Abstract
Protein-protein interaction is one of the key regulatory mechanisms for controlling protein function in various cellular processes. Chemical cross-linking coupled with mass spectrometry has proven to be a powerful method not only for mapping protein-protein interactions of all natures including weak and transient ones, but also for determining their interaction interfaces. One critical challenge remaining in this approach is how to effectively isolate and identify cross-linked products from a complex peptide mixture. In this work, we have developed a novel strategy using conjugation chemistry for selective enrichment of cross-linked products. An azide-tagged cross-linker along with two biotinylated conjugation reagents were designed and synthesized. Cross-linking of model peptides and cytochrome c as well as enrichment of the resulting cross-linked peptides has been assessed. Selective conjugation of azide-tagged cross-linked peptides has been demonstrated using two strategies: copper catalyzed cycloaddition and Staudinger ligation. While both methods are effective, Staudinger ligation is better suited for enriching the cross-linked peptides since there are fewer issues with sample handling. LC MSn analysis coupled with database searching using the Protein Prospector software package allowed identification of fifty-eight cytochrome c cross-linked peptides after enrichment and affinity purification. The new enrichment strategy developed in this work provides useful tools for facilitating identification of cross-linked peptides in a peptide mixture by MS, thus presenting a step forward in future studies of protein-protein interactions of protein complexes by cross-linking and mass spectrometry.
INTRODUCTION
Proteins form stable and/or dynamic multi-subunit protein complexes under different physiological conditions to maintain cell viability and normal cell homeostasis. A thorough understanding of protein interactions and structures of protein complexes is fundamental to the understanding of protein function and regulation. Chemical cross-linking coupled with mass spectrometry (MS) is a powerful method that can be used to study protein-protein interactions [1–6]. The unique capability of chemical cross-linking to stabilize protein interactions through covalent bond formation allows not only the structural elucidation [7–17], but also the detection of stable, weak and/or transient protein-protein interactions in native cells or tissues [18–25].
In addition to capturing protein interacting partners, many studies have shown that chemical cross-linking can yield low-resolution structural information about the constraints within a molecule [2–4]. Traditional methods such as NMR analysis and X-ray crystallography can yield detailed information on protein structure, however NMR spectroscopy requires large quantities of pure protein in a specific solvent, and X-ray crystallography is often limited by the crystallization process. The combination of chemical cross-linking, enzymatic digestion, and subsequent mass spectrometric analysis for the elucidation of three dimensional protein structures offers distinct advantages over these traditional methods due to its speed, sensitivity, and versatility [2–4]. This strategy has been successfully employed for unraveling protein complex topology and protein-protein interacting interface [7–17, 26].
Due to the inherent complexity in cross-linking reactions and the abundance of non-cross-linked peptides, detection of cross-linked peptides in a complex mixture is quite challenging [2, 4]. The digested mixture of cross-linked protein complexes typically contains four types of peptides: 1) unmodified peptides of the complex components, 2) dead end modified peptides, 3) intramolecular (inter-linked and intra-linked) cross-linking products between peptides originated from one protein, and 4) intermolecular cross-linking products (inter-linked) between peptides from different proteins [3, 8]. In order to facilitate the identification process, various methods using biotinylated [27–31], isotope-labeled [32–34], fluorescently labeled [35, 36], or mass-tag labeled cross-linking reagents [28, 37] have been reported. Among them, biotinylated cross-linkers are advantageous due to their specificity and efficiency. However, these linkers have the drawback of being bulky and therefore not very effective or ideal for in vivo protein cross-linking [4, 38]. Recently, new strategies have also been developed based on click chemistry with chemical conjugation of azide and alkyne groups using alkyne-tagged [39] or azide tagged [40, 41] cross-linkers. Applications using click chemistry for enrichment of other types of biomolecules have also been reported [42–49]. The marked advantage of this approach is the complete orthogonality to functional groups in normal biomolecules since azides and alkynes are not present in peptides, proteins, nucleic acids or polysaccharides, and are very rare in all other biological materials and natural products. With the absence of significant competing reactions [43], click chemistry shows great promise for protein cross-linking studies.
Although both alkyne-tagged [39] and azide-tagged cross-linkers [41] can be used for the same purpose, azide-tagged reagents have some advantages that would be preferred. Azides present a high chemical stability under physiological conditions and unlike alkynes, their reactivity enables several types of mild and selective organic transformations, including Cu(I)-catalyzed [3+2] cycloaddition [42, 50], strain-promoted [3+2]-cycloaddition [49, 51, 52], and Staudinger ligation [53–55]. The Staudinger ligation has been widely used in biological applications, especially for selective enrichment of chemically modified peptides [56–61], but its applicability in the study of cross-linked peptides has not been evaluated. Due to complications in sample handling using copper-catalyzed click chemistry and challenges in synthesizing strained alkynes, alternative strategy using Staudinger ligation may be advantageous for isolation of cross-linked peptides. In this work, we have explored azide-based conjugation chemistry using Staudinger ligation and developed a new enrichment strategy for cross-linked peptides using a newly designed azide-tagged cross-linker and biotin-phosphine. These new reagents not only allow efficient cross-linking of model peptides and cytochrome c and subsequent enrichment of cross-linked peptides, but also provide diagnostic ions in MS/MS analysis to facilitate MS identification of cross-linked peptides with high confidence. In combination with various software tools (i.e. Batch-tag, MS-bridge, MS-product and Search compare) in Protein Prospector [62], LC MS/MS analyses allowed identification of fifty-eight enriched cytochrome c cross-linked peptides.
MATERIALS AND METHODS
Materials and Reagents
All general chemicals for buffers were purchased from Fisher Scientific or VWR International. Bovine heart cytochrome c (98% purity) was purchased from Sigma-Aldrich. Ac-myelin peptide (Ac-ASQKRPSQRHG, 92.7% purity) was purchased from American Peptide (Sunnyvale, CA) and IR-7 peptide (Ac-IEAEKGR, 98.1% purity) was synthesized by GL Biochem (Shanghai, China).
Synthesis and characterization of Azide-DSG, biotin-alkyne and biotin-phosphine
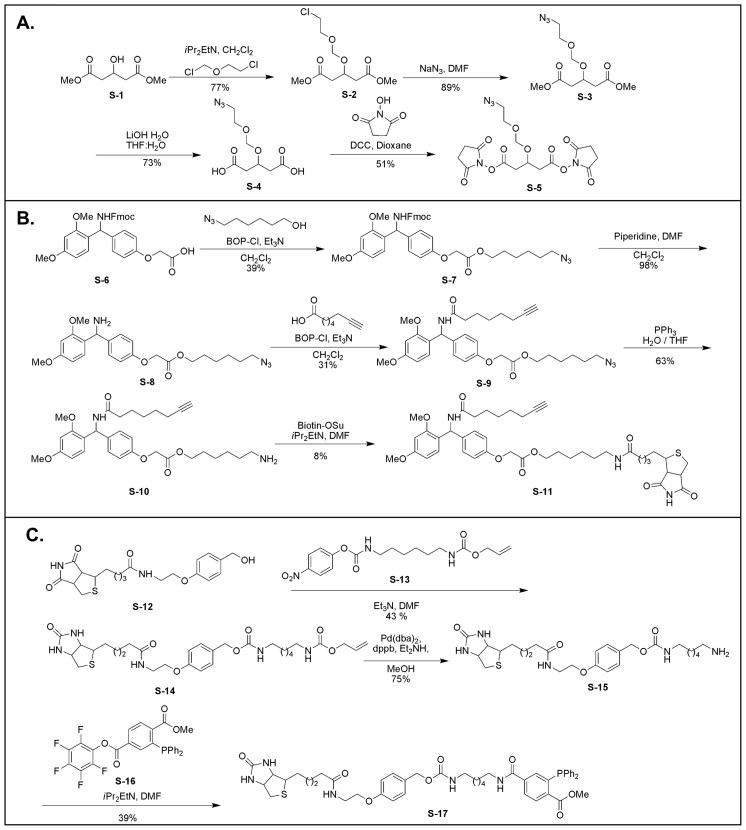
The synthesis schemes for Azide-DSG (S-5) (spacer length 7.7 Å), biotin-alkyne (S-11) and biotin-phosphine (S-17) are displayed in Fig. 1. As shown, Azide-DSG was synthesized from commercially available dimethyl 1,3-acetonedicaboxylate (S-1) in four steps. Biotin-alkyne was synthesized from S-6 in five steps. Biotin-phosphine was synthesized from S-12 in three steps based on Bertozzi’s protocols [53, 63]. The details in synthesis and chemical characterization of each product have been summarized in Supplemental methods.
