Abstract
Stachybotrys is a hydrophilic fungal genus that is well known for its ability to colonize water-damaged building materials in indoor environments. Personal exposure to Stachybotrys chartarum allergens, mycotoxins, cytolytic peptides, and other immunostimulatory macromolecules has been proposed to exacerbate respiratory morbidity. To date, advances in Stachybotrys detection have focused on the identification of unique biomarkers that can be detected in human serum; however, the availability of immunodiagnostic reagents to Stachybotrys species have been limited. In this study, we report the initial characterization of monoclonal antibodies (MAbs) against a semi-purified cytolytic S. chlorohalonata preparation (cScp) derived from hyphae. BALB/c mice were immunized with cScp and hybridomas were screened against the cScp using an antigen-mediated indirect ELISA. Eight immunoglobulin M MAbs were produced and four were specifically identified in the capture ELISA to react with the cScp. Cross-reactivity of the MAbs was tested against crude hyphal extracts derived from 15 Stachybotrys isolates representing nine Stachybotrys species as well as 39 other environmentally abundant fungi using a capture ELISA. MAb reactivity to spore and hyphal antigens was also tested by a capture ELISA and by fluorescent halogen immunoassay (fHIA). ELISA analysis demonstrated that all MAbs strongly reacted with extracts of S. chartarum but not with extracts of 39 other fungi. However, four MAbs showed cross-reactivity to the phylogenetically related genus Memnoniella. fHIA analysis confirmed that greatest MAb reactivity was ultrastructurally localized in hyphae and phialides. The results of this study further demonstrate the feasibility of specific MAb-based immunoassays for the detection of S. chartarum.
Introduction
Awidely recognized fungal genus, Stachybotrys is capable of contaminating water-infiltrated cellulose-based building materials. The genus is characterized by septate hyphae and conidiophores that bear clusters of phialides where chains of dematiaceaous conidia emerge. Identification of Stachybotrys conidia in tape lift or air samples in indoor environments is considered a biomarker of indoor fungal contamination by various federal, state, and academic institutions. Stachybotrys conidia and hyphae contain mycotoxins, allergens, proteases, and other immunostimulatory molecules.(1,2) Personal exposure to S. chartarum is also considered an etiological agent for respiratory disease. Mycological expertise is required to confirm the presence of Stachybotrys conidia in indoor environments; however, morphologically indiscernible hyphae and fragments that are equally important biomarkers of contamination, remain overlooked and are not quantified.(3) Therefore, the development of standardized methods for the detection of Stachybotrys is required for more precise quantification of this species in indoor environments.
Cytolytic proteins such as the fungal hemolysin, stachylysin, have been reported in the inner wall of spores and hyphae of S. chartarum.(4) Stachylysin has been proposed to form pores from the outside of membranes and facilitate the lysis of cells.(5) This cytolytic activity is suggested to enhance the uptake of nutrients for growth,(6) protect against insect predation,(7) or help to evade macrophages and polymorphonuclear cells in the respiratory mucosa. To date the production of stachylysin has been shown to be highest in strains of S. chartarum associated with exacerbations of respiratory disease.(8) Animal exposure studies have also shown stachylysin to diffuse from spores into surrounding lung tissue following inhalation.(9) As a result of these experimental observations, stachylysin has been proposed as a potential biomarker of personal exposure to S. chartarum.
Polyclonal antibodies (pAb) toward stachylysin have been previously utilized in an inhibition ELISA to quantify the levels of stachylysin in environmental samples as well as from the sera of Stachybotrys exposed rats and humans.(9) Since, pAbs often lack specificity and are cross reactive, we aimed to develop monoclonal antibodies (MAb) that were specific for Stachybotrys species.(10–13) Compared to other detection methodologies, MAbs are highly specific and can be used in the development of standardized immunoassays. In our laboratory, we have previously developed a species-specific MAb against S. chartarum phialides and conidia but not hyphae.(11,13,14) Given the presence of morphologically indiscernible Stachybotrys hyphae and fragments in indoor air samples and the potential health effects associated with personal exposure, the development of MAbs that recognize this overlooked fraction is an important step that will improve the quantification of these particulates. Recent studies have also identified a new Stachybotrys species, S. chlorohalonata, that has the same phenotypic features as S. chartarum chemotype A, but morphologically is characterized by green extracellular pigmentation.(15).The strain of S. chlorohalonata (ATCC 201863; IBT 9825) that was used in this study to produce the cytolytic S. chlorohalonata preparation (cScp) was originally designated S. chartarum and isolated from the home of an infant diagnosed with idiopathic pulmonary hemorrhage (IPH).(8) In this manuscript, we describe the production of MAbs that recognize antigens derived from the cScp.
Materials and Methods
Semi-purified cytolytic Stachybotrys preparation
Stachybotrys chlorohalonata (ATCC 201863) cytolytic antigens were semi-purified from tryptic soy broth (TSB, Becton Dickinson, Sparks, MD) culture supernatants as previously described.(16) Briefly, S. chlorohalonata conidia (1 × 105) were used to inoculate 500 mL of TSB in a 1 L flask placed on an incubator shaker for 7 days.(16) Cellular debris was removed from the TSB culture supernatant by centrifugation for 15 min at 5000 g. The supernatant was then centrifuged in a Centricon plus 80 filter apparatus with a molecular mass cut-off of 50 kDa (Millipore, Bedford, MA) at 4000 g for 15 min. The concentrate was then subjected to gel filtration as previously described.(16) Fractions were collected and plated onto sheep blood agar to determine hemolytic activity. The five most hemolytic fractions were pooled, desalted, and lyophilized as previously described.(16) The lyophilized pellet was resuspended in sterile water for further analysis or to use in other experiments.
