Abstract
Cancer cells show alterations in metabolism that support malignancy and disease progression. Prominent among these metabolic changes is elevations in neutral ether lipids (NELs). We have previously shown that the hydrolytic enzyme KIAA1363 (or AADACL1) is highly elevated in aggressive cancer cells, where it plays a key role in generating the monoalkylglycerol ether (MAGE) class of NELs. Here, we use activity-based protein profiling-guided medicinal chemistry to discover a highly potent and selective inhibitor of KIAA1363, the carbamate JW480. We show that JW480, and an shRNA probe that targets KIAA1363, reduce MAGEs and impair the migration, invasion, survival, and in vivo tumor growth of human prostate cancer cell lines. These findings indicate that the KIAA1363-MAGE pathway is important for prostate cancer pathogenesis and designate JW480 as a versatile pharmacological probe for disrupting this pro-tumorigenic metabolic pathway.
For more than 40 years, it has been known that tumor cells show dramatic elevations in their neutral ether lipid (NEL) content. Seminal work by Snyder and colleagues in the 1960s first reported that rodent and human tumors possess significantly higher levels of NELs relative to normal tissue (Snyder and Wood, 1969; Wood and Snyder, 1967). Over the ensuing decades, this finding has been confirmed for a wide range of cancer cells and primary tumors from several tissues of origin (Albert and Anderson, 1977; Lin et al., 1978; Roos and Choppin, 1984). Provocative evidence has also emerged to suggest a pro-tumorigenic function for NELs, including a study where the levels of these lipids were found to correlate closely with tumorigenicity across a panel of mouse fibroblast cell lines (Roos and Choppin, 1984). Nonetheless, whether elevated NELs are causally linked, or merely associated with cancer has remained unclear. Moreover, the enzymes responsible for regulating NEL metabolism in cancer cells are, for the most part, poorly understood.
We have recently determined that the previously uncharacterized transmembrane enzyme KIAA1363 (also called AADACL1) controls the production of the monoalkylglycerol ether (MAGE) class of NELs in cancer cells (Chiang et al., 2006). KIAA1363 acts as a 2-acetyl MAGE hydrolase (Chiang et al., 2006) and is likely the principal source for this activity in tumor cells, which was originally detected by Snyder’s group in the early 1990s (Blank et al., 1990). MAGEs can be further converted by cancer cells into the bioactive lysophospholipids alkyl-lysophosphatidyl choline (alkyl-LPC) and alkyl-lysophosphatidic acid (alkyl-LPA) (Chiang et al., 2006). Stable knockdown of KIAA1363 expression impaired tumor cell migration and tumor growth in vivo, suggesting a potentially key role for this enzyme in promoting cancer pathogenesis. Consistent with this premise, we, and others, have found that KIAA1363 is highly elevated in aggressive breast, melanoma, ovarian (Chiang et al., 2006; Jessani et al., 2002), and pancreatic (Iacobuzio-Donahue et al., 2002) cancer cells, as well as primary breast (Ferguson et al., 2005; Jessani et al., 2005) and ovarian (Haverty et al., 2009) tumors.
While the aforementioned discoveries suggest that KIAA1363 could represent an interesting pharmacological target for impairing cancer malignancy, selective inhibitors for this enzyme are lacking. Here, we report the results of an activity-based protein profiling [ABPP (Berger et al., 2004; Cravatt et al., 2008; Liu et al., 1999)]-guided medicinal chemistry study aimed at optimizing inhibitors for KIAA1363. This effort identified an O-aryl carbamate JW480 that acts as a potent and selective inhibitor of KIAA1363. JW480 is orally active and capable of fully inhibiting KIAA1363 in mice for at least 24 hrs following a single administration. We show that JW480, as well as an shRNA probe that targets KIAA1363, reduce MAGE levels in human prostate cancer cells and impair their migration, invasion, survival, and in vivo tumor growth.
Results
Aggressive human prostate cancer cells show high levels of KIAA1363 activity
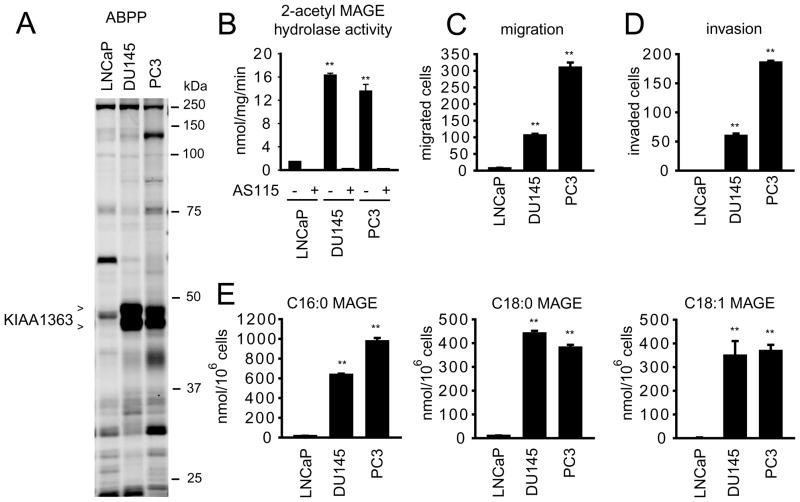
In previous ABPP studies, we found that KIAA1363 activity was elevated in aggressive human breast, melanoma, and ovarian cancer cells (Chiang et al., 2006; Jessani et al., 2002), as well as primary human breast tumors (Jessani et al., 2005). Here, we analyzed a panel of human prostate cancer cell lines by ABPP using the serine hydrolase-directed activity-based probe fluorophosphonate-rhodamine (Jessani et al., 2002; Patricelli et al., 2001) and observed that KIAA1363 activity, detectable as a ~45 kDa FP-rhodamine-reactive doublet (Figure 1A), was much higher in the androgen-independent human prostate cancer lines PC3 and DU145 compared to the androgen-dependent human prostate cancer line LNCaP (Figure 1A and B). This difference in activity was also detected using the KIAA1363 substrate 2-acetyl MAGE (Figure 1B). PC3 and DU145 cells showed much greater migratory (Figure 1C) and invasive (Figure 1D) activity compared to LNCaP cells, consistent with the reported differences in aggressiveness among these cancer lines (Hoosein et al., 1991). Finally, PC3 and DU145 cells possessed higher levels of the KIAA1363-regulated NELs C16:0, C18:0, and C18:1 MAGE compared to LNCaP cells (Figure 1E). These data, together, indicate that aggressive prostate cancer cells contain a hyperactive KIAA1363-MAGE pathway.
Figure 1. KIAA1363 is elevated in androgen-independent human prostate cancer cell lines.