Figure 1.
Synthetic schemes of A) Azide-DSG (S-5); B) Biotin-alkyne (S-11); C) Biotin-Phosphine (S-17).
Cross-linking of synthetic peptides with Azide-DSG
A solution of tryptic-like synthetic peptides Ac-myelin or Ac-IR7 (0.31 μmol) in DMF (400 μL) was mixed with 20 μl 8 mmole/μL Azide-DSG cross-linker in DMF, and the reaction mixture was treated with 20 μl 8 mmole/μL diisopropylethylamine in DMF. The reaction mixture was stirred for 16 h at ambient temperature. The solution was then dried by SpeedVac and resuspended in PBS buffer or 20% ACN.
Cross-linking of Cytochrome C with Azide-DSG
200 μM bovine cytochrome c in 1× PBS (pH 7.5) was reacted with Azide-DSG in DMSO at 20:1 (linker:protein) molar ratio and incubated at room temperature for one hour with agitation. After quenching with glycine, it was then digested with 1% trypsin (w/w) overnight at 37°C prior to enrichment.
Enrichment of azide-tagged cross-linked peptides using copper catalyzed click chemistry
A solution of Azide-DSG cross-linked Ac-myelin peptide (20 μL, 2.6 ×10−8 mol) in PBS buffer was treated with 20 μL of a 13 mM solution of biotin-alkyne in DMSO. Subsequently, 40 μL of 6.5 mM CuSO4•5H2O, 4 μL of 193 mM sodium ascorbate, and 8 μL of 6.3 mM TBTA solution were added [64]. The reaction mixture was gently stirred for 18 h. The solution was then desalted by a C18 ZipTip, dried by SpeedVac, and resuspended in 1.2 mL of 4% ACN/0.1% formic acid. Following desalting, 5 μL of the mixture was incubated with 400 μL of Dynabeads® M-280 Streptavidin (Invitrogen). After 30 min incubation, the beads were washed with PBS buffer and 20% ACN. Peptides were eluted from the beads with 90% TFA. The TFA solution was dried by SpeedVac and resuspended in 4 % ACN/0.1 % formic acid for analysis.
Enrichment of azide-tagged cross-linked peptides using Staudinger ligation
Staudinger ligation was performed with a similar procedure as described [53]. Briefly, 10 μL of Azide-DSG cross-linked Ac-myelin peptide (7.73 × 10−9 mol) was treated with 7 μL of a 25 μM stock solution of biotin-phosphine (S-17 in Fig. 1) in DMSO. The reaction mixture was heated to 60 °C. After 16 h, 1.5 μL of the reaction mixture was incubated with 200 μL of Dynabeads® M-280 Streptavidin (Invitrogen). After 30 min, the beads were washed with 200 μL of 20% ACN, 200 μL of PBS buffer, and 200 μL of 25% isopropanol. The peptides were then eluted with 85% TFA. The TFA solution was dried by SpeedVac and the sample was resuspended in 4% ACN/0.1% formic acid for LC MS analysis.
LC MSn analysis
LC MSn analysis of cross-linked peptides was carried out using an LTQ-Orbitrap XL MS (Thermo Scientific, San Jose, CA) coupled with an Eksigent NanoLC system (Eksigent, Dublin, CA). The LC analysis was performed using a capillary column (100 μm ID × 150 mm long) packed with C18 resins (GL Sciences, Torrance, CA) and the peptides were eluted using a linear gradient of 2–40 % B in 35 min; (solvent A: 100 % H2O/0.1 % formic acid; solvent B: 100 % ACN/0.1 % formic acid). For LC MS/MS analysis, a cycle of one full FT MS scan mass spectrum (350–1800 m/z, resolution of 60,000 at m/z 400) was followed by ten data-dependent MS/MS acquired in the linear ion trap with normalized collision energy (setting of 35%). Target ions selected for MS/MS were dynamically excluded for 30 s. For LC MS3 analysis, one full FT MS scan was followed by two cycles of one MS/MS scan and three MS3 scans acquired in the linear ion trap. MS3 scan was operated in data-dependent mode to allow sequencing the top three most abundant fragment ions observed in a MS/MS spectrum with exclusion of 447.2, 508.2 and 520.2 ions, which are diagnostic ions of cross-linked peptides due to fragmentation in the conjugated cross-linker region in MS/MS.
Identification of cross-linked peptides by database searching
Monoisotopic masses of parent ions and corresponding fragment ions, parent ion charge states and ion intensities from LC MS/MS spectra were extracted using in-house software based on Raw_Extract script from Xcalibur v2.4 (Thermo Scientific). Database searching was performed with a developmental version of Protein Prospector (v. 5.3.2., http://proteinprospector.ucsf.edu) using its software suite, i.e. Batch-tag, MS-bridge, MS-product, MS-tag and Search Compare, similarly as described [62]. It has been shown that Batch-tag with mass modification search can be applied for identifying cross-linked peptides [62]. Therefore, the MS/MS data were first searched using Batch-Tag with mass modification against bovine cytochrome c sequence (SwissProt accession #: P62894). Mass modification up to 4000 Da was set on uncleaved lysine residues and protein free N-terminus. The mass accuracy for parent ions and fragment ions were set as ± 0.25 Da and 0.8 Da, respectively. The other search parameters included trypsin as the enzyme, two maximum missed cleavages, and three maximum variable modifications. Variable modifications chosen were protein N-terminal acetylation, methionine oxidation, and N-terminal conversion of glutamine to pyroglutamic acid. For all of the ions being sequenced in MS/MS, their parent monoisotopic masses and charges were extracted from Search Compare and submitted to MS-bridge program to identify putative cross-linked peptides by mass fitting against theoretical masses of cross-linked cytochrome c peptides with the given cross-linker. The parent mass error for MS-bridge search was set as ± 20 ppm and only one bridge was allowed in the cross-linked peptides. All of the three types of cross-linked peptides [8], i.e. inter-linked (type 2), intra-linked (type 1) and dead-end modified (type 0) peptides, can be computed and matched in MS-bridge. MS-tag was used to identify the additional peptide sequence in the inter-linked peptides, and MS-product was applied to validate the identified sequences. Due to difficulties in the analysis of the complex fragmentation of cross-linked peptides by automated database searching, MS3 spectra interpretation was performed manually.
RESULTS AND DISCUSSION
Selective isolation of cross-linked peptides
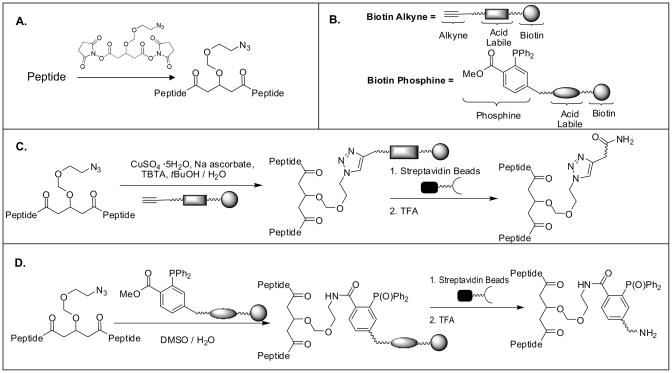
In this work, a new cross-linking and enrichment strategy was developed to enable effective isolation of cross-linked peptides from a complex mixture. Fig. 2 shows the overview of the cross-linking reaction and enrichment protocols. A novel azide-tagged NHS ester based cross-linker (i.e. Azide-DSG) was designed and synthesized (S-5 in Fig. 1A) such that the azide tag could be used as a chemical handle to allow selective isolation of azide-tagged cross-linked peptides using azide-based conjugation chemistry including azide-alkyne cycloaddition and Staudinger ligation chemistry [42, 49–55]. Azide-DSG was chosen for this work due to its desirable water solubility, spacer length, membrane permeability, expected stability, and ease of synthesis. To determine the chemistry best suited for enrichment of cross-linked peptides from complex mixtures, we have designed and synthesized two new biotinylated conjugation reagents (S-11 and S-17 in Figs. 1B & C) for enrichment strategies based on either copper catalyzed click chemistry (Fig. 2C) or Staudinger ligation (Fig. 2D) and their effectiveness has been evaluated using model peptides as described below.
Figure 2.