Preparation of fungal hyphal extracts
Fungi were grown in standard unsealed Petri plates containing 5 mL of malt extract agar (MEA; 2% dextrose, 0.1% peptone, 2% malt extract, 2% agar; Difco, Becton Dickinson). After 2 weeks of incubation at room temperature (RT), conidia were collected from cultures into TSB and 10 mL of the spore suspension (1 × 106/mL) were transferred into a 125 mL Corning flask containing 50 mL of TSB. The flasks were rotated at 120 rpm at 37°C for 3–4 days before the hyphae were harvested by filtration using a cell strainer (70 μm, Becton Dickinson). The collected hyphae were washed two times in 50 mL phosphate-buffered saline (pH 7.4) containing 0.05% (v/v) Tween-20 (PBST) before being homogenized in the cell strainer using the plunger from a 10 mL syringe. The homogenate was centrifuged at 4100 g for 5 min at 4°C and aliquots of the supernatants were stored at −80°C. The total protein concentration in the hyphal extracts was determined using a BCA protein assay kit (Pierce, Rockford, IL).
Production of monoclonal and polyclonal antibodies against cScp
Five 10–14 week old BALB/c female mice were housed under controlled environmental conditions in HEPA-filtered ventilated polycarbonate cages on autoclaved hardwood Beta-chip bedding and were provided feed (Teklad 7913 rodent chow, Madison, WI) and autoclaved tap water ad libitum. Sentinel mice were free of viral pathogens, parasites, mycoplasma, and Helicobacter spp. The animal facility is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
Mice were immunized intraperitoneally at bi-weekly intervals. Mice were primed with 50 μg of the cScp emulsified in equal volumes of TiterMax (TiterMax USA, Norcross, GA). The antigen concentration was reduced by half for each of the five subsequent booster immunizations. A final boost of 50 μg was given 3 weeks after the sixth immunization and mice were sacrificed 3 days later for hybridoma production.
Hybridomas were produced by standard polyethylene glycol-based cell fusion techniques using SP2/0-AG14 myleoma cells (ATCC# CRL-1581). Cell cultures were maintained in Dulbecco's Modified Eagle Medium (Life Technologies, Rockville, MD), supplemented with 1 mM sodium pyruvate, 100 U/mL penicillin, 100 μg/mL streptomycin, 0.292 mg/mL L-glutamine, 100 mM sodium hypoxanthine, 16 mM thymidine, 10% fetal calf serum (HyClone, Logan, UT), and 100 U/mL IL-6 (Boehringer, Mannheim, Germany). Positive clones were identified using 1 μg/mL of the cScp in an indirect ELISA (see below). Positive colonies were cloned twice by limiting dilution and the stable hybridomas were grown in bulk, aliquoted, and stored in liquid nitrogen.
Rabbit pAbs against the cScp were custom-produced and affinity-purified by Bethyl Laboratories (Montgomery, TX) using standard laboratory protocols as previously described.
Screening ELISA format for the analysis of hybridoma culture supernatants
Hybridoma tissue culture supernatants (CSN) were screened using an indirect ELISA as previously described.(11) In brief, ELISA plate wells were coated with 100 μL of the cScp (1 μg/mL) in carbonate coating buffer (CCB, 60 mM sodium carbonate, 140 mM sodium bicarbonate [pH 9.6]) and incubated at room temperature (RT) overnight. Wells were washed three times by incubating 200 μL/well of phosphate buffered-saline containing 0.5% Tween-20 (PBST) for 10 min. The plates were then blocked for 1 h at RT with 200 μL/well of PBST containing 1% non-fat dry milk powder (PBSTM). Hybridoma culture supernatants were incubated for 1 h at 37°C with 100 μL of MAb culture supernatant diluted 1:5 (v/v) in PBSTM. Bound antibodies were labeled with 100 μL of biotin-SP-conjugated AffiPure goat anti-mouse IgG and IgM secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 h at 37°C at a dilution of 1:5000 (v/v) in PBSTM. Bound biotin was detected with 100 μL of alkaline phosphatase-conjugated streptavidin (Jackson ImmunoResearch Laboratories) by incubation for 1 h at 37°C at a dilution of 1:5000 (v/v) in PBSTM. The reaction product was produced by incubating 100 μL per well of p-nitrophenyl phosphate (1 mg/mL) in alkaline phosphatase substrate buffer (1 M diethanolamine [pH 9.5], 5 mM MgCl2) at RT and the optical density (OD) was determined at 405 nm after 30 min using an UltraMicroplate Reader (Model ELx800, Bio-Tek Instruments, Winooski, VT). Negative control values were obtained by substituting plain culture supernatant for MAb culture.
Capture ELISA format for the analysis of hyphal extracts
The specificity of MAbs was tested against hyphal extracts of 7 S. chartarum isolates, 8 isolates of other Stachybotrys species, as well as 39 related and non-related fungi commonly found in indoor environments. All extracts were tested in a capture ELISA using rabbit pAbs as solid-phase capture reagent. In brief, ELISA plate wells were coated with 100 μL of pAb (1 μg/mL) in CCB overnight at RT. The plates were washed and blocked as described above, and 100 μL/well of each fungal extract (50 μg/mL) in PBST were added and allowed to react for 1 h at 37°C. After washing the plates, MAb CSNs diluted 4-fold in PBST were incubated for 1 h at 37°C and the plates were processed as described for the screening ELISA. The ODs of plain culture supernatant negative controls ranged from 0 to 0.06. An OD ≥0.2 (negative control +3 standard deviations) was considered to be a positive result. The sensitivity of the MAbs was measured with the same capture ELISA format except that the pAb was used at 2 μg/mL and the MAb CSN were diluted 5-fold.