(A) ABPP of serine hydrolase activities in the androgen-dependent LNCaP and androgen-independent PC3 and DU145 cells line. Serine hydrolase activities were labeled in whole cell proteomes with the activity-based probe FP-rhodamine (1 μM, 30 min) (Jessani et al., 2002; Patricelli et al., 2001) and detected by SDS-PAGE and in-gel fluorescence scanning (fluorescent gel shown in grayscale). KIAA1363 activity is much higher in PC3 and DU145 cells compared to LNCaP cells. Note that KIAA1363 migrates as two distinct glycoforms in cancer cells, as described previously (Chiang et al., 2006; Jessani et al., 2002). (B) PC3 and DU145 cells also show much higher 2-acetyl MAGE hydrolytic activity catalytic compared to LNCaP cells. Note that virtually all 2-acetyl MAGE hydrolase activity in prostate cancer cells is eliminated by the KIAA1363 inhibitor AS115 (10 μM, 4 h treatment, in situ). (C and D) PC3 and DU145 cells show elevated migratory (C) and invasive (D) activity compared to LNCaP cells. (E) PC3 and DU145 cells have much higher levels of MAGEs (C16:0, 1-O-palmityl MAGE; C18:0, 1-O-stearyl MAGE; C18:1 1-O-oleyl MAGE) compared to LNCaP cells. ** p <0.01 for comparisons between PC3 and DU145 cells versus LNCaP cells. Data are presented as means ± standard error of the mean (SEM); n = 4–5/group.
Development of JW480, a potent, selective, and in vivo active KIAA1363 inhibitor
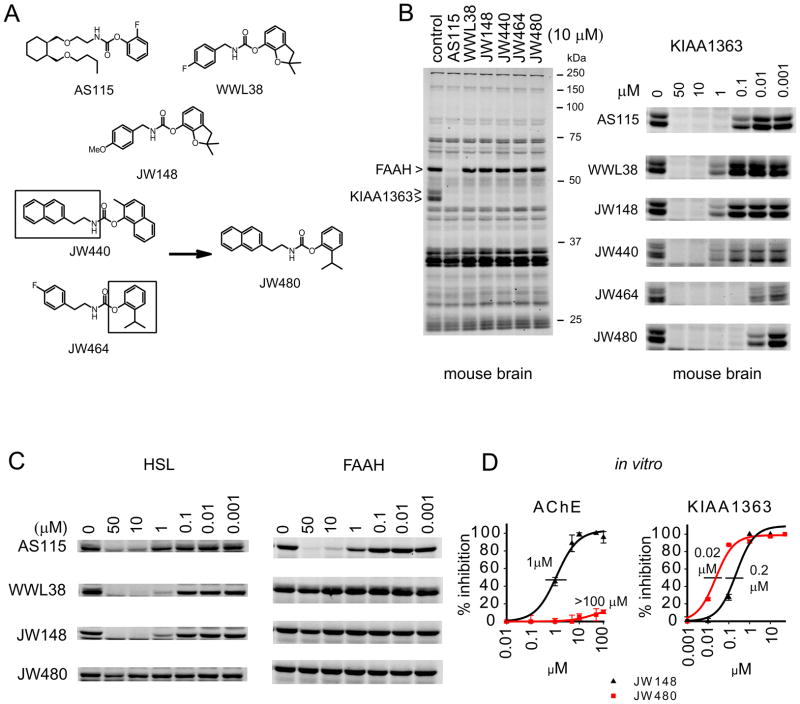
We next wanted to examine the function of KIAA1363 in prostate cancer cells. While lead inhibitors, such as trifluoromethyl ketones (Leung et al., 2003) and the carbamate AS115 (Chiang et al., 2006) (Figure 2A), have been created for KIAA1363, these reagents lack the desired combination of selectivity and in vivo activity suitable for extensive biological studies. In a recent large-scale screen (Bachovchin et al., 2010), we discovered a new structural class of carbamates, representative members of which include WWL38 and JW148 (Figure 2A), that inhibited KIAA1363 with good selectivity compared to common off-target enzymes such as fatty acid amide hydrolase (FAAH), as judged by gel-based competitive ABPP of a mouse brain proteome (Figure 2B). This enhanced selectivity appeared to be imparted by the O-2,3-dihydrobenzofuran leaving group, which is well-tolerated by KIAA1363 but not most other serine hydrolases (see also Figure S1 and Table S1). However, WWL38 and JW148 showed only moderate potency for KIAA1363 (IC50 values ~200 nM) and still exhibited cross-reactivity with two other serine hydrolases – hormone-sensitive lipase (HSL) (Figure 2C) and acetylcholinesterase (AChE) (Figure 2D). We sought to enhance the potency and selectivity of these lead carbamates through ABPP-guided medicinal chemistry.
Figure 2. Development of JW480 – a potent and selective KIAA1363 inhibitor.
(A) Structures of various KIAA1363 inhibitors, including first-generation inhibitors, such as AS115, WWL38, and JW148, along with second-generation inhibitors JW440 and JW464, which showed improved selectivity and potency for KIAA1363, respectively. Combining key structural features of JW440 and JW464 (boxed) provided JW480, which showed improved potency and selectivity for KIAA1363. (B) The left panel shows a competitive ABPP gel for various KIAA1363 inhibitors with the mouse brain membrane proteome (10 μM inhibitor concentration). Note that AS115, but not other agents, also inhibits FAAH. The right panel shows concentration-dependent inhibition of KIAA1363 in mouse brain proteome. (C) Competitive ABPP gels comparing the activity of KIAA1363 inhibitors against the common off-target enzymes HSL and FAAH. HSL activity was measured in a rosiglitazone-differentiated 3T3-10T1/2 adipocyte proteome, while FAAH activity was measured in mouse brain proteome. (D) Concentration-dependent inhibition of AChE and KIAA1363 by JW148 and JW480. KIAA1363 activity was measured by competitive ABPP in a mouse brain proteome. AChE activity was measured in a mouse brain proteome using the substrate acetylthiocholine (Ellman et al., 1961) because this enzyme is too low in abundance for detection by gel-based ABPP. Note, however, that AChE activity can be measured by competitive ABPP-MudPIT (see Figure 4C), which confirmed negligible cross-reactivity for JW480 with this enzyme. The following IC50 values, also shown in the figure, were measured: JW148, IC50 for KIAA1363 of 0.2 μM (95 % confidence limit, 0.1–0.3 μM); IC50 for AChE of 1 μM (1.09–1.3 μM); JW480 IC50 for KIAA1363 of 0.02 μM (0.021–0.025 μM); IC50 for AChE > 100 μM. Gels are representative images from n = 3; Data in (D) are presented as means ± standard error of the mean (SEM); n=3/group. See also Table S1 and Figures S1 and S2.