Schematic overview of general cross-linking and enrichment strategies. A) Cross-linking of a peptide using Azide-DSG; B) Cartoon structures of biotin-alkyne and biotin-phosphine (see detailed structures in Fig. 1); Enrichment strategies based on C) copper catalyzed click chemistry; D) Staudinger ligation.
1). Enrichment of azide-tagged cross-linked peptides using copper catalyzed click chemistry
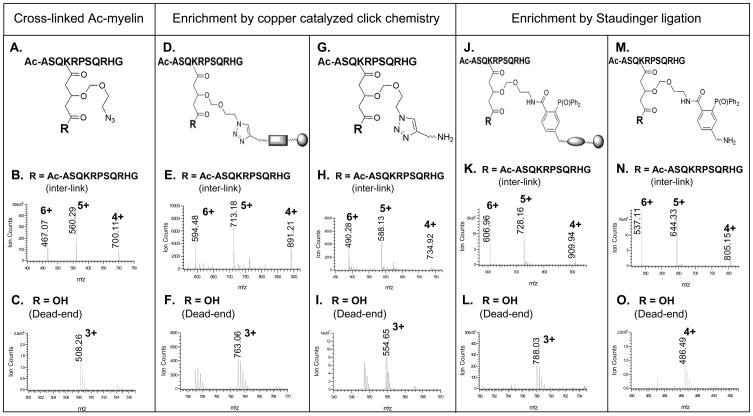
Click chemistry based strategy (Fig. 2C) was first evaluated using a model peptide Ac-myelin. Cross-linking of Ac-myelin was carried out in organic solvents with about equal molar amount of Azide-DSG and the resulting products were analyzed by LC MS/MS. The cross-linking reaction appeared to be highly effective as no unmodified peptide was detected in MS spectra after the reaction. Only two types of cross-linking products, i.e. inter-linked (type 2) and dead-end (type 0) modified peptides, were detected with the inter-linked product as the major product under our experimental conditions. This can be attributed to N-acetylation of Ac-myelin, one lysine residue in the sequence, and the limited hydrolysis of the cross-linker. The structural illustration and MS spectra of the azide-tagged cross-linked products of Ac-myelin are displayed in Figs. 3A–C. A series of ions ([M+6H]6+ 467.07, [M+5H]5+ 560.29, [M+4H]4+ 700.11) were detected as the Ac-myelin homodimer and a triply charged ion ([M+3H]3+ 508.26) was measured as the dead-end modified Ac-myelin. The mass addition to the cross-linked peptides was 211 Da for the cross-link (inter- or intra-link) and 229 Da for the dead-end modification. To isolate the cross-linked peptides, the azide-tagged cross-linked Ac-myelin products were subjected to conjugation with a biotin-alkyne (S-11) (Fig. 1B). An acid cleavable site was incorporated into the new biotin-alkyne in order to allow subsequent elution after streptavidin-biotin binding during affinity purification of enriched cross-linked peptides. Upon cycloaddition, the conjugated products containing a triazole was formed for both inter-linked and dead-end modified Ac-myelin peptides and measured by MS analysis as displayed in Figs. 3D–F. The azide-alkyne cycloaddition reaction was very efficient since no detectable non-conjugated cross-linked peptides were detected. After conjugation, the cycloaddition products were isolated by binding to magnetic streptavidin beads followed by TFA cleavage (Fig. 2C). The structural illustration and MS spectra of final Ac-myelin cross-linked products are shown in Figs. 3G–I. A series of ions ([M+6H]6+ 490.28, [M+5H]5+ 588.13, [M+4H]4+ 734.92) were observed for interlinked Ac-myelin, and a triply charged ion (MH33+ 554.65) was detected for the dead-end product. Although isolation of azide-tagged Ac-myelin cross-linked products using copper catalyzed click chemistry was successful, we found that addition of CuSO4 and sodium ascorbate to the reaction mixture often caused peptide precipitation, leading to problems in sample handling and sample losses. In addition, it has been noted that copper (I) may induce peptide oxidation and thus hamper identification of the cross-linked peptides by MS [41].
Figure 3.
Enrichment of Azide-DSG cross-linked Ac-myelin peptides by chemical conjugation. A–C) Cross-linked products of Ac-myelin prior to enrichment: A) structural illustration of cross-linked peptides, R represents either an additional peptide for inter-linked or a hydroxyl group for a dead-end modified Ac-myelin peptides; MS spectra of B) inter-link (observed: [M+6H]6+ 467.07, [M+5H]5+ 560.29, [M+4H]4+ 700.11; theoretical: [M+6H]6+ 467.07, [M+5H]5+ 560.29, [M+4H]4+ 700.11) and C) dead-end (observed: [M+3H]3+ 508.26; theoretical: [M+3H]3+ 508.25) modified Ac-myelin. D–I) Cross-linked products of Ac-myelin after copper catalyzed click chemistry (D–F), and affinity purification/TFA cleavage (H–I): D) structural illustration of conjugated products after click chemistry; MS spectra of E) inter-linked (observed: [M+6H]6+ 594.48, [M+5H]5+ 713.18, [M+4H]4+ 891.21; theoretical: [M+6H]6+ 594.47, [M+5H]5+ 713.16, [M+4H]4+ 891.20;) and F) dead-end (observed: [M+3H]3+ 763.06; theoretical: [M+3H]3+ 763.05) products; G) structural illustration of final cross-linked products after affinity purification and TFA cleavage; MS spectra of H) inter-linked (observed: [M+6H]6+ 490.28, [M+5H]5+ 588.13, [M+4H]4+ 734.92; theoretical: [M+6H]6+ 490.26, [M+5H]5+ 588.11, [M+4H]4+ 734.88;) and I) dead-end (observed: [M+3H]3+ 554.65; theoretical: [M+3H]3+ 554.62) modified peptides. J–O) Cross-linked products of Ac-myelin after Staudinger ligation (J–L) and affinity purification/TFA cleavage (M–O): J) structural illustration of Staudinger ligation conjugates; MS spectra of E) inter-linked (observed: [M+6H]6+ 609.96, [M+5H]5+ 728.16, [M+4H]4+ 909.94; theoretical: [M+6H]6+ 609.96, [M+5H]5+ 728.15, [M+4H]4+ 909.94) and F) dead-end (observed: [M+3H]3+ 788.03; theoretical: [M+3H]3+ 788.03) products; M) structural illustration of the final cross-linked peptides after affinity purification and TFA cleavage; MS spectra of N) inter-linked (observed: [M+6H]6+ 537.11, [M+5H]5+ 644.33, [M+4H]4+ 805.15; theoretical: [M+6H]6+ 537.10, [M+5H]5+ 644.32, [M+4H]4+ 805.15) and O) dead-end (observed: [M+4H]4+ 486.49; theoretical: [M+4H]4+ 486.49) modified peptides. Note: E, F, H and I were acquired by QSTAR MS. The rest of MS spectra were acquired by LTQ-Orbitrap XL MS.
2). Enrichment of azide tagged cross-linked peptides using Staudinger ligation
To avoid the problems associated with copper catalyzed click reaction, we have investigated a strategy based on Staudinger ligation using a biotin-phosphine with an acid cleavage site (S-17) (Fig. 1C). The Azide-DSG cross-linked Ac-myelin products were then subjected to Staudinger ligation and the resulting conjugates were affinity purified by binding to magnetic streptavidin beads followed by TFA cleavage. The products at each step were monitored by LC MS/MS. Optimization of reaction temperature was carried out by comparing LC MS signal intensity of the cross-linked and non-cross-linked Ac-myelin before and after the reaction. All of the cross-linked Ac-myelin can be conjugated with biotin-phosphine even at room temperature since no unconjugated Ac-myelin cross-linking product was detected, similar to that of copper catalyzed click reaction. However the conversion rate appears to be much faster at elevated temperature (~60°C), and this could be advantageous for real protein samples with limited concentrations to obtain efficient conjugation. The structural illustrations and MS spectra of Staudinger products of cross-linked Ac-myelin before and after affinity purification/TFA cleavage are displayed in Figs. 3J–O. As shown, a series of multiply charged ions ([M+6H]6+ 606.96, [M+5H]5+ 728.16, [M+4H]4+, 909.94) were detected as the conjugate of the inter-linked Ac-myelin (Fig. 3K). TFA cleavage of the affinity purified conjugates resulted in a loss of 419.15 Da, and the final product of inter-link Ac-myelin was measured as [M+6H]6+ 537.11, [M+5H]5+ 644.33, and [M+4H]4+, 805.15 (Fig. 3N). Similarly, Staudinger products of the dead-end Ac-myelin before and after TFA cleavage were detected as [M+3H]3+ 788.03 (Fig. 3L) and [M+4H]4+ 486.49 (Fig. 3O), respectively. The final mass addition on cross-linked peptides attributed to the conjugated cross-linker after TFA cleavage are 649.26 Da (i.e. C34H40N3O8P) for dead-end modification, and 631.24 Da (i.e. C34H38N3O7P) for both intra-linked and inter-linked ones.