Western blot analysis of MAb reactivity
The cScp was separated by SDS-PAGE on a 10% acrylamide gel and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA). The membranes were blocked with 3% bovine serum albumin for 1 h at RT and incubated with MAbs (diluted 1:5 in PBST) for 1 h at RT. After incubation for 1 h with a 1:5000 (v/v) dilution of biotin-conjugated goat anti-mouse IgM antibodies (Jackson Immuno Research Laboratories), the membranes were incubated with alkaline phosphatase-conjugated streptavidin (1:5000, v/v) for 1 h at RT. Blots were developed with the nitroblue tetrazolium and bromo-chloro-indolyl phosphate system (NBT/BCIP, Promega, Madison, WI).
Fluorescent halogen immunoassay
S. chartarum conidia, phialides, and hyphae were aerosolized by directing a jet of air across 2-week-old sporulating cultures. Aerosolized particles were collected by suction onto mixed cellulose ester (MCE) protein-binding membranes (0.45 μm pore size; Millipore) and immunostained using the fluorescent halogen immunoassay (fHIA) as previously described.(13) Briefly, a clear adhesive and glass coverslip was used to laminate samples and the antigens were extracted in 0.2 M borate buffer (pH 8.2) at RT for 3 h. The samples were blocked with 5% BSA in PBS for 90 min and then incubated overnight at 4°C with the MAb 6D4 diluted 1:50 (v/v) in PBST containing 5% BSA (PBSTB). Negative control treatments were processed in parallel by substituting the MAbs with hybridoma tissue culture medium diluted 1:50 (v/v) or control MAb 1B9 (IgM) diluted 1:50. The membranes were rinsed three times in PBST and incubated for 1.5 h with Alexa Fluor 488-conjugated goat anti-mouse IgM (Molecular Probes, Eugene, OR) diluted 1:500 (v/v) in PBSTB. The membranes were rinsed three times in distilled H2O and mounted on a microscope slide in ProLong Gold (Molecular Probes) anti-fade reagent. Confocal laser scanning images were captured using a Zeiss LSM 510 laser scanning confocal system (Carl Zeiss, Thornwood, NY). The images of MAb-labeled fungal particles were captured using 488 nm excitation and a narrow emission filter bandwidth (505–550 nm). Fluorescent and differential interference contrast images (DIC) were captured using Zeiss software, v. 3.2. All settings on the confocal laser microscope remained constant in the analysis.
Results
MAb reactivity to S. chlorohalonata
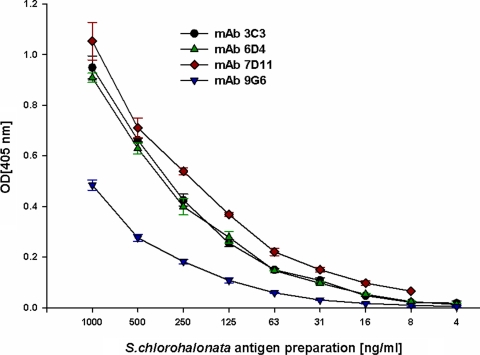
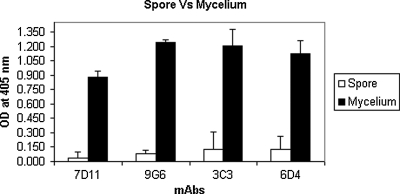
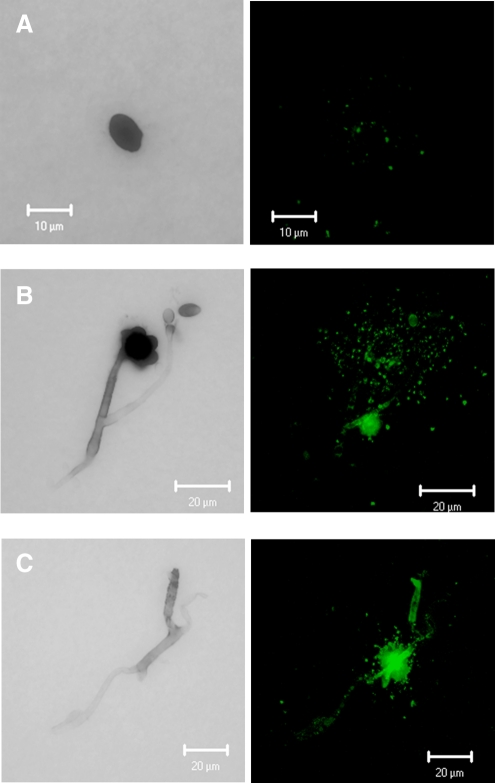
Immunization with the cScp resulted in the production of eight IgM isotype antibodies (3C3, 6D4, 7D11, 9G6, 24D11, 27C10, 27E2, and 29E5). The MAbs 3C3, 6D4, 7D11, and 9G6 were selected for preliminary studies based on their reactivity to S. chlorohalonata. These antibodies showed varying degrees of reactivity towards the semi-purified cytolytic preparation (Fig. 1). MAbs 7D11, 3C3, and 6D4 showed highest reactivity, while MAb 9G6 showed the lowest reactivity (Fig. 1). The same pattern of MAb reactivity was also found against the hyphal extract of S. chlorohalonata when analyzed by capture ELISA (Fig. 2). Reduced reactivity was observed in spore extracts compared to mycelium (Fig. 2). To confirm the reactivity of the MAbs, we used the fHIA to ultrastructurally locate the antibody binding sites of MAb 6D4 in S. chlorohalonata spores and hyphae (Fig. 3). MAb 6D4 immunostaining was primarily localized around phialides and sterigmata (Fig. 3), with highest concentrations primarily localized around hyphal septal junctions (Fig. 3B). In contrast, the staining of S. chlorohalonata conidia was less intense and mostly restricted to the surface of the conidia.
FIG. 1.