We noted that replacing the O-2,3-dihydrobenzofuran group of WWL38 and JW148 with an O-2-isopropylphenyl leaving group created a carbamate JW464 (Figure 2A) with much improved potency for KIAA1363 (Figure 2B), but which still showed cross-reactivity with AChE (Figure S2). On the other hand, O-aryl carbamates with a bulkier N-ethylnaphthalene substituent, such as JW440 (Figure 2B), showed little or no activity against AChE or HSL (Figure S2), while maintaining good potency for KIAA1363 (Figure 2B). Combining these two structural features engendered a carbamate JW480 that inhibited mouse brain KIAA1363 with exceptional potency (IC50 value = 20 nM; Figure 2B, D), while showing negligible cross-reactivity with HSL (Figure 2C), ACHE (Figure 2D), or other mouse brain serine hydrolases (Figure 2B).
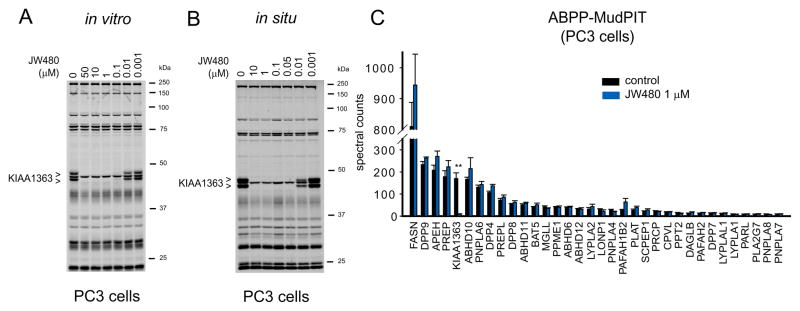
We next confirmed that JW480 also inhibited human KIAA1363 in PC3 cell proteomes (in vitro treatment), showing an IC50 value of 12 nM in competitive ABPP assays (Figure 3A). Similar inhibition was observed in living PC3 cells in the presence of absence of 10% fetal calf serum (in situ treatment), where JW480 inactivated KIAA1363 with an IC50 values of 6–12 nM (Figure 3B and Figure S3). We also confirmed inhibition of KIAA1363 by JW480 using a 2-acetyl MAGE substrate assay (Figure S3). The in situ inhibition of KIAA1363 by JW480 (1 μM) was maintained for at least 48 hr (Figure S3). JW480 showed excellent selectivity for KIAA1363 in PC3 cells (Figure 3A, B), as well as in DU145 and LNCaP cells (Figure S3), as judged by gel-based competitive ABPP. This selectivity was confirmed by competitive ABPP-MudPIT (Li et al., 2007; Long et al., 2009a), a mass-spectrometry-based method that displays higher resolution than gel-based ABPP and revealed no off-target activity for JW480 (1 uM in situ treatment, 4 hr) across the > 30 serine hydrolase activities detected in PC3 cells (Figure 3C).
Figure 3. JW480 selectively inhibits KIAA1363 in human prostate cancer cells.
(A and B) Gel-based competitive ABPP profiles of proteomes from PC3 cells treated with a range of concentrations of JW480 in vitro (A) or in situ (B). The following IC50 values, also shown in the figure, were measured for KIAA1363: in vitro: 0.012 μM (95% confidence limits, 0.009–0.014 μM); in situ: 0.006 μM (0.005–0.007 μM). Note that other serine hydrolase activities detected by gel-based ABPP were not affected by JW480. Gels are representative images from n=3. (C) Competitive ABPP-MudPIT results for PC3 cells treated with JW480 (1 μM, 48 hr). Among the ~30 serine hydrolase activities measured by ABPP-MudPIT in whole PC3 cell proteomes, only KIAA1363 was inhibited by JW480. Only serine hydrolases that showed an average spectral count value ≥ 8 were subjected to quantitative analysis. **p < 0.01 for JW480- versus DMSO-treated control groups. Data are presented as means ± standard error of the mean (SEM); n=4/group. See also Figure S3.
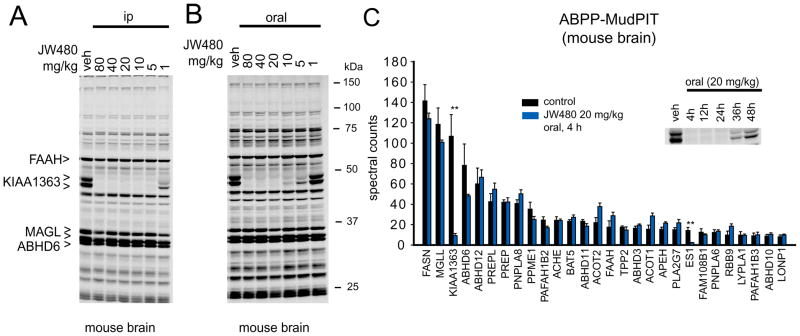
We next asked whether JW480 could inhibit KIAA1363 in vivo by treating mice with varying quantities of this inhibitor (1–80 mg/kg, i.p. or oral administration, 4 hr) and then sacrificing the animals and analyzing their brain proteomes by competitive ABPP. JW480 proved to be highly active in vivo, showing complete inhibition of brain KIAA1363 at doses of 5 and 20 mg/kg following i.p. (Figure 4A) and oral (Figure 4B) routes of administration, respectively. As was observed in PC3 cells, JW480 showed excellent selectivity for KIAA1363 in brain proteomes from inhibitor-treated mice as judged by gel-based competitive ABPP (Figure 4A, B). This selectivity was confirmed by competitive ABPP-MudPIT, where only a single off-target was observed among the ~30 serine hydrolase activities detected in the brain proteome – the carboxylesterase ES1 (Figure 4C). ES1 is not expressed in the brain (Krishnasamy et al., 1998), but rather likely originates from contaminating blood in the brain tissue preparation [this enzyme is secreted into the blood by the liver (Krishnasamy et al., 1998)]. The selectivity profile of JW480 thus matches those of other carbamate inhibitors of serine hydrolases, such as the FAAH inhibitor URB597 (Kathuria et al., 2003) or the monoacylglycerol lipase inhibitor JZL184 (Long et al., 2009a), which also show excellent specificity for their target enzymes with the exception of some cross-reactivity with carboxylesterases (Alexander and Cravatt, 2005; Long et al., 2009b). Finally, time course studies confirmed that inhibition of KIAA1363 was maintained in vivo for up to 24 hr following a single administration of JW480 (20 mg/kg, oral; Figure 4C, inset).
