synthesis of full-length DNA with purified proteins. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4266–4270. doi: 10.1073/pnas.80.14.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Hurwitz J. Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6177–6181. doi: 10.1073/pnas.80.20.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowock J., Borgmeyer U., Püschel A. W., Rupp R. A., Sippel A. E. The TGGCA protein binds to the MMTV-LTR, the adenovirus origin of replication, and the BK virus enhancer. Nucleic Acids Res. 1985 Mar 25;13(6):2045–2061. doi: 10.1093/nar/13.6.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R., Shapiro B., Lipkin L. E., Maizel J. V., Jr DNAase I hypersensitive sites may be correlated with genomic regions of large structural variation. J Mol Biol. 1984 Aug 25;177(4):591–607. doi: 10.1016/0022-2836(84)90039-1. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins D. R., Rosenfeld P. J., Wides R. J., Challberg M. D., Kelly T. J., Jr Structure and function of the adenovirus origin of replication. Cell. 1984 May;37(1):309–319. doi: 10.1016/0092-8674(84)90327-1. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Ross W., Landy A., Kikuchi Y., Nash H. Interaction of int protein with specific sites on lambda att DNA. Cell. 1979 Oct;18(2):297–307. doi: 10.1016/0092-8674(79)90049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler J. R., Sasmor H., Betz J. L. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., Gander I., Müller U., Mertz R., Winnacker E. L. A sensitive and rapid gel retention assay for nuclear factor I and other DNA-binding proteins in crude nuclear extracts. Nucleic Acids Res. 1986 Feb 11;14(3):1303–1317. doi: 10.1093/nar/14.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Stillman B. W., Tamanoi F., Mathews M. B. Purification of an adenovirus-coded DNA polymerase that is required for initiation of DNA replication. Cell. 1982 Dec;31(3 Pt 2):613–623. doi: 10.1016/0092-8674(82)90317-8. [DOI] [PubMed] [Google Scholar]
- Stillman B. W. The replication of adenovirus DNA with purified proteins. Cell. 1983 Nov;35(1):7–9. doi: 10.1016/0092-8674(83)90201-5. [DOI] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Vliet P. C., Zandberg J., Jansz H. S. Evidence for a function of the adenovirus DNA-binding protein in initiation in DNA synthesis as well as in elongation of nascent DNA chains. Virology. 1977 Jul 1;80(1):98–110. doi: 10.1016/0042-6822(77)90383-x. [DOI] [PubMed] [Google Scholar]
- Vocke C., Bastia D. DNA-protein interaction at the origin of DNA replication of the plasmid pSC101. Cell. 1983 Dec;35(2 Pt 1):495–502. doi: 10.1016/0092-8674(83)90183-6. [DOI] [PubMed] [Google Scholar]
- Wang K., Pearson G. D. Adenovirus sequences required for replication in vivo. Nucleic Acids Res. 1985 Jul 25;13(14):5173–5187. doi: 10.1093/nar/13.14.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobbe C. R., Dean F., Weissbach L., Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985 Oct 3;317(6036):451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]
- de Vries E., van Driel W., Tromp M., van Boom J., van der Vliet P. C. Adenovirus DNA replication in vitro: site-directed mutagenesis of the nuclear factor I binding site of the Ad2 origin. Nucleic Acids Res. 1985 Jul 11;13(13):4935–4952. doi: 10.1093/nar/13.13.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]