MAb reactivity against the cScp protein preparation. Purified antigens from S. chlorohalonata were diluted in PBSTM, and the reactivity of MAbs 3C3, 6D4, 7D11, and 9G6 were analyzed by capture ELISA. Optical densities (OD) were measured at 405 nm after 30 min of incubation with substrate.
FIG. 2.
Reactivity of the four MAbs against spores and S. chlorohalonata mycelial extracts. Extracts from spores or mycelium of S. chlorohalonata were analyzed with MAbs 3C3, 6D4, 7D11, and 9G6 using the capture ELISA. Optical densities (OD) were measured at 405 nm after 30 min incubation with substrate.
FIG. 3.
Fluorescent halogen immunostaining. (A) Conidia, (B) phialides, and (C) hyphae. Green dots identify the localization of MAb 6D4 specific antigens in the distinct morphological structures of S. chlorohalonata.
Cross-reactivity
Hyphal extracts obtained from 48 different fungi were tested in the capture ELISA for cross-reactivity with eight different MAbs developed against S. chlorohalonata hyphal antigens (Table 1), including hyphal extracts from seven different strains of S. chartarum. All MAbs identified various antigens in most of the strains with varying degrees of reactivity. However, MAb 29E5 failed to react with four of the seven (57%) S. chartarum strains. All MAbs demonstrated limited cross-reactivity with different species of the genus Stachybotrys. The most common cross-reactivity was observed for S. chartarum, S. bisbyi, and S. parvispora. However, none of the MAbs reacted with hyphal extracts of S. albipes, S. kampalensis, or S. oenanthes. MAbs 27E2 and 29E5 did not react with hyphal antigens derived from S. chlorohalonata and MAb 7D11, as well as MAbs 27E2 and 6D4 were the only MAbs to react with S. cylindropora or S. nephrospora, respectively. MAbs 7D11, 9G6, and 27E2 also displayed minimal cross-reactivity with Memnoniella echinata, a species that is phylogenetically related to S. chartarum. MAbs 9G6 and 27E2 also cross-reacted with M. subsimplex. No cross-reactivity was observed with any of the other 39 tested fungal species (Table 1).
Table 1.
Cross-reactivity Profiles of IgM MAbs Analyzed by Capture ELISA
| Fungal species | MAb 3C3 | MAb 6D4 | MAb 7D11 | MAb 9G6 | MAb 24D11 | MAb 27C10 | MAb 27E2 | MAb 29E5 |
|---|---|---|---|---|---|---|---|---|
| Stachybotrys chartarum IBT 7711 | 1.886 | 1.806 | 2.411 | 1.841 | 1.419 | 1.285 | 0.519 | 0.187 |
| Stachybotrys chartarum IBT 9460 | 1.888 | 1.636 | 2.185 | 1.717 | 1.302 | 1.511 | 0.437 | 0.241 |
| Stachybotrys chartarum IBT 9466 | 1.572 | 1.484 | 0.307 | 1.649 | 0.925 | 1.932 | 0.736 | 0.399 |
| Stachybotrys chartarum IBT 9631 | 1.307 | 1.149 | 1.537 | 1.499 | 0.598 | 1.241 | 0.591 | 0.356 |
| Stachybotrys chartarum IBT 9633 | 1.113 | 1.139 | 1.244 | 1.134 | 0.694 | 1.208 | 0.382 | 0.211 |
| Stachybotrys chartarum IBT 14915 | 2.305 | 2.244 | 0.930 | 1.993 | 1.541 | 1.736 | 0.862 | 0.335 |
| Stachybotrys chartarum IBT 14916 | 1.702 | 1.799 | 2.013 | 2.520 | 0.786 | 1.649 | 0.655 | 0.207 |
| Stachybotrys chlorohalonata ATCC 201863 (IBT 9825) | 1.856 | 1.761 | 1.951 | 0.795 | 1.333 | 1.565 | 0.043 | 0.017 |
| Stachybotrys albipes ATCC 18873 | 0.031 | 0.014 | 0.041 | 0.025 | 0.031 | 0.050 | 0.023 | 0.018 |
| Stachybotrys bisbyi ATCC 18825 | 2.790 | 2.575 | 0.320 | 2.618 | 1.783 | 2.052 | 0.938 | 0.578 |
| Stachybotrys cylindrospora ATCC 16276 | 0.010 | 0.009 | 1.039 | 0.068 | 0.026 | 0.410 | 0.012 | 0.003 |
| Stachybotrys kampalensis ATCC 22705 | 0.084 | 0.285 | 0.000 | 0.007 | 0.009 | 0.025 | 0.136 | 0.077 |
| Stachybotrys nephrospora ATCC 18839 | 0.027 | 0.952 | 0.047 | 0.042 | 0.033 | 0.064 | 0.455 | 0.022 |
| Stachybotrys oenanthes CBS 252.76 | 0.024 | 0.016 | 0.236 | 0.046 | 0.059 | 0.121 | 0.039 | 0.034 |
| Stachybotrys parvispora CBS 100155 | 2.156 | 2.332 | 3.355 | 0.017 | 1.528 | 2.768 | 0.008 | 0.004 |
| Memnoniella echinata NRRL 2373 | 0.217 | 0.265 | 1.623 | 1.185 | 0.054 | 0.033 | 0.439 | 0.199 |