Similar experiments were carried out using another model peptide Ac-IR-7 in organic solvent in order to limit cross-linker hydrolysis. Cross-linking of the Ac-IR-7 peptide, selective enrichment and release of the cross-linked Ac-IR-7 peptides were as efficient as Ac-myelin (Supplemental Fig. 1). In comparison to the copper catalyzed click chemistry, this approach has similar conversion efficiency, but precipitation can be completely avoided and background reactions were largely suppressed. Therefore the Staudinger ligation has been selected as the method for enrichment of azide-containing cross-linked peptides in the sections described below.
Capture of a cross-linked model peptide from a protein digest mixture
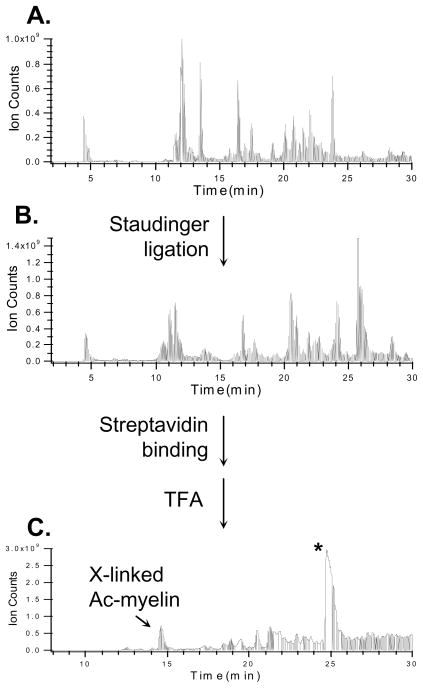
In order to test the effectiveness of the enrichment of Azide-DSG cross-linked peptides in a mixture, we mixed Azide-DSG cross-linked Ac-myelin with chicken lysozyme tryptic digest in a 1:1 ratio. The peptide mixture was subjected to Staudinger conjugation, then affinity purified and eluted by TFA. This process was monitored by LC MS analysis at each step as seen in Fig. 4. As shown, the Ac-myelin cross-linked peptides (i.e. inter-linked and dead-end) were not the most abundant species either before (Fig. 4A) or after Staudinger ligation (Fig. 4B). However, after affinity purification and acid cleavage elution, both inter-linked and dead-end products were successfully isolated and the inter-linked Ac-myelin became the 2nd most abundant signal in the purified sample (Fig. 4C). This result demonstrates the effectiveness of the selective isolation of azide-tagged cross-linked peptides from a peptide mixture. Due to the presence of the acid cleavage site in the biotin-phosphine, the excess biotin-phosphine used in the reaction that bound to the streptavidin beads was also present in the final elution, thus leading to undesirable signals including the most abundant peak (labeled with “*”) in the sample (Fig. 4C). Since the excess biotin-phosphine elutes at a particular time point as a distinct peak at the end of the gradient (Fig. 4C), its possible interference would be limited to its co-eluting peptides. However no obvious hindrance has been observed for MS analysis of cross-linked peptides due to excess biotin-phosphine. Ultimately, elimination of these impurities is desirable and can be accomplished by incorporating an acid cleavage site in the cross-linkers. This is currently under investigation.
Figure 4.
Enrichment of cross-linked Ac-myelin peptides after its mixing with a Lysozyme digest (1:1) using Staudinger ligation. LC MS traces of peptide mixtures: A) before (inter-linked and dead-end modified Ac-myelin eluted at 11.6 and 11.2 min, respectively) and B) after (interlinked and dead-end modified Ac-myelin eluted at 20.1 and 26.4 min, respectively) Staudinger ligation; C) after affinity purification and TFA cleavage elution. X-linked Ac-myelin (14.4 min) indicates the enriched inter-linked peptide. *: cleaved biotin-phosphine products. The dead-end modified Ac-myelin eluted at 16.8 min.
MSn analysis to facilitate identification of cross-linked peptides
To confirm the cross-linked products, MS/MS analyses were first carried out. As an example, the MS/MS spectrum of an inter-linked Ac-myelin product ([M+5H]5+ 644.33) after enrichment, purification and elution is illustrated in Fig. 5A, in which two sets of major ions were observed. One set of the ions (i.e. [M+4H]4+ 670.85, 675.36, 678.36) were the fragment ions resulting from the parent ion ([M+5H]5+ 644.33) with a loss of 538.2, 520.2, and 508.2 Da respectively. Two of the corresponding losses (i.e. MH+ 520.24 and 508.24) and another fragment ion with MH+ 447.19 were observed as the second set of ions (labeled as I, II & III) in Fig. 5A. The fragment ion with MH+ 538.2 was often not detected due to its further fragmentation into its water loss product ion (MH+ 520.2). Similarly, in the MS/MS spectrum of the inter-linked Ac-IR-7 after enrichment and purification (Fig. 5B), two sets of fragment ions were observed including one set of ions with MH+ 447.28, 508.23 and 520.18, and another set of fragment ions with [M+2H]2+ 891.83, 900.54, 906.47 corresponding to the parent ion ([M+3H]3+ 773.73) with a loss of 538.2, 520.2, or 508.2 Da respectively. Similar fragmentation pattern was detected in both dead-end modified Ac-myelin and Ac-IR-7 peptides (Supplemental Fig. 2). Taken together, the results suggest that this type of fragmentation is unique to the cross-linked peptides, and detection of a group of the fragment ions (i.e. MH+ 447.2, 508.2, and 520.2) is independent of the type and sequence of the cross-linked peptides. This prompted us to believe that such fragmentation is due to unique bond cleavages at the conjugated cross-linker region during CID analysis. The proposed cleavage sites in the conjugated cross-linker region and resultant structures of the fragmentation ions are depicted in Supplemental Fig. 3. Therefore, these ions (MH+ 447.2, 508.2, 520.2) are characteristic fragmentation ions to the cross-linked products after Staudinger ligation and affinity purification/TFA cleavage and can be used as diagnostic markers to facilitate identifying the cross-linked peptides by MS/MS.
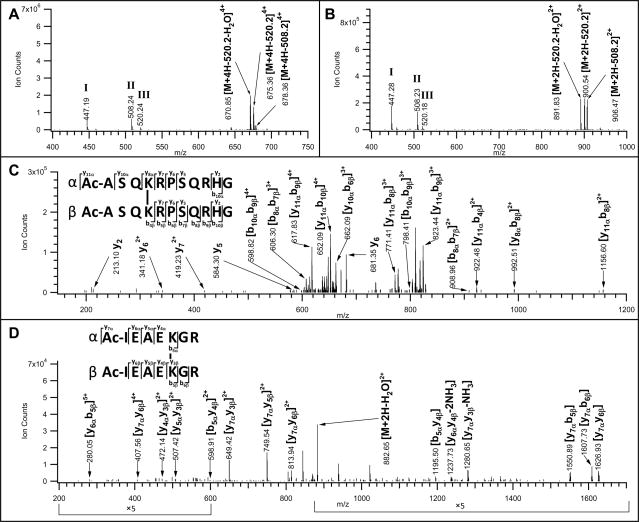
Figure 5.
MSn analyses of inter-linked Ac-myelin and IR-7 after Staudinger ligation and affinity purification/TFA cleavage. MS/MS spectra of A) inter-linked Ac-myelin ([M+5H]5+ 644.33); and B) inter-linked Ac-IR-7 ([M+3H]3+ 773.73) peptides. MS3 spectra of C) a fragment ion ([M+4H]4+, 670.85) after a loss of 538.2 Da from the parent ion ([M+5H]5+ 644.34) observed in A); and D) a fragment ion ([M+2H]2+ 891.83) after a loss of 538.2 Da from the parent ion ([M+3H]3+ 773.73) observed in B). Characteristic fragment ions in MS/MS spectra, i.e. 447.2, 508.2 and 520.2 were detected due to fragmentation in the cross-linker region and their structures are displayed in Supplemental Figure 2.