Figure 4. JW480 selectively inhibits KIAA1363 in vivo.
(A and B) Gel-based competitive ABPP profiles of brain membrane proteomes from mice treated with a range of doses of JW480 by intraperitoneal (i.p.) (A) or oral (B) administration (4 hr treatment). Note that among the serine hydrolase activities detected by gel-based ABPP, only KIAA1363 was inhibited by JW480. Representative other brain serine hydrolases are marked for comparison. (C) Competitive ABPP-MudPIT results for brain membrane proteomes from mice treated with JW480 (20 mg/kg, oral gavage, 4 hr). Among the ~30 serine hydrolase activities measured by ABPP-MudPIT, only KIAA1363 and the carboxylesterase ES1 were inhibited by JW480. Only serine hydrolases that showed an average spectral count value ≥ 8 were subjected to quantitative analysis. Inset shows the time course for KIAA1363 inhibition by a single administration of JW480 (20 mg/kg, oral). Gels are representative images from n = 3. **p < 0.01 for JW480- versus vehicle-treated control groups. Data in (C) are presented as means ± standard error of the mean (SEM); n = 4/group.
These data, taken together indicate that JW480 is a potent and selective inhibitor of both human and mouse KIAA1363 that can be used to inactivate this enzyme in living cells or animals.
JW480 lowers MAGEs and impairs pathogenicity of prostate cancer cells
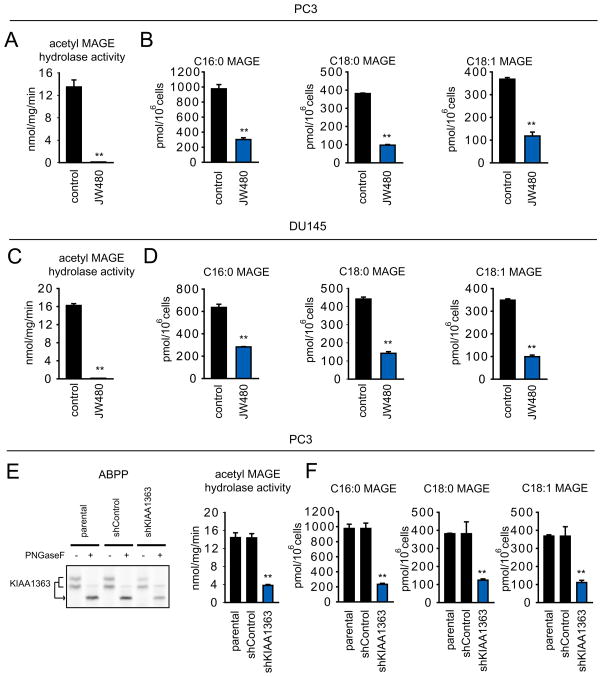
We next tested the effects of JW480 on the KIAA1363-MAGE pathway in prostate cancer cells. JW480 treatment (1 μM) completely blocked 2-acetyl MAGE hydrolase activity (Figure 5A, C) and caused significant reductions in MAGE lipids (Figure 5B, D) in both PC3 (Figure 5B) and DU145 (Figure 5D) cells. Similar effects were observed in a PC3 line where KIAA1363 was stably knocked down by a small hairpin (sh) RNA (shKIAA1363 cells; Figure 5E, F). Other KIAA1363 inhibitors, such as AS115 and JW148, but not the FAAH inhibitor URB597, also caused reductions in MAGE lipids in prostate cancer cells (Figure S4).
Figure 5. Disruption of KIAA1363 activity reduces MAGE lipids in prostate cancer cells.
(A and C) JW480 ablates KIAA1363 activity as assessed by 2-acetyl MAGE hydrolytic activity (1 μM, 48 h) in PC3 (A) and DU145 (C) cells. (B and D) JW480 significantly reduces MAGE levels in PC3 (B) and DU145 (D) (1 μM, 48 hr). (E) Stable knockdown of KIAA1363 using a short-hairpin oligonucleotide (shKIAA1363) reduces KIAA1363 activity by > 70 % in PC3 cells as assessed by ABPP (left) or 2-acetyl MAGE hydrolytic activity (right). (F) shKIAA1363 PC3 cells have significantly lower levels of MAGEs. **p < 0.01 for JW480-treated or shKIAA1363 cells compared to their respective control groups (DMSO-treated and parental/shControl cells). Data are presented as means ± standard error of the mean (SEM); n = 4–5/group. See also Figure S4.
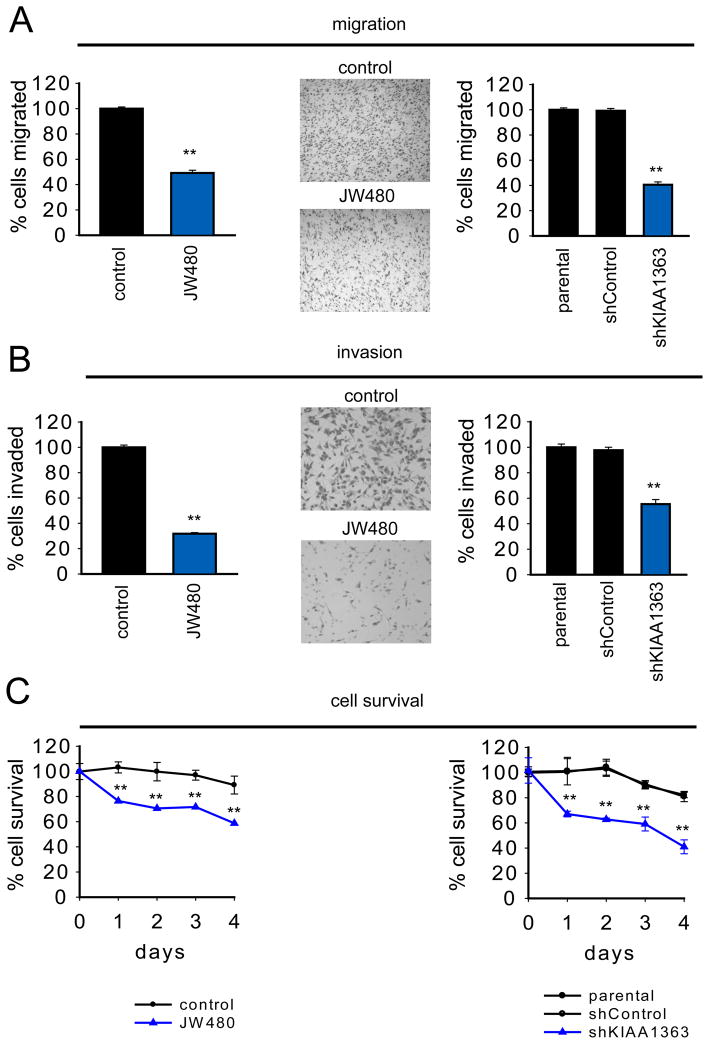
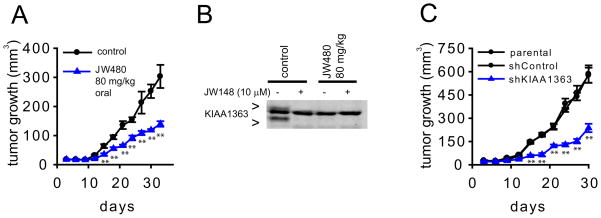
Additional studies provided evidence that the KIAA1363-MAGE pathway is important for prostate cancer aggressiveness. For instance, JW480-treated and shKIAA1363 prostate cancer cells displayed reductions in migration (Figure 6A), invasion (Figure 6B), and survival in serum-free media (Figure 6C). These effects were also observed with other KIAA1363 inhibitors (AS115 and JW148), but not with the FAAH inhibitor URB597 (Figure S5). JW480 did not affect the survival in LNCaP cells, which express low levels of KIAA1363 (Figure S5). Finally, we took advantage of JW480’s exceptional in vivo activity to test whether pharmacological blockade of KIAA1363 affected PC3 tumor growth in a mouse xenograft model. We found that PC3 tumor growth was significantly impaired in mice treated daily with JW480 (80 mg/kg, oral) compared to control mice treated with vehicle (Figure 7A). We confirmed by competitive ABPP that KIAA1363 activity was completely ablated in tumors explanted from mice treated with JW480 (Figure 7B). We observed a similar reduction in tumor growth for shKIAA1363 PC3 cells compared to control cells (Figure 7C).