| Memnoniella subsimplex ATCC 32888 | 0.107 | 0.103 | 0.043 | 1.099 | 0.057 | 0.090 | 0.346 | 0.130 |
| Aspergillus chevalieri NRRL 78 | 0.130 | 0.052 | 0.091 | 0.126 | 0.094 | 0.132 | 0.069 | 0.062 |
| Aspergillus clavatus NIOSH 6-22-78 | 0.024 | 0.016 | 0.067 | 0.062 | 0.058 | 0.098 | 0.042 | 0.035 |
| Aspergillus flavus ATCC 24689 | 0.026 | 0.013 | 0.069 | 0.059 | 0.060 | 0.116 | 0.047 | 0.041 |
| Aspergillus fumigatus FGSC A1100 | 0.023 | 0.011 | 0.031 | 0.029 | 0.038 | 0.053 | 0.016 | 0.013 |
| Aspergillus nidulans NIOSH 15-22-08 | 0.016 | 0.013 | 0.029 | 0.042 | 0.057 | 0.018 | 0.031 | 0.021 |
| Aspergillus niger FGSC A1144 | 0.014 | 0.003 | 0.044 | 0.054 | 0.065 | 0.060 | 0.019 | 0.010 |
| Aspergillus parasiticus ATCC 26691 | 0.023 | 0.014 | 0.053 | 0.087 | 0.021 | 0.054 | 0.027 | 0.015 |
| Aspergillus repens NRRL 13 | 0.061 | 0.061 | 0.097 | 0.137 | 0.091 | 0.181 | 0.097 | 0.080 |
| Aspergillus sydowii ATCC 9507 | 0.102 | 0.053 | 0.098 | 0.120 | 0.094 | 0.109 | 0.073 | 0.067 |
| Aspergillus terreus ATCC 1012 | 0.019 | 0.016 | 0.056 | 0.058 | 0.041 | 0.095 | 0.028 | 0.025 |
| Aspergillus ustus NRRL 275 | 0.018 | 0.000 | 0.043 | 0.052 | 0.036 | 0.019 | 0.005 | 0.000 |
| Aspergillus versicolor ATCC 44408 | 0.023 | 0.017 | 0.037 | 0.054 | 0.067 | 0.106 | 0.043 | 0.034 |
| Penicillium aurantiogriseum NRRL 971 | 0.029 | 0.010 | 0.052 | 0.099 | 0.029 | 0.068 | 0.040 | 0.031 |
| Penicillium expansum NRRL 973 | 0.015 | 0.012 | 0.038 | 0.027 | 0.039 | 0.060 | 0.025 | 0.013 |
| Penicillium fellutanum NRRL 746 | 0.032 | 0.005 | 0.045 | 0.054 | 0.025 | 0.023 | 0.012 | 0.002 |
| Penicillium purpurogenum NRRL 1062 | 0.104 | 0.026 | 0.017 | 0.084 | 0.003 | 0.043 | 0.012 | 0.000 |
| Penicillium roqueforti NRRL 844 | 0.014 | 0.012 | 0.026 | 0.016 | 0.051 | 0.034 | 0.012 | 0.011 |
| Alternaria alternata ATCC 11612 | 0.065 | 0.044 | 0.097 | 0.095 | 0.070 | 0.156 | 0.084 | 0.085 |
| Alternaria brassicicola ATCC 96836 | 0.046 | 0.039 | 0.064 | 0.091 | 0.034 | 0.088 | 0.053 | 0.044 |
| Wallemia sebi NIOSH 26-41-01 | 0.015 | 0.017 | 0.040 | 0.027 | 0.026 | 0.049 | 0.027 | 0.023 |
| Acremonium strictum ATCC 46646 | 0.011 | 0.005 | 0.027 | 0.017 | 0.032 | 0.030 | 0.009 | 0.007 |
| Stemphylium botryosum ATCC 26881 | 0.014 | 0.028 | 0.069 | 0.052 | 0.056 | 0.093 | 0.060 | 0.050 |
| Trichoderma viride ATCC 16640 | 0.026 | 0.009 | 0.037 | 0.021 | 0.031 | 0.033 | 0.017 | 0.009 |
| Ulocladium chartarum UAMH 5703 | 0.016 | 0.007 | 0.039 | 0.031 | 0.031 | 0.053 | 0.019 | 0.014 |
| Cladosporium herbarum ATCC 6506 | 0.018 | 0.006 | 0.027 | 0.014 | 0.028 | 0.021 | 0.020 | 0.013 |
| Cladosporium sphaerospermum ATCC 11288 | 0.027 | 0.019 | 0.037 | 0.075 | 0.021 | 0.060 | 0.038 | 0.033 |
| Paecilomyces variotii ATCC 66705 | 0.104 | 0.064 | 0.102 | 0.093 | 0.081 | 0.141 | 0.082 | 0.073 |
| Chaetomium globosum ATCC 6205 | 0.009 | 0.025 | 0.015 | 0.035 | 0.003 | 0.036 | 0.018 | 0.007 |
| Botrytis cinerea ATCC 11542 | 0.026 | 0.022 | 0.042 | 0.031 | 0.032 | 0.048 | 0.017 | 0.010 |
| Geotrichum candidum UAMH 7863 | 0.018 | 0.012 | 0.041 | 0.031 | 0.036 | 0.085 | 0.035 | 0.017 |
| Epicoccum nigrum ATCC 34929 | 0.041 | 0.026 | 0.052 | 0.037 | 0.041 | 0.068 | 0.038 | 0.029 |
| Eurotium amstelodami NIOSH 19-23-3 | 0.004 | 0.003 | 0.013 | 0.000 | 0.000 | 0.002 | 0.010 | 0.013 |
| Exserohilum rostratum ATCC 26856 | 0.008 | 0.007 | 0.026 | 0.015 | 0.046 | 0.037 | 0.015 | 0.012 |
| Fusarium moniliforme Penn State M6131 | 0.025 | 0.034 | 0.040 | 0.059 | 0.072 | 0.098 | 0.027 | 0.018 |
| Myrothecium verrucaria NRRL 2003 | 0.014 | 0.013 | 0.044 | 0.034 | 0.053 | 0.096 | 0.041 | 0.034 |
| Rhizopus stolonifer NIOSH 17-59-14 | 0.021 | 0.007 | 0.040 | 0.035 | 0.037 | 0.077 | 0.027 | 0.019 |
| Scopulariosis brumptii ATCC 16278 | 0.083 | 0.047 | 0.087 | 0.057 | 0.102 | 0.131 | 0.076 | 0.060 |
Optical densities (OD) were measured at 405 nm after 30 min incubation with substrate. Bold values identify reactivity of MAbs against species.