Due to the preferential bond cleavages at the linker region, limited fragmentation on the peptide backbone was observed in MS/MS spectra. In order to obtain detailed sequence information, MS3 analyses of the parent ions after their characteristic losses were further performed. Fig. 5C displays the MS3 spectrum of the fragment ion ([M+4H]4+ 670.85) which resulted from the parent inter-linked Ac-myelin ([M+5H]5+ 644.33) after a loss of 538.2 Da as measured in Fig. 5A. Based on the previously described terminology of inter-linked peptides and their fragment ions [8], we have labeled one sequence of the inter-linked Ac-myelin as the α chain and the other as the β chain. In addition to fragment ions belonging to α or β chain sequence, fragment ions containing both chains were detected. The masses of all possible fragment ions of the inter-linked Ac-myelin were calculated, which were then manually matched to the observed ions in Fig. 5C for unambiguous identification of the inter-linked Ac-myelin sequence. Similarly, MS3 analysis of the fragment ion ([M+2H]2+ 891.83) of the parent interlinked Ac-IR-7 ion ([M+3H]3+ 773.73) after a loss of 538.2 Da (Fig. 5B) confirmed its sequence as illustrated in Fig. 5D.
Enrichment and identification of Cytochrome C cross-linked peptides
To determine whether the cross-linking enrichment strategy can be applied to more complex samples, cytochrome c was used as a model protein since it has been widely used in other cross-linking experiments [3, 8, 40, 41, 65–68]. Cytochrome c was cross-linked using Azide-DSG and the cross-linked products were separated by 1-D SDS-PAGE. Various molar ratios of Azide-DSG and cytochrome c (10:1, 15:1; 20:1, 50:1) were tested to optimize the cross-linking efficiency with 10-fold excess of Azide-DSG determined as the optimal ratio for final analyses. After cross-linking, the cross-linked cytochrome c was digested in-solution with trypsin and the tryptic peptides were subjected to Staudinger ligation using the biotin-phosphine (S-17) as described above (Figs. 1 & 2). The resulting conjugates were then affinity purified, eluted with TFA, and analyzed by LC MS/MS. In comparison to model peptides, data processing of cross-linked cytochrome c peptides was much more complicated due to the presence of a large number of heterogeneous and unknown cross-linked peptides. To facilitate the data interpretation, we have employed a suite of software (i.e. Batch-tag, MS-bridge, MS-tag, MS-product and Search Compare) in Protein Prospector to identify cytochrome c peptides via a method similar to that previously described [62]. MS/MS spectra were first searched against cytochrome c sequence using Batch-tag with mass modification search to identify unmodified peptides as well as peptides with various mass modifications. Our cross-linker targets at lysine residues and the free protein N-terminus, therefore we considered these residues as potential sites for mass modifications [62]. For dead-end (type 0) modified and intra-linked (type 1) peptides, defined mass modifications on lysine residues, i.e. 649.26 and 631.24 Da, would be observed respectively. These masses are attributed to the addition of cross-links on the peptides due to the cross-linking nature of the type 0 and 1 products [8], which can be used for their identification. However, for inter-linked peptides, the mass modification would be undefined since it contains both the cross-link (i.e. 631.24 Da) and an additional covalently attached peptide. Even with mass modification search, most of the cross-linked peptides were not identified by Batch-tag search. This is a result of the complicated fragmentation in MS/MS spectra caused by preferred cleavage at the conjugated linker region and the presence of an additional sequence in the interlinked peptides.
In order to determine the cross-linked peptide identities, three additional steps were carried out. First, the monoisotopic masses and charges of all parent ions selected for MS/MS were subjected to the MS-bridge program to identify putative Azide-DSG cross-linked peptides of cytochrome c by peptide mass mapping with high mass accuracy (<20 ppm) [62]. Second, we manually examined MS/MS spectra of the putative cross-linked cytochrome c peptides identified in MS-bridge to determine whether the diagnostic fragment ions (MH+, 447.2, 508.2, 520.2) of the conjugated cross-link were observed. Thirdly, MS-tag, MS-product and manual inspection were performed to identify and confirm the cross-linked peptide sequences and correlate them with MS-bridge search results. For instance, based on MS-bridge search results, a tryptic peptide of [M+4H]4+ 542.54 matched to an inter-linked peptide [Ac-GDVEKGK inter-linked to KIFVQK], a peptide of [M+3H]3+ 526.96 matched to an intra-linked peptide [GK(631.24)KIFVQK], and a peptide of [M+3H]3+ 745.34 matched to a dead-end product [K(649.26)TGQAPGFSYTDANK]. Consistent with cross-linked model peptides (Figs. 5A&B and Supplemental Fig. 2), all of the putative cross-linked cytochrome c peptides gave same characteristic fragment ions (MH+ 447.2, 508.2, and 520.2) in their MS/MS spectra as illustrated in Figs. 6A–C, suggesting that these peptides are indeed enriched cross-linked products. Additionally, the corresponding fragment ions of parent ions with a loss of 508.2 or 520.2 Da were observed in their MS/MS spectra (Figs. 6A–C).
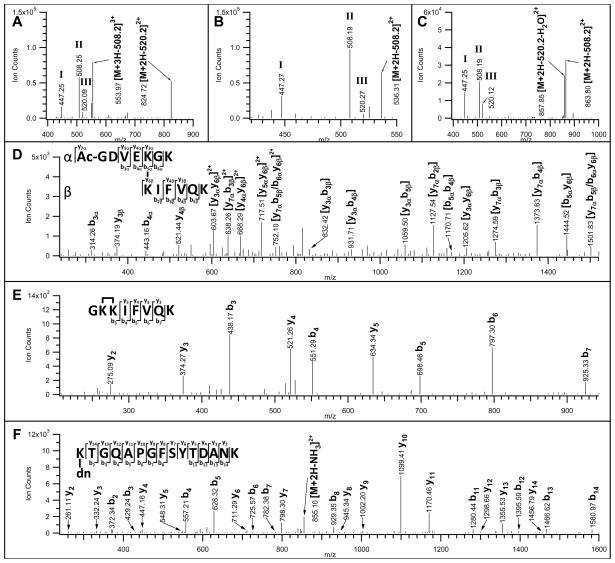
Figure 6.
MSn analyses of three Azide-DSG cross-linked peptides of cytochrome c after Staudinger ligation and affinity purification/TFA cleavage. MS/MS spectra of the selected A) inter-linked ([M+4H]4+ 542.54); B) intra-linked ([M+3H]3+ 526.96); and C) dead-end ([M+3H]3+ 745.35) modified peptides. Characteristic fragment ions (MH+ 447.2, 508.2, 520.2) were detected in all spectra. MS3 spectra of D) a fragment ion ([M+2H]2+ 824.72) observed in A), after a loss of 508.2 Da from the parent ion ([M+4H]4+ 542.54); E) a fragment ion ([M+2H]2+ 536.31) observed in B), after a loss of 508.2 Da from the parent ion ([M+3H]3+ 526.96); and F) a fragment ion ([M+2H]2+ 863.80) observed in C), after a loss of 508.2 Da from the parent ion ([M+3H]3+ 745.35). The identified cross-linked peptide sequences are displayed in D-F. dn: dead-end modification.
In addition to MS/MS experiments, MS3 analyses of selected fragment ions observed in MS/MS spectra of these three cross-linked cytochrome c peptides (Figs. 6A–C) were carried out to further confirm their identities. For the inter-linked peptide ([M+4H]4+ 542.54), its fragment ion with [M+2H]2+ 824.72 (Fig. 6A) was selected for MS3 analysis. The loss of 520.2 Da from the parent ion ([M+4H]4+ 542.54) led to a mass modification of 112 Da on the inter-linked fragment ion ([M+2H]2+ 824.72) due to the remnant of the conjugated link (See Supplemental Fig. 3). As shown in Fig. 6D, a series of b and y ions for either a single or both peptide chains were observed in the MS3 spectrum of [M+2H]2+ 824.72, further confirming the inter-linked peptide sequences identified by MS-bridge. For the intra-linked peptide ([M+3H]3+ 526.96), the fragment ion with [M+2H]2+ 536.31 (Fig. 6B) was analyzed by MS3, which has a mass modification of 124 Da due to the loss of 508.2 Da from its intra-linked parent ion. In the MS3 spectrum of [M+2H]2+ 536.31 (Fig. 6E), detection of y2-y5 and b3-b7 ion series allowed unambiguous identification of the intra-linked peptide as GKKIFVQK, in which the two adjacent lysine residues at the N-terminus were determined to be cross-linked. Similar fragmentation was also observed in the MS3 spectrum (Fig. 6F) of a MS/MS fragment ion with [M+2H]2+ 863.80 (Fig. 6C) from the dead-end modified parent ion ([M+3H]3+ 745.35) after a loss of 508.2 Da. As shown, y2-y14 and b2-b14 ions confirmed the dead-end modified peptide with a sequence of K(dn)TGQAPGFSYTDANK, in which the N-terminal lysine was modified.