Figure 6. Disruption of KIAA1363 activity impairs prostate cancer cell pathogenicity.
JW480-treated and shKIAA1363 PC3 cells show reduced migration (A), invasion (B), and serum-free cell survival (C). Cells were treated with JW480 (1 μM, 48 h in situ) for 48 hr prior to biological measurements. **p < 0.01 for JW480-treated or shKIAA1363 cells compared to their respective control groups (DMSO-treated and parental/shControl cells). Data are presented as means ± standard error of the mean (SEM); n = 4–5/group. See also Figure S5.
Figure 7. Disruption of KIAA1363 activity Impairs prostate tumor growth in vivo.
(A) JW480 treatment (80 mg/kg, oral gavage, one dose per day, initiated on the day of tumor cell implantation) significantly reduces PC3 tumor xenograft growth in immune-deficient SCID mice. We used a dose of 80 mg/kg JW480 because we found that a higher quantity of this compound was required to completely block KIAA1363 in SCID mice compared to normal mice (Figure S6). (B) Tumors from mice treated with JW480, removed 4 hr after the final administered dose of JW480 (day 33, part A), show complete loss of KIAA1363 activity compared to control (vehicle-treated) tumors as determined by ABPP. Ex vivo treatment with JW148 (10 μM, 30 min) was used to confirm that the upper and lower FP-rhodamine-reactive bands correspond to KIAA1363, while the middle FP-rhodamine-reactive band is another (JW148-insensitive) serine hydrolase. (C) shKIAA1363 PC3 cells also show significantly reduce tumor growth compared to parental and shControl PC3 cells in a SCID mouse xenograft model. **p<0.01 for JW480-treated or shKIAA1363 cells compared to their respective control groups (vehicle-treated and parental/shControl cells). Data are presented as means ± standard error of the mean (SEM); n = 6–8/group. See also Figure S6.
These results, taken together, show that pharmacological or shRNA-mediated disruption of KIAA1363 reduces MAGE levels and impairs the pathogenic properties of human prostate cancer cells.
Discussion
Serine hydrolases are an exceptionally large and diverse class of enzymes that play important roles in virtually all biological processes in mammals (Simon and Cravatt, 2010). Many serine hydrolases, however, remain poorly characterized with respect to their biochemical and physiological functions. Using ABPP, we previously identified one such serine hydrolase, the integral membrane enzyme KIAA1363, as being highly expressed by aggressive human cancer cells (Jessani et al., 2002) and primary tumors (Jessani et al., 2005). We also succeeded in identifying lead inhibitors for KIAA1363, which helped to define a role for this enzyme in NEL metabolism in cancer cells (Chiang et al., 2006). These findings, when integrated with the large historical body of work designating NEL metabolism as a prominent biochemical pathway dysregulated in cancer cells (Albert and Anderson, 1977; Lin et al., 1978; Roos and Choppin, 1984; Snyder and Wood, 1969; Wood and Snyder, 1967), suggest that KIAA1363 might play an important role in tumorigenesis. Testing this hypothesis, however, required more advanced KIAA1363 inhibitors that possess the potency and selectivity needed for extensive pharmacological studies.
Here, we have used competitive ABPP-guided medicinal chemistry to create JW480 – a low-nM, selective, and in vivo-active inhibitor for KIAA1363. We believe that JW480 likely acts as an irreversible inhibitor of KIAA1363 that carbamoylates the enzyme’s serine nucleophile, which is consistent with the known mechanism of action for other carbamate inhibitors of serine hydrolases (Alexander and Cravatt, 2005; Long et al., 2009b). Confirmation of an irreversible mode of inhibition for JW480 will require further studies, such as direct mass-spectrometry characterization of the carbamoylated inhibitor-enzyme adduct as has been measured for other serine hydrolases (Alexander and Cravatt, 2005). But, an irreversible mechanism of action for JW480 is supported by our competitive ABPP results that show sustained inhibition of KIAA1363 in tissue extracts from JW480-treated mice. Assuming that JW480 is an irreversible inhibitor of KIAA1363, future experiments could include: 1) appending an alkyne onto JW480 to convert it into a ‘clickable’ probe for assessing proteome-wide target interactions beyond the serine hydrolases that are counterscreened by competitive ABPP, as has been shown for other enzymes (Ahn et al., 2009; Alexander and Cravatt, 2005), and 2) incorporation of a fluorophore into the JW480 structure for creating a probe to image KIAA1363 activity in vivo, as has been achieved for other enzymes (Blum et al., 2005).
The starting scaffold for JW480 originated from a recent ‘library-versus-library’ competitive ABPP screen where we assayed 70+ serine hydrolases against 150+ carbamate small molecules (Bachovchin et al., 2010). The breadth of this screen pointed to areas for improvement of lead KIAA1363 inhibitors, most notably, in designating HSL and AChE as common off-targets for these compounds. Interestingly, and as noted previously (Bachovchin et al., 2010), neither of these enzymes share much sequence homology with KIAA1363, underscoring the value of proteomic profiling methods like competitive ABPP that can identify ‘pharmacological homology’ among distantly related enzymes. We were able to minimize cross-reactivity with HSL and AChE by incorporating a bulky naphthalene group into the N-alkyl substituent of JW480.