ATCC, American Type Culture Collection; NRRL, Agricultural Research Service Culture Collection; NIOSH, National Institute for Occupational Safety and Health; FGSC, Fungal Genetics Stock Center; UAMH, University of Alberta Microfungus Collection and Herbarium; Penn State, Pennsylvania State University; IBT, Instituttet for Bioteknologi, Denmark; CBS; Centraalbureau voor Schimmelcultures, Netherlands.
Western blot analysis
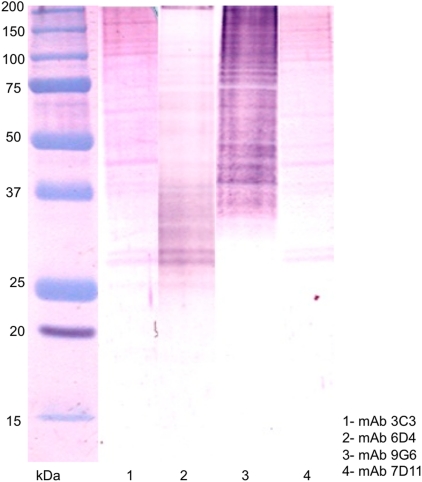
Western blot analysis following SDS–PAGE of S. chlorohalonata hyphal extracts indicated that the MAbs recognized multiple bands. MAbs 3C3, 6D4, and7D11 recognized two doublet bands at ∼30 kDa and ∼39 kDa and single band at ∼48 kDa, ∼70 kDa, and ∼110 kDa (Fig. 4), while MAb 9G6 recognized a streak of high molecular bands. Since the antibodies were cloned by limiting dilution, the antigens identified by these antibodies may be highly processed proteins or possibly be components of protein complexes of varying molecular weight.
FIG. 4.
Western blot reactivity patterns. Mycelial extract from S. chlorohalonata were developed in Western blot analysis using MAb 3C3 (lane 1); MAb 6D4 (lane 2); MAb 9G6 (lane 3); MAb 7D11 (lane 4).
Discussion
The fungal genus Stachybotrys is a tertiary colonizer of moisture-infiltrated cellulose-based building materials(17) and an indoor air contaminant.(18) Personal exposure to Stachybotrys conidia has been associated with respiratory disease(5,8,19–23); however, the scientific basis is not fully understood. The identification of Stachybotrys in indoor environments requires macroscopic and microscopic identification of conidia by a certified indoor air quality professional. The contribution of Stachybotrys hyphae to indoor contamination is recognized,(3) but is not reported in indoor investigations. Fragments derived from hyphae and spores also contain mycotoxins and other immunostimulatory antigens(24); however, the health effects associated with personal exposure remain uncharacterized. Recent developments in molecular and immunodiagnostic detection methodologies have improved the detection and quantification of S. chartarum. These studies have provided new insight into potential biomarkers of personal exposure,(11,25–27) including the cytolytic protein stachylysin.(4,9,28,29)
The utility of antibody-based immunoassays for the quantification of personal exposure to S. chartarum has recently been explored.(9) A polyclonal antibody has been utilized in an indirect ELISA for the quantification of stachylysin in environmental samples. Stachylysin has been detected in the serum of S. chartarum exposed rats. Similarly, stachylysin has also been detected in pooled serum derived from S. chartarum exposed workers but not in control subjects that had no a priori S. chartarum exposure.(9) Although polyclonal antibodies have been developed against other S. chartarum exoantigens for indoor environment exposure assessment studies, pAbs often lack specificity and cross-react with other fungal species.(9) Recently, several laboratories have developed MAbs towards S. chartarum.(30,31) Compared to pAbs, MAbs have potentially improved immunoassay specificity, sensitivity, and have been utilized in rapid immunodiagnostic tests for the detection of Stachybotrys in indoor environments.(11) However, hyphae are another principle source of contamination,(3) and these fungal particulates may aerosolize in higher concentrations than spores.(32) We did not specifically identify these antigens, and to our knowledge, MAbs that recognize Stachybotrys hyphae have yet to be developed.
In the present study, we immunized mice with a semi-purified cytolytic preparation from S. chlorohalonata hyphal cultures. The cScp contained cytolytic exoantigens and other secreted proteins that have been proposed to be potential biomarkers for Stachybotrys personal exposure.(9) Eight IgM isotype MAbs were produced and showed various degrees of reactivity to the original semi-purified cytolytic preparation. Four MAbs, which showed greatest reactivity toward hyphal extracts derived from Stachybotrys species, were selected for further characterization. Immunolocalization studies utilizing the fHIA confirmed that greatest reactivity of the MAbs were toward antigens released from phialides and hyphae. Although the MAbs showed limited reactivity with conidia, the results demonstrate the potential utility of these MAbs for the development of immunodetection methods for the quantification of Stachybotrys hyphae in indoor environments. Current research is also directed at the development of immunoassays for the detection of Stachybotrys antigens in clinical samples such as blood or bronchoalveolar lavage fluid.