In this work, although all of the cross-linked peptides have given characteristic losses in MS/MS for their identification, we were only able to interpret four additional MS3 spectra, including two inter-linked and two intra-linked cytochrome c peptides as displayed in Supplemental Figs. 4 & 5. This is due to lower sensitivity in MS3 and lack of software for effective searching of MS3 spectra with complex fragmentation which result from cross-linked peptides. Further software development which allows automated interpretation of MS3 spectra of enriched cross-linked peptides will significantly facilitate the identification process by peptide sequencing and increase the confidence of sequence assignments.
As summarized in Table 1, 14 dead-end modified (type 0), 24 intra-linked (type 1) and 20 inter-linked (type 2) peptides of cytochrome c have been identified from our new enrichment strategy using Staudinger ligation. This represents 35 unique cytochrome c cross-linked sequences, among which 19 of them have been previously reported using other NHS ester cross-linking reagents [3, 8, 40, 41, 65–68], and 16 of them were first identified in this study. In order to have a better comparison, we have summarized a combined non-redundant list of inter-linked, intra-linked and dead-end modified lysine residues of bovine cytochrome c identified in this work and other reports [3, 8, 65, 66, 68] (Supplemental Table 1). As shown, a total of 19 putative inter-linked, 8 intra-linked and 10 dead-end modified lysine residues have been identified by MS analyses using various cross-linkers. Among them, our study has identified 12 inter-linked lysines including 8 new ones, 8 intra-linked lysines including 4 new ones, and 10 dead-end modified lysines including 9 new ones. The absence of a few previously reported linkages may be due to the facts that we have only listed out the cross-linked peptides with characteristic diagnostic ions in Table 1, which do not include peptides identified based solely on the m/z and charge state as other papers had in the past. In addition, different cross-linkers with various spacer lengths and experimental conditions may result in identification of different peptides.
Table 1.
Summary of Identified Cross-linked Peptides of Cytochrome C after Enrichment.
| Type | Peptide Sequence | AA location | m/z(Observed) | z | Δ(ppm) | Seen Before Enrichment | References |
|---|---|---|---|---|---|---|---|
| 0 | Ac-GDVEKGK | G1-K7 | 712.3342 | 2 | 5 | Yes | |
| 0 | Ac-GDVEKGKK | G1-K8 | 517.9262 | 3 | 9 | Yes | |
| 0 | KIFVQK | K8-K13 | 706.3799 | 2 | 7 | Yes | 40,41,67 |
| 0 | GGKHK | G23-K27 | 588.2895 | 2 | 6 | Yes | |
| 0 | KTGQAPGFSYTDANK | K39-K53 | 745.3507* | 3 | 7 | Yes | 40,41,66,67 |
| 559.2618 | 4 | 2 | Yes | ||||
| 0 | KTGQAPGFSYTDANKNK | K39-K55 | 826.0621 | 3 | 5 | Yes | 40,65,66,67 |
| 619.7963 | 4 | 1 | Yes | ||||
| 0 | KYIPGTK | K73-K79 | 728.3737 | 2 | 5 | Yes | 41,67 |
| 485.9183 | 3 | 5 | Yes | ||||
| 0 | KGER | K88-R91 | 569.7707 | 2 | 1 | Yes | |
| 0 | EDLIAYLKK | E92-K100 | 871.4472 | 2 | 1 | Yes | 67 |
| 581.3033 | 3 | 5 | Yes | ||||
| 0 | KATNE | K100-E104 | 606.2740 | 2 | 2 | Yes | |
| 1 | Ac-GDVEKGKK | G1-K8 | 767.3693 | 2 | −5 | No | |
| 1 | Ac-GDVEKGKKIFVQK | G1-K13 | 717.0482 | 3 | 7 | Yes | |
| 1 | GKKIFVQK | G6-K13 | 789.9327 | 2 | 6 | Yes | 41,66,67 |
| 526.957* | 3 | 5 | Yes | ||||
| 395.4689 | 4 | 3 | Yes | ||||
| 1 | GGKHKTGPNLHGLFGR | G23-R38 | 769.7238 | 3 | −1 | Yes | 8,40,41,66,67 |
| 577.5500* | 4 | 8 | Yes | ||||
| 462.2401 | 5 | 5 | Yes | ||||
| 1 | GGKHKTGPNLHGLFGRK | G23-K39 | 609.5687 | 4 | −1 | Yes | 66,67 |
| 487.8605 | 5 | 7 | Yes | ||||
| 406.7162 | 6 | 3 | Yes | ||||
| 1 | KTGQAPGFSYTDANKNK | K39-K55 | 1229.5850 | 2 | 6 | Yes | 8,40,41,66,68 |
| 820.0587 | 3 | 5 | Yes | ||||
| 615.2941 | 4 | 2 | Yes | ||||
| 1 | MIFAGIKKK | M80-K88 | 833.9532 | 2 | 10 | Yes | 8,40,41,66,67 |
| 556.2999 | 3 | 1 | Yes | ||||
| 1 | MoxIFAGIKKK | M80-K88 | 841.9451 | 2 | 3 | Yes | |
| 561.6340* | 3 | 5 | Yes | ||||
| 1 | KKGER | K87-R91 | 624.8149 | 2 | 4 | Yes | 41 |
| 416.8803 | 3 | 6 | Yes | ||||
| 1 | GEREDLIAYLKKATNE | G89-E104 | 827.7458 | 3 | 5 | Yes | |
| 621.0619 | 4 | 6 | Yes | ||||
| 1 | EDLIAYLKKATNE | E92-E104 | 1070.0338 | 2 | 7 | Yes | 67 |
| 713.6908 | 3 | 6 | Yes | ||||
| 2 | Ac-GDVEKGK KIFVQK |
G1-K7 ~ K8-K13 | 723.0504 | 3 | 6 | Yes | 3,40,41,67 |
| 542.5392* | 4 | 5 | Yes | ||||
| 2 | Ac-GDVEKGK KK |
G1-K7 ~ K87-K88 | 560.6229 | 3 | 6 | Yes | 67,68 |
| 420.7197 | 4 | 7 | Yes | ||||
| 2 | Ac-GDVEKGK KKGER |
G1-K7 ~ K87-R91 | 506.2615 | 4 | 7 | Yes | 67,68 |
| 2 | Ac-GDVEKGKK KK |
G1-K8 ~ K87-K88 | 452.7451 | 4 | 10 | Yes | 67 |
| 2 | Ac-GDVEKGKK KKGER |
G1-K8 ~ K87-R91 | 538.2844 | 4 | 5 | Yes | 67,68 |
| 2 | GKK KGER |
G6-K8 ~ K88-R91 | 726.3632 | 2 | −17 | Yes | |
| 2 | GKK KATNE |
G6-K8 ~ K100-E104 | 762.8826* | 2 | 6 | Yes | 66,68 |
| 508.9219 | 3 | 1 | Yes | ||||
| 381.9450 | 4 | 5 | Yes | ||||
| 2 | KIFVQK KATNE |
K8-K13 ~ K100-E104 | 652.3468 | 3 | 9 | Yes | |
| 489.5106 | 4 | 6 | Yes | ||||
| 2 | GGKHK KATNE |
K23-K27 ~ K100-E104 | 573.6183* | 3 | 6 | Yes | |
| 2 | KTGQAPGFSYTDANK MIFAGIKKK |
K39-K53 ~ M80-K88 | 813.4261 | 4 | 13 | No | |
| 650.9429 | 5 | 13 | No | ||||
| 2 | KTGQAPGFSYTDANK KATNE |
K39-K53 ~ K100-E104 | 695.0784 | 4 | 2 | Yes | 67,68 |
| 2 | KKGER KATNE |
K87-R91 ~ K100-E104 | 453.2339 | 4 | 11 | Yes | |
| 2 | KGER KATNE |
K88-R91 ~ K100-E104 | 561.2752 | 3 | 7 | Yes | |
| 2 | KATNE KATNE |
K100-E104 ~ K100-E104 | 585.6121 | 3 | 10 | Yes |
Note: All of the cross-linked peptides have displayed characteristic losses in MS/MS.