Eliminating AChE cross-reactivity was of obvious importance, given that potent inhibitors of this enzyme are neurotoxic (Casida and Quistad, 2005). Removing HSL cross-reactivity may also be valuable because both KIAA1363 and HSL are expressed at high levels in macrophages, where each enzyme has been suggested to play a role in neutral cholesterol ester hydrolysis (Buchebner et al., 2010; Igarashi et al., 2010; Okazaki et al., 2008). The relative contribution that KIAA1363 and HSL make to cholesterol ester metabolism in macrophages remains unclear, and we anticipate that JW480 should offer a valuable pharmacological tool to investigate this question. Among the more than 40 serine hydrolases counterscreened by competitive ABPP in our combined analyses of brain and cancer cell proteomes, only a single off-target for JW480 was detected – the carboxylesterase ES1. As has been discussed previously (Bachovchin et al., 2010), CEs are promiscuous enzymes involved in xenobiotic metabolism in tissues such as the liver. They are common off-targets for mechanism-based serine hydrolase inhibitors, including carbamates (Alexander and Cravatt, 2005; Bachovchin et al., 2010; Long et al., 2009b). We do not believe, however, that such cross-reactivity with ES1 (and possibly other CEs) is a major problem for using JW480 as a pharmacological tool to investigate KIAA1363 function, especially for studies in the nervous system and cancer, where CE expression is low. Furthermore, one can use carbamates that do not inhibit KIAA1363, but still show CE cross-reactivity as ‘negative control’ compounds, as we have shown in this study with the FAAH inhibitor URB597.
Using JW480 (and an shRNA probe that targets KIAA1363), we found that disrupting KIAA1363 reduces MAGE levels in human prostate cancer cells and impairs several of their pro-tumorigenic properties, including migration, invasion, and serum-free survival. These metabolic and cell biological effects were correlated with significant reductions in tumor growth in mouse xenograft models treated with JW480 (or using shKIAA1363 prostate cancer cells). These data collectively support a pro-tumorigenic function for KIAA1363. We should note, however, that tumors treated with JW480 or from shKIAA1363 prostate cancer cells continued to grow in vivo, indicating that blockade of KIAA1363 slows, but does not completely block tumor progression. This outcome was not due to incomplete inhibition of KIAA1363, as we were able to confirm full inactivation of KIAA1363 in explanted tumors from JW480-treated mice. In future studies, it would be interesting to test whether KIAA1363 inhibitors show additive or synergistic anti-tumor activity when combined with other chemotherapeutic agents. Also, the strong anti-invasive effects of JW480 suggest that blockade of KIAA1363 could impede cancer metastasis in vivo. Finally, more extensive biochemical and cell biological studies are required to understand the mechanism by which KIAA1363-MAGE pathway supports prostate cancer pathogenicity. Previous work showed that this pathway is coupled to the production of pro-tumorigenic lipids, such as alkyl-LPA, in ovarian cancer cells (Chiang et al., 2006). Broader metabolomic experiments should reveal whether changes in LPA or other bioactive lipids are also observed in JW480-treated prostate cancer cells.
In closing, we believe that JW480 possesses an impressive array of features that qualify it as a frontline pharmacological probe for KIAA1363, including high potency against both the human and mouse orthologues of this enzyme, minimal cross-reactivity with other serine hydrolases, and excellent activity in living cells and mice. We anticipate that future studies with JW480 will help to illuminate the role that KIAA1363 plays in many (patho)physiological processes, including cancer, macrophage biology, and the nervous system. From a methodological perspective, our success in converting lead carbamates originating from a large-scale screen (Bachovchin et al., 2010) into a KIAA1363 inhibitor that displays greatly improved potency and selectivity can be attributed, at least in part, to the information content garnered by competitive ABPP, which assays inhibitors against numerous enzymes in parallel directly in native proteomes. This type of “proteomic medicinal chemistry”, which has also impacted other inhibitor development programs (Arastu-Kapur et al., 2008; Deu et al.; Li et al., 2007; Long et al., 2009a; Staub and Sieber, 2009), should continue to provide an efficient means to create versatile pharmacological probes for a wide range of enzymes.
Significance
Mapping dysregulated metabolic pathways in cancer cells has the potential to uncover biochemical mechanisms that support tumorigenesis and identify new drug targets. The serine hydrolase KIAA1363 has been found to be highly elevated in aggressive cancer cells from multiple tumors of origin, where it plays a role in ether lipid metabolism. Attempts to further characterize KIAA1363 function in cancer have, however, been thwarted by a lack of pharmacological tools to study this enzyme. Here, we have addressed this challenge by creating a potent, selective, and in vivo-active inhibitor of KIAA1363. Key to the development of this compound (termed JW480) was the use of competitive activity-based profiling assays to simultaneously refine potency for KIAA1363 and selectivity across the larger serine hydrolase class. JW480 reduces monoalkylglycerol ether levels in androgen-independent prostate cancer cells and impairs their migration, invasion, survival, and in vivo tumor growth. These studies thus support a pro-tumorigenic function for KIAA1363 and report a versatile inhibitor for pharmacological characterization of this enzyme in living systems.
Experimental Procedures
Preparation of Mouse Tissue Proteomes
Tissues were Dounce-homogenized in PBS, pH 7.4, followed by a low-speed spin (1,400 g, 5 min) to remove debris. The supernatant was then subjected to centrifugation (100,000 × g, 45 min) to provide the cytosolic fraction in the supernatant and the unsoluble fraction as a pellet. The pellet was washed and resuspended in PBS buffer by sonication. Total protein concentration in each fraction was determined using a protein assay kit (Bio-Rad). Samples were stored at −80 °C until use.
Inhibitor treatments of cells
Inhibitors were dissolved in DMSO and diluted into media or buffer prior to cell or proteome treatment, respectively. For in vitro treatment, final DMSO concentration was 4%. For in situ treatments of cells for ABPP, lipid measurements, migration, cell-survival, and invasion assays, 2 × 106 cells were seeded in 6 cm dishes (~100% confluency) 24 h prior to inhibitor pre-treatment (in DMSO at 0.1 % final concentration) in serum-free media (3 mL) for the designated time before harvesting cells for ABPP or lipid measurements or before the initiation of migration, cell-survival, and invasion assays.