Recent taxonomic studies have segregated S. chartarum into two separate chemotypes based on the production of metabolites.(15,33) Chemotype A comprises atranone producing strains (IBT 9466, IBT 9633, IBT 14915), and chemotype S consists of satratoxin and other macrocyclic trichothecene producing strains (IBT 7711, IBT 9460, IBT 9631, IBT 14916). In the present study, all MAbs with the exception of MAb 29E5, reacted to antigens released from both chemotype A and S strains. All MAbs except 7D11, 9G6, 27E2, and 29E5 that were developed in this study identified S. chlorohalonata and did not cross-react with other closely related species belonging to the genus Memnoniella. Recent studies showed that other commercially available detection methodologies could also not differentiate between S. chartarum and S. chlorohalonata.(34) The reduced cross-reactivity observed for these MAbs compared to the MAbs previously produced in our laboratory against Penicillium brevicompactum(10) could be due to the source of the antigen. Spore antigens can be shared between various fungal species and may show less antigenic variation compared to antigens localized in hyphae. We have observed similar results in Aspergillus versicolor, where MAbs produced against spores were cross-reactive.(12)
In this study, four IgM monoclonal antibodies have been developed and partially characterized that were directed against antigens derived from a semi-purified cytolytic S. chlorohalonata preparation. In addition to reacting with S. chlorohalonata hyphae, the MAbs additionally identified extracts derived from multiple S. chartarum isolates.
Acknowledgments
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health. This work was supported in part by an InterAgency agreement with the National Institute of Environmental Health Sciences (Y1-ES0001-06).
Author Disclosure Statement
The authors have no financial conflicts to declare.
References
- 1.Pestka JJ. Yike I. Dearborn D. Ward MDW. Harkema JR. Stachybotrys chartarum, trichothecene, mycotoxins, and damp building-related illness: new insights into a public health enigma. Toxicol Sci. 2008;104:4–26. doi: 10.1093/toxsci/kfm284. [DOI] [PubMed] [Google Scholar]
- 2.Brasel TL. Martin JM. Carriker CG. Wilson SC. Straus DC. Detection of airborne Stachybotrys chartarum macrocyclic trichothecene mycotoxins in the indoor environment. Appl Environ Microbiol. 2005;71:7376–7388. doi: 10.1128/AEM.71.11.7376-7388.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang CS. Heinsohn PA. Sampling, Analysis of Indoor Organisms. Wiley; Hoboken, NJ: 2003. [Google Scholar]
- 4.Gregory L. Rand TG. Dearborn D. Yike I. Vesper S. Immunocytochemical localization of stachylysin in Stachybotrys chartarum spores and spore-impacted mouse and rat lung tissue. Mycopathologia. 2003;156:109–117. doi: 10.1023/a:1022968121285. [DOI] [PubMed] [Google Scholar]
- 5.Vesper SJ. Vesper MJ. Possible role of fungal hemolysins in sick building syndrome. Adv Appl Microbiol. 2004;55:191–213. doi: 10.1016/S0065-2164(04)55007-4. [DOI] [PubMed] [Google Scholar]
- 6.Vesper SJ. Dearborn DG. Elidemir O. Haugland RA. Quantification of siderophore and hemolysin from Stachybotrys chartarum strains, including a strain isolated from the lung of a child with pulmonary hemorrhage and hemosiderosis. Appl Environ Microbiol. 2000;66:2678–2681. doi: 10.1128/aem.66.6.2678-2681.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M. Trigueros V. Paquereau L. Chavant L. Fournier D. Proteins as active compounds involved in insecticidal activity of mushroom fruitbodies. J Econ Entomol. 2002;95:603–607. doi: 10.1603/0022-0493-95.3.603. [DOI] [PubMed] [Google Scholar]
- 8.Vesper SJ. Dearborn DG. Yike I. Sorenson WG. Haugland RA. Hemolysis, toxicity, and randomly amplified polymorphic DNA analysis of Stachybotrys chartarum strains. Appl Environ Microbiol. 1999;65:3175–3181. doi: 10.1128/aem.65.7.3175-3181.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Emon JM. Reed AW. Yike I. Vesper SJ. ELISA measurement of stachylysin in serum to quantify human exposures to the indoor mold Stachybotrys chartarum. J Occup Environ Med. 2003;45:582–591. doi: 10.1097/01.jom.0000071503.96740.65. [DOI] [PubMed] [Google Scholar]
- 10.Schmechel D. Gorny RL. Simpson JP. Reponen T. Grinshpun SA. Lewis DM. Limitations of monoclonal antibodies for monitoring of fungal aerosols using Penicillium brevicompactum as a model fungus. J Immunol Methods. 2003;283:235–245. doi: 10.1016/j.jim.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Schmechel D. Simpson JP. Beezhold D. Lewis DM. The development of species-specific immunodiagnostics for Stachybotrys chartarum: the role of cross-reactivity. J Immunol Methods. 2006;309:150–159. doi: 10.1016/j.jim.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Schmechel D. Simpson JP. Lewis DM. The production and characterization of monoclonal antibodies to the fungus Aspergillus versicolor. Indoor Air. 2005;15(Suppl 9):11–19. doi: 10.1111/j.1600-0668.2005.00340.x. [DOI] [PubMed] [Google Scholar]