These cross-linked peptides have been further confirmed by MS3.
In comparison to the results prior to enrichment, the Staudinger ligation based enrichment strategy has demonstrated its effectiveness in isolating cross-linked products. Based on the three dimensional crystal structure of bovine heart cytochrome c (PDB ID; 2B4Z) [69], we have calculated the alpha carbon distances between the cross-linked lysine residues identified in the cross-linked peptides. As shown in Supplemental Table 2, among 16 different combinations of type 1 or 2 cross-linked lysines in cytochrome c identified in this work (not considering linkages between two adjacent lysines), 13 linkages have the distances between their alpha carbons within 20 Å, and 3 of them have the distances ~ 21–25 Å, which was measured using Jmol (http://jmol.org/). This is consistent not only with the length of a fully expanded Azide-DSG (7.7 AǺ spacer length) and two lysine side chains, but also with the previous results using similar lengths of NHS ester cross-linkers [39, 67, 70]. The results suggest that our cross-linking condition did not induce significant disturbance to cytochrome c structural conformations.
CONCLUSION
In this study, we have designed, synthesized, and evaluated a novel azide-tagged cross-linker, Azide-DSG, which enables selective and effective enrichment of cross-linked peptides using Staudinger ligation. In comparison to other types of enrichment strategies using alkyne tags [39], the azide tag is more resistant to oxidative side reactions that could reduce the capture efficiency. Although azide-alkyne chemistry has been applied to capture azide-tagged cross-linked peptides [40, 41], we demonstrate that the new Staudinger ligation based strategy is efficient for selective enrichment of the cross-linked peptides from complex mixtures and better suited for peptide analysis. In addition, the diagnostic ions (MH+ 447.2, 508.2, 520.2) detected in MS/MS spectra due to characteristic fragmentation in the conjugated link after Staudinger reaction and affinity purification offer a unique means for MS identification with increased confidence. This set of diagnostic ions is easy to detect and would be more reliable for identifying cross-linked peptides by MS in comparison to a diagnostic fragmentation with a neutral loss as employed in alkyne tagged cross-linked peptides [39]. Moreover, MS3 analysis permitted additional fragmentation information for unambiguous peptide identification. In comparison to immobilized cyclic alkyne [41], synthesis of biotin-phosphine is much simpler with only three steps. Taken together, the novel enrichment strategy developed in this work helps facilitate MS identification of cross-linked peptides from complex mixtures. With the development of bioinformatics tools, we anticipate that this approach can be easily generalized for capturing and identifying cross-linked peptides of protein complexes.
Supplementary Material
Supplemental Figure 1. Enrichment of Azide-DSG cross-linked IR-7 peptides by Staudinger ligation. A–C) Cross-linked products of IR-7 prior to enrichment: A) structural illustration of cross-linked peptides, R represents either an additional peptide for inter-linked or a hydroxyl group for a dead-end modified Ac-IR-7 peptides; MS spectra of B) inter-link ([M+3H]3+ 633.66, [M+2H]2+ 949.98) and C) dead-end (MH+ 1073.52) modified Ac-IR-7. D-I) Cross-linked products of Ac-IR-7 after Staudinger ligation (D-F), and affinity purification/TFA cleavage (HI): D) structural illustration of conjugated products after Staudinger ligation; MS spectra of E) inter-linked ([M+3H]3+ 913.77, [M+2H]2+ 1370.15) and F) dead-end ([M+3H]3+ 638.29) products; G) structural illustration of final cross-linked products after affinity purification and TFA cleavage; MS spectra of H) inter-linked ([M+4H]4+ 580.79, [M+3H]3+ 774.05) and I) dead-end ([M+3H]3+ 498.25) modified peptides.
Supplemental Figure 2. MS/MS spectra of the dead-end modified A) Ac-myelin ([M+3H]3+ 648.31); B) Ac-IR-7 ([M+2H]2+ 747.37) peptides. Characteristic fragment ions (i.e. MH+ 447.2, 508.2, 520.2) and corresponding parent ions after these losses were detected.
Supplemental Figure 3. Chemical structures of fragmentation ions of a cross-linked peptide after Staudinger ligation, affinity purification and TFA elution observed in MS/MS. There are three most commonly observed cleavage sites (i.e. I, II, III), resulting in three unique fragment ions, MH+ 447.2, 508.2, 520.2. Structures of the remnant chemical moieties left on the cross-linked peptides are presented.
Supplemental Figure 4. MSn analyses of two inter-linked peptides of cytochrome c after Staudinger ligation and affinity purification/TFA cleavage. MS/MS spectra of inter-linked peptides with A) [M+2H]2+ 762.88 (KATNE interlinked with GKK); B) [M+3H]3+ 573.62 (KATNE interlinked with GGKHK). Characteristic fragment ions (i.e. MH+ 447.2, 508.2, 520.2) were detected in both MS/MS spectra. MS3 spectra of D) a fragment ion (MH+ 1017.49 observed in A), after loss of 508.2 Da from the parent ion ([M+2H]2+ 762.88); E) a fragment ion ([M+2H]2+ 606.54) observed in B), after loss of 508.2 Da from the parent ion ([M+3H]3+ 573.62).
Supplemental Figure 5. MSn analyses of two intra-linked peptides of cytochrome c after Staudinger ligation and affinity purification/TFA cleavage. MS/MS spectra of intra-linked peptides with A) [M+4H]4+ 577.55; B) [M+3H]3+ 561.63. Characteristic fragment ions (i.e. MH+ 447.2, 508.2, 520.2) were detected in both MS/MS spectra. MS3 spectra of D) a fragment ion ([M+3H]3+ 597.08) observed in A), after loss of 520.2 Da from the parent ion ([M+4H]4+ 577.55); E) a fragment ion ([M+2H]2+ 582.28) observed in B), after loss of 520.2 Da from the parent ion ([M+3H]3+ 561.63).
Summary of inter-linked, intra-linked and dead-end modified lysines identified in bovine cytochrome c.