2-Acetyl MAGE Hydrolytic Activity Assays
Cells were pretreated with KIAA1363 inhibitors in situ (for 4 hr in serum-free F-12K media before harvesting cells by scraping) or in vitro (for 30 min at 37 °C in PBS) before addition of 2-acetyl MAGE (100 μM) to cell lysates at room temperature for 30 min in a volume of 200 μL. Reactions were quenched with 600 μL 2:1 chloroform:methanol and 10 nmol of C12:0 MAGE internal standard was added. The organic layer was extracted and 30 μL injected into an Agilent 1100-MSD LC-MS. LC-MS settings were as previously described (Chiang et al., 2006). Product levels (C16:0 MAGE) were quantified in relation to the internal standard. Specific activity was determined during the linear phase of enzymatic reactions (i.e., less than 20% substrate utilized).
AChE Activity Assays
AChE activity was measured using a method similar to that described previously (Ellman et al., 1961). Briefly, 50 μL of 10 mM acetylthiocholine was added to 200 μL of PBS containing 2 mM DTNB and 20 μg of cell lysate or mouse brain membrane proteome. Absorbance was measured at 412 nm over 5 min, and the rate of product accumulation was calculated from the slope of the absorbance over time. For assays involving preincubation with inhibitors, the reactions were prepared without acetylthiocholine and JW480 was incubated at the indicated concentration for 30 min at 37 °C. Acetylthiocholine was then added and the assay was carried out exactly as described above.
Competitive ABPP Experiments
For ABPP experiments, cell lysate and tissue proteomes were treated with 1 μM FP-rhodamine for 30 min at room temperature (50 μL total reaction volume) as described previously (Nomura et al., 2010). Reactions were quenched with one volume of standard 4x SDS/PAGE loading buffer (reducing), separated by SDS/PAGE (10% acrylamide), and visualized in-gel with a Hitachi FMBio IIe flatbed fluorescence scanner (MiraiBio). Inhibitors were preincubated prior (30 min or 4 h at 37 °C in vitro or in situ, respectively) to the addition of FP-rhodamine.
Competitive ABPP-MudPIT Analysis of Serine Hydrolase Activities in Proteomes
Competitive ABPP-MudPIT experiments were performed as previously described (Nomura et al., 2010). Briefly, 1 mg of proteome was labeled with 5 μM FP-biotin, followed by solubilization with 1% Triton X-100, denaturation by SDS and heating, avidin precipitation of labeled proteins, and on-bead tryptic digest. Tryptic peptides were then loaded on to a biphasic (strong cation exchange/reverse phase) capillary column and analyzed by two-dimensional liquid chromatography (2D-LC) separation in combination with tandem mass spectrometry using an Agilent 1100 LC system coupled with a ThermoFisher LTQ linear ion trap mass spectrometer. Spectral counts are reported as the average of three samples with the standard error of the mean (SEM).
Generation of KIAA1363 Knockdown Cells
Stable knockdown of KIAA1363 in PC3 cells was achieved using a targeted short-hairpin oligonucleotide described previously (Chiang et al., 2006).
Cell Migration, Cell Survival, and Invasion Studies
Cell migration, cell survival, and invasion studies were performed as previously described (Nomura et al., 2010). Briefly, migration assays were performed in Transwell chambers (Corning) with 8 μm pore-sized membranes coated with 10 μg/mL collagen at 37 °C for LNCaP (24 h), PC3 (4 h) and DU145 cells (24 h), respectively. Cell survival assays were performed using the Cell Proliferation Reagent WST-1 (Roche) Invasion assays were conducted using the BD Matrigel Invasion Chambers per the manufacturer’s protocol. Inhibitors were preincubated for the stated duration before seeding cells into migration, cell-survival, or invasion chambers. Prior to seeding the cells in these chambers, cells were serum-starved for 4 h. Inhibitors were also present during the migration, cell-survival, and invasion assays.
In Vivo Studies with JW480
JW480 was administered by oral gavage (in PEG300, 4 μL/g mouse) or intraperitoneally (in 18:1:1 v/v/v solution of saline:ethanol:emulphor, 10 μL/g). After the indicated amount of time, mice were anesthetized with isoflurane and killed by decapitation. Tissues were removed and then flash frozen in liquid N2. Tissues were stored at −80 °C until use. Animal experiments were conducted in accordance with the guidelines of the institutional Animal Care and Use Committee of The Scripps Research Institute.
Tumor Xenograft Studies
Human cancer xenografts were established by transplanting cancer cell lines ectopically into the flank of C.B17 SCID mice (Taconic Farms). Briefly, cells were washed two times with PBS, trypsinized, and harvested in serum-containing medium. Next, the harvested cells were washed two times with serum-free medium and resuspended at a concentration of 2.0 × 104 cells/μL and 100 μL was injected. Growth of the tumors was measured every 3 days with calipers. For chronic JW480 treatment studies, mice were treated with JW480 or vehicle once daily (at approximately the same time everyday) by oral gavage in PEG300 (4 μL/g mouse). The treatments were initiated immediately after ectopic injection of cancer cells.
Lipid Measurements in Cancer Cells
Lipid measurements were performed in cancer cells as previously described (Chiang et al., 2006). Briefly, frozen cell pellets from cells harvested after 4 h serum starvation were extracted in 2:1:1 chloroform:methanol:Tris buffer pH 8.0 by dounce homogenization with 10 nmol of internal standard C12 MAGE. The organic layer was removed, dried under N2, and resuspended in 120 μL of chloroform, and 30 μL was injected into an Agilent 1100-MSD LC-MS. MAGE levels were quantified by measuring the area under the peak and were normalized to the C12 MAGE internal standard.
Supplementary Material
Highlights.