- 13.Green BJ. Millecchia LL. Blachere FM. Tovey ER. Beezhold DH. Schmechel D. Dual fluorescent halogen immunoassay for bioaerosols using confocal microscopy. Anal Biochem. 2006;354:151–153. doi: 10.1016/j.ab.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 14.Schmechel D. Lewis DM. The production of species-specific monoclonal antibodies (Mabs) against the allergenic and toxigenic fungus Stachybotrys chartarum. FASEB J. 2001;15:A662. [Google Scholar]
- 15.Andersen B. Nielsen KF. Thrane U. Szaro T. Taylor JW. Jarvis BB. Molecular and phenotypic descriptions of Stachybotrys chlorohalonata sp. nov. and two chemotypes of Stachybotrys chartarum found in water-damaged buildings. Mycologia. 2003;95:1227–1238. doi: 10.1080/15572536.2004.11833031. [DOI] [PubMed] [Google Scholar]
- 16.Vesper SJ. Magnuson ML. Dearborn DG. Yike I. Haugland RA. Initial characterization of the hemolysin stachylysin from Stachybotrys chartarum. Infect Immun. 2001;69:912–916. doi: 10.1128/IAI.69.2.912-916.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn DM. Ghannoum MA. Indoor mold, toxigenic fungi, and Stachybotrys chartarum: infectious disease perspective. Clin Microbiol Rev. 2003;16:144–172. doi: 10.1128/CMR.16.1.144-172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vesper S. McKinstry C. Ashley P. Haugland R. Yeatts K. Bradham K. Svendsen E. Quantitative PCR analysis of molds in the dust from homes of asthmatic children in North Carolina. J Environ Monit. 2007;9:826–830. doi: 10.1039/b704359g. [DOI] [PubMed] [Google Scholar]
- 19.Mahmoudi M. Gershwin ME. Sick building syndrome. III. Stachybotrys chartarum. J Asthma. 2000;37:191–198. doi: 10.3109/02770900009055442. [DOI] [PubMed] [Google Scholar]
- 20.Etzel RA. Montana E. Sorenson WG. Kullman GJ. Allan TM. Dearborn DG. Olson DR. Jarvis BB. Miller JD. Acute pulmonary hemorrhage in infants associated with exposure to Stachybotrys atra and other fungi. Arch Pediatr Adolesc Med. 1998;152:757–762. doi: 10.1001/archpedi.152.8.757. [DOI] [PubMed] [Google Scholar]
- 21.Hodgson MJ. Morey P. Leung WY. Morrow L. Miller D. Jarvis BB. Robbins H. Halsey JF. Storey E. Building-associated pulmonary disease from exposure to Stachybotrys chartarum and Aspergillus versicolor. J Occup Environ Med. 1998;40:241–249. doi: 10.1097/00043764-199803000-00006. [DOI] [PubMed] [Google Scholar]
- 22.McGinnis MR. Pathogenesis of indoor fungal diseases. Med Mycol. 2004;42:107–117. doi: 10.1080/13693780410001661473. [DOI] [PubMed] [Google Scholar]
- 23.Vesper SJ. McKinstry C. Yang C. Haugland RA. Kercsmar CM. Yike I. Schluchter MD. Kirchner HL. Sobolewski J. Allan TM. Dearborn DG. Specific molds associated with asthma in water-damaged homes. J Occup Environ Med. 2006;48:852–858. doi: 10.1097/01.jom.0000224736.52780.2f. [DOI] [PubMed] [Google Scholar]
- 24.Brasel TL. Douglas DR. Wilson SC. Straus SC. Detection of airborne Stachybotrys chartarum macrocyclic trichothecene mycotoxins on particulates smaller than conidia. Appl Environ Microbiol. 2005;71:114–122. doi: 10.1128/AEM.71.1.114-122.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haugland RA. Heckman JL. Identification of putative sequence specific PCR primers for detection of the toxigenic fungal species Stachybotrys chartarum. Mol Cell Probes. 1998;12:387–396. doi: 10.1006/mcpr.1998.0197. [DOI] [PubMed] [Google Scholar]
- 26.Haugland RA. Vesper SJ. Wymer LJ. Quantitative measurement of Stachybotrys chartarum conidia using real time detection of PCR products with the TaqMan(TM) fluorogenic probe system. Mol Cell Probes. 1999;13:329–340. doi: 10.1006/mcpr.1999.0258. [DOI] [PubMed] [Google Scholar]
- 27.Cruz-Perez P. Buttner MP. Stetzenbach LD. Specific detection of Stachybotrys chartarum in pure culture using quantitative polymerase chain reaction. Mol Cell Probes. 2001;15:129–138. doi: 10.1006/mcpr.2001.0347. [DOI] [PubMed] [Google Scholar]
- 28.Vojdani A. Antibodies against Stachybotrys chartarum extract and its antigenic components, Stachyhemolysin and Stachyrase-A: a new clinical biomarker. Med Sci Monit. 2005;11:Br139–145. [PubMed] [Google Scholar]
- 29.Page E. Biagini RE. Beezhold DH. Methodologic issues concerning Stachyhemolysin and Stachyrase-A as clinical biomarkers. Med Sci Monit. 2005;11:LE7–8. [PubMed] [Google Scholar]
- 30.Xu J. Jensen JT. Liang Y. Belisle D. Miller JD. The biology and immunogenicity of a 34-kDa antigen of Stachybotrys chartarum sensu lato. Int Biodeterior Biodegradation. 2007;60:308–318. [Google Scholar]
- 31.Xu J. Liang Y. Belisle D. Miller JD. Characterization of monoclonal antibodies to an antigenic protein from Stachybotrys chartarum and its measurement in house dust. J Immunol Methods. 2008;332:121–128. doi: 10.1016/j.jim.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 32.Gorny RL. Reponen T. Willeke K. Schmechel D. Robine E. Boissier M. Grinshpun SA. Fungal fragments as indoor air biocontaminants. Appl Environ Microbiol. 2002;68:3522–3531. doi: 10.1128/AEM.68.7.3522-3531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersen B. Nielsen KF. Jarvis BB. Characterization of Stachybotrys from water-damaged buildings based on morphology, growth, and metabolite production. Mycologia. 2002;94:392–403. [PubMed] [Google Scholar]
- 34.Li DW. Yang CS. Taxonomic history and current status of Stachybotrys chartarum and related species. Indoor Air. 2005;15(Suppl 9):5–10. doi: 10.1111/j.1600-0668.2005.00339.x. [DOI] [PubMed] [Google Scholar]