KIAA1363 is a hydrolytic enzyme highly expressed in aggressive cancer cells
JW480 is a potent, selective, and in vivo-active inhibitor of KIAA1363
JW480 disrupts neutral ether lipid metabolism in cancer cells
JW480 impairs cancer cell migration and in vivo tumor growth
Acknowledgments
We thank the Cravatt lab and, in particular Alexander Adibekian and Jonathan Long, for helpful discussions and critical reading of the manuscript. This work was supported by the NIH [CA087660, DA025285 (B.F.C.), DA030908 (D.K.N.)], an American Cancer Society Postdoctoral Fellowship (D.K.N.), a California Institute for Regenerative Medicine Predoctoral Fellowship (J.W.C.), and the Skaggs Institute for Chemical Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn K, Johnson DS, Mileni M, Beidler D, Long JZ, McKinney MK, Weerapana E, Sadagopan N, Liimatta M, Smith SE, et al. Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem Biol. 2009;16:411–420. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert DH, Anderson CE. Ether-linked glycerolipids in human brain tumors. Lipids. 1977;12:188–192. doi: 10.1007/BF02533292. [DOI] [PubMed] [Google Scholar]
- Alexander JP, Cravatt BF. Mechanism of carbamate inactivation of FAAH: implications for the design of covalent inhibitors and in vivo functional probes for enzymes. Chem Biol. 2005;12:1179–1187. doi: 10.1016/j.chembiol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arastu-Kapur S, Ponder EL, Fonovic UP, Yeoh S, Yuan F, Fonovic M, Grainger M, Phillips CI, Powers JC, Bogyo M. Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nat Chem Biol. 2008;4:203–213. doi: 10.1038/nchembio.70. [DOI] [PubMed] [Google Scholar]
- Bachovchin DA, Ji T, Li W, Simon GM, Hoover H, Niessen S, Cravatt BF. A superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc Natl Acad Sci U S A. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AB, Vitorino PM, Bogyo M. Activity-based protein profiling: applications to biomarker discovery, in vivo imaging and drug discovery. Am J Pharmacogenomics. 2004;4:371–381. doi: 10.2165/00129785-200404060-00004. [DOI] [PubMed] [Google Scholar]
- Blank ML, Smith ZL, Cress EA, Snyder F. Characterization of the enzymatic hydrolysis of acetate from alkylacetylglycerols in the de novo pathway of PAF biosynthesis. Biochim Biophys Acta. 1990;1042:153–158. doi: 10.1016/0005-2760(90)90001-e. [DOI] [PubMed] [Google Scholar]
- Blum G, Mullins SR, Kernen K, Fonovic M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nat Chem Biol. 2005;1:203–209. doi: 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- Buchebner M, Pfeifer T, Rathke N, Chandak PG, Lass A, Schreiber R, Kratzer A, Zimmermann R, Sattler W, Koefeler H, et al. Cholesteryl ester hydrolase activity is abolished in HSL−/− macrophages but unchanged in macrophages lacking KIAA1363. J Lipid Res. 2010;51:2896–2908. doi: 10.1194/jlr.M004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casida JE, Quistad GB. Serine hydrolase targets of organophosphorus toxicants. Chem Biol Interact. 2005:157–158. 277–283. doi: 10.1016/j.cbi.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Deu E, Leyva MJ, Albrow VE, Rice MJ, Ellman JA, Bogyo M. Functional studies of Plasmodium falciparum dipeptidyl aminopeptidase I using small molecule inhibitors and active site probes. Chem Biol. 17:808–819. doi: 10.1016/j.chembiol.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellman GL, Courtney KD, Andres V, Jr, Feather-Stone RM. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- Ferguson DA, Muenster MR, Zang Q, Spencer JA, Schageman JJ, Lian Y, Garner HR, Gaynor RB, Huff JW, Pertsemlidis A, et al. Selective identification of secreted and transmembrane breast cancer markers using Escherichia coli ampicillin secretion trap. Cancer Res. 2005;65:8209–8217. doi: 10.1158/0008-5472.CAN-04-3726. [DOI] [PubMed] [Google Scholar]
- Haverty PM, Hon LS, Kaminker JS, Chant J, Zhang Z. High-resolution analysis of copy number alterations and associated expression changes in ovarian tumors. BMC Med Genomics. 2009;2:21. doi: 10.1186/1755-8794-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoosein NM, Boyd DD, Hollas WJ, Mazar A, Henkin J, Chung LW. Involvement of urokinase and its receptor in the invasiveness of human prostatic carcinoma cell lines. Cancer Commun. 1991;3:255–264. doi: 10.3727/095535491820873146. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, et al. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi M, Osuga J, Uozaki H, Sekiya M, Nagashima S, Takahashi M, Takase S, Takanashi M, Li Y, Ohta K, et al. The critical role of neutral cholesterol ester hydrolase 1 in cholesterol removal from human macrophages. Circ Res. 2010;107:1387–1395. doi: 10.1161/CIRCRESAHA.110.226613. [DOI] [PubMed] [Google Scholar]
- Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer invasiveness. Proc Natl Acad Sci USA. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- Kathuria S, Gaetani S, Fegley D, Valino F, Duranti A, Tontini A, Mor M, Tarzia G, La Rana G, Calignano A, et al. Modulation of anxiety through blockade of anandamide hydrolysis. Nat Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- Krishnasamy S, Teng AL, Dhand R, Schultz RM, Gross NJ. Molecular cloning, characterization, and differential expression pattern of mouse lung surfactant convertase. Am J Physiol. 1998;275:L969–975. doi: 10.1152/ajplung.1998.275.5.L969. [DOI] [PubMed] [Google Scholar]
- Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat Biotechnol. 2003;21:687–691. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- Li W, Blankman JL, Cravatt BF. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. J Am Chem Soc. 2007;129:9594–9595. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- Lin HJ, Ho FC, Lee CL. Abnormal distribution of O-alkyl groups in the neutral glycerolipids from human hepatocellular carcinomas. Cancer Res. 1978;38:946–949. [PubMed] [Google Scholar]
- Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavon FJ, Serrano AM, Selley DE, Parsons LH, et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol. 2009a;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Nomura DK, Cravatt BF. Characterization of monoacylglycerol lipase inhibition reveals differences in central and peripheral endocannabinoid metabolism. Chem Biol. 2009b;16:744–753. doi: 10.1016/j.chembiol.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki H, Igarashi M, Nishi M, Sekiya M, Tajima M, Takase S, Takanashi M, Ohta K, Tamura Y, Okazaki S, et al. Identification of neutral cholesterol ester hydrolase, a key enzyme removing cholesterol from macrophages. J Biol Chem. 2008;283:33357–33364. doi: 10.1074/jbc.M802686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteome using fluorescent active site-directed probes. Proteomics. 2001;1:1067–1071. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Roos DS, Choppin PW. Tumorigenicity of cell lines with altered lipid composition. Proc Natl Acad Sci U S A. 1984;81:7622–7626. doi: 10.1073/pnas.81.23.7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J Biol Chem. 2010;285:11051–11055. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder F, Wood R. Alkyl and alk-1-enyl ethers of glycerol in lipids from normal and neoplastic human tissues. Cancer Res. 1969;29:251–257. [PubMed] [Google Scholar]
- Staub I, Sieber SA. Beta-lactam probes as selective chemical-proteomic tools for the identification and functional characterization of resistance associated enzymes in MRSA. J Am Chem Soc. 2009;131:6271–6276. doi: 10.1021/ja901304n. [DOI] [PubMed] [Google Scholar]
- Wood R, Snyder F. Characterization and identification of glyceryl ether diesters present in tumor cells. J Lipid Res. 1967;8:494–500. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.