Distances between alpha carbons of the identified cross-linked lysine residues in cytochrome c
Acknowledgments
We wish to thank Drs. Christian Tagwerker and Lei Fang for initial analysis and members of the Huang and Rychnovsky laboratory for their help during this study. We would like to thank Prof. A.L. Burlingame, Peter Baker and Aenoch Lynn at UCSF for using Protein Prospector. This work was supported by National Institutes of Health grants (GM-74830 to L.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Agou F, Ye F, Veron M. In vivo protein cross-linking. Methods Mol Biol. 2004;261:427–42. doi: 10.1385/1-59259-762-9:427. [DOI] [PubMed] [Google Scholar]
- 2.Back JW, de Jong L, Muijsers AO, de Koster CG. Chemical cross-linking and mass spectrometry for protein structural modeling. J Mol Biol. 2003;331(2):303–13. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 3.Sinz A. Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J Mass Spectrom. 2003;38(12):1225–37. doi: 10.1002/jms.559. [DOI] [PubMed] [Google Scholar]
- 4.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom Rev. 2006;25(4):663–82. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 5.Sinz A. Investigation of protein-protein interactions in living cells by chemical crosslinking and mass spectrometry. Anal Bioanal Chem. 2010 Jan 15; doi: 10.1007/s00216-009-3405-5. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Mouradov D, King G, Ross IL, Forwood JK, et al. Protein structure determination using a combination of cross-linking, mass spectrometry, and molecular modeling. Methods Mol Biol. 2008;426:459–74. doi: 10.1007/978-1-60327-058-8_31. [DOI] [PubMed] [Google Scholar]
- 7.Young MM, Tang N, Hempel JC, Oshiro CM, et al. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc Natl Acad Sci U S A. 2000;97(11):5802–6. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schilling B, Row RH, Gibson BW, Guo X, Young MM. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J Am Soc Mass Spectrom. 2003;14(8):834–50. doi: 10.1016/S1044-0305(03)00327-1. [DOI] [PubMed] [Google Scholar]
- 9.Back JW, Sanz MA, De Jong L, De Koning LJ, et al. A structure for the yeast prohibitin complex: Structure prediction and evidence from chemical crosslinking and mass spectrometry. Protein Sci. 2002;11(10):2471–8. doi: 10.1110/ps.0212602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shih CL, Chen MJ, Linse K, Wang K. Molecular Contacts between Nebulin and Actin: Cross-Linking of Nebulin Modules to the N-Terminus of Actin. Biochemistry. 1997;36:1814–1825. doi: 10.1021/bi961236b. [DOI] [PubMed] [Google Scholar]
- 11.Chen T, Jaffe JD, Church GM. Algorithms for Identifying Protein Cross-Links Via Tandem Mass Spectrometry. J Comput Biol. 2001;8:571–583. doi: 10.1089/106652701753307494. [DOI] [PubMed] [Google Scholar]
- 12.Lacroix M, Rossi V, Gaboriaud C, Chevallier S, et al. Structure and assembly of the catalytic region of human complement protease C1r: a three-dimensional model based on chemical cross-linking and homology modeling. Biochemistry. 1997;36(21):6270–82. doi: 10.1021/bi962719i. [DOI] [PubMed] [Google Scholar]
- 13.Taverner T, Hall NE, O’Hair RA, Simpson RJ. Characterization of an Antagonist Interleukin-6 Dimer by Stable Isotope Labeling, Cross-linking, and Mass Spectrometry. J Biol Chem. 2002;277(8):46487–46492. doi: 10.1074/jbc.M207370200. [DOI] [PubMed] [Google Scholar]
- 14.Rappsilber J, Siniossoglou S, Hurt EC, Mann M. A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Anal Chem. 2000;72(2):267–75. doi: 10.1021/ac991081o. [DOI] [PubMed] [Google Scholar]
- 15.Chen ZA, Jawhari A, Fischer L, Buchen C, et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. Embo J. 2010 Jan 21; doi: 10.1038/emboj.2009.401. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu F, Shan SO, Moustakas DT, Alber F, et al. Unraveling the interface of signal recognition particle and its receptor by using chemical cross-linking and tandem mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(47):16454–9. doi: 10.1073/pnas.0407456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu F, Maynard JC, Chiosis G, Nicchitta CV, Burlingame AL. Identification of novel quaternary domain interactions in the Hsp90 chaperone, GRP94. Protein Sci. 2006;15(6):1260–9. doi: 10.1110/ps.052065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt-Ulms G, Legname G, Baldwin MA, Ball HL, et al. Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J Mol Biol. 2001;314(5):1209–25. doi: 10.1006/jmbi.2000.5183. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt-Ulms G, Hansen K, Liu J, Cowdrey C, et al. Time-controlled transcardiac perfusion cross-linking for the study of protein interactions in complex tissues. Nat Biotechnol. 2004;22(6):724–31. doi: 10.1038/nbt969. [DOI] [PubMed] [Google Scholar]
- 20.Vasilescu J, Guo X, Kast J. Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics. 2004;4:3845–3854. doi: 10.1002/pmic.200400856. [DOI] [PubMed] [Google Scholar]
- 21.Guerrero C, Tagwerker C, Kaiser P, Huang L. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol Cell Proteomics. 2006;5(2):366–78. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci U S A. 2008;105(36):13333–8. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bousquet-Dubouch MP, Baudelet E, Guerin F, Matondo M, et al. Affinity purification strategy to capture human endogenous proteasome complexes diversity and to identify proteasome-interacting proteins. Mol Cell Proteomics. 2009;8(5):1150–64. doi: 10.1074/mcp.M800193-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tardiff DF, Abruzzi KC, Rosbash M. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc Natl Acad Sci U S A. 2007;104(50):19948–53. doi: 10.1073/pnas.0710179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chowdhury SM, Shi L, Yoon H, Ansong C, et al. A method for investigating protein-protein interactions related to salmonella typhimurium pathogenesis. J Proteome Res. 2009;8(3):1504–14. doi: 10.1021/pr800865d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallon G, Rappsilber J, Mann M, Serrano L. Model for stathmin/OP18 binding to tubulin. EMBO J. 2000;19(2):213–22. doi: 10.1093/emboj/19.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trester-Zedlitz M, Kamada K, Burley SK, Fenyo D, Chait BT, Muir TW. A modular cross-linking approach for exploring protein interactions. J Am Chem Soc. 2003;125(9):2416–25. doi: 10.1021/ja026917a. [DOI] [PubMed] [Google Scholar]
- 28.Tang X, Munske GR, Siems WF, Bruce JE. Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal Chem. 2005;77(1):311–8. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Tang X, Munske GR, Tolic N, Anderson GA, Bruce JE. Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol Cell Proteomics. 2009;8(3):409–20. doi: 10.1074/mcp.M800232-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu F, Mahrus S, Craik CS, Burlingame AL. Isotope-coded and affinity-tagged cross-linking (ICATXL): an efficient strategy to probe protein interaction surfaces. J Am Chem Soc. 2006;128(32):10362–3. doi: 10.1021/ja0614159. [DOI] [PubMed] [Google Scholar]
- 31.Kang S, Mou L, Lanman J, Velu S, Brouillette WJ, Prevelige PE., Jr Synthesis of biotin-tagged chemical cross-linkers and their applications for mass spectrometry. Rapid Commun Mass Spectrom. 2009;23(11):1719–26. doi: 10.1002/rcm.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller DR, Schindler P, Towbin H, Wirth U, et al. Isotope-tagged cross-linking reagents, a new tool in mass spectrometric protein interaction analysis. Anal Chem. 2001;73:1927–1934. doi: 10.1021/ac001379a. [DOI] [PubMed] [Google Scholar]
- 33.Collins CJ, Schilling B, Young M, Dollinger G, Guy RK. Isotopically labeled crosslinking reagents: resolution of mass degeneracy in the identification of crosslinked peptides. Bioorg Med Chem Lett. 2003;13(22):4023–6. doi: 10.1016/j.bmcl.2003.08.053. [DOI] [PubMed] [Google Scholar]
- 34.Petrotchenko EV, Olkhovik VK, Borchers CH. Isotopically coded cleavable cross-linker for studying protein-protein interaction and protein complexes. Mol Cell Proteomics. 2005;4(8):1167–79. doi: 10.1074/mcp.T400016-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Sinz A, Wang K. Mapping protein interfaces with a fluorogenic cross-linker and mass spectrometry: application to nebulin-calmodulin complexes. Biochemistry. 2001;40:7903–13. doi: 10.1021/bi010259+. [DOI] [PubMed] [Google Scholar]
- 36.Sinz A, Wang K. Mapping spatial proximities of sulfhydryl groups in proteins using a fluorogenic cross-linker and mass spectrometry. Anal Biochem. 2004;331:27–32. doi: 10.1016/j.ab.2004.03.075. [DOI] [PubMed] [Google Scholar]
- 37.Back JW, Hartog AF, Dekker HL, Muijsers AO, de Koning LJ, de Jong L. A New Crosslinker for Mass Spectrometric Analysis of the Quaternary Structure of Protein Complexes. J Am Soc Mass Spectrom. 2001;12:222–227. doi: 10.1016/S1044-0305(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Tang X, Munske GR, Zakharova N, et al. In vivo identification of the outer membrane protein OmcA-MtrC interaction network in Shewanella oneidensis MR-1 cells using novel hydrophobic chemical cross-linkers. J Proteome Res. 2008;7(4):1712–20. doi: 10.1021/pr7007658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chowdhury SM, Du X, Tolic N, Wu S, et al. Identification of cross-linked peptides after click-based enrichment using sequential collision-induced dissociation and electron transfer dissociation tandem mass spectrometry. Anal Chem. 2009;81(13):5524–32. doi: 10.1021/ac900853k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasper PT, Back JW, Vitale M, Hartog AF, et al. An aptly positioned azido group in the spacer of a protein cross-linker for facile mapping of lysines in close proximity. Chembiochem. 2007;8(11):1281–92. doi: 10.1002/cbic.200700150. [DOI] [PubMed] [Google Scholar]
- 41.Nessen MA, Kramer G, Back J, Baskin JM, et al. Selective Enrichment of Azide-Containing Peptides from Complex Mixtures. J Proteome Res. 2009;8(7):3702–3711. doi: 10.1021/pr900257z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41(14):2596–9. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 43.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc Natl Acad Sci U S A. 2007;104(4):1171–6. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sieber SA, Cravatt BF. Analytical platforms for activity-based protein profiling--exploiting the versatility of chemistry for functional proteomics. Chem Commun (Camb) 2006;(22):2311–9. doi: 10.1039/b600653c. [DOI] [PubMed] [Google Scholar]
- 45.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11(4):535–46. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 46.Speers AE, Cravatt BF. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J Am Chem Soc. 2005;127(28):10018–9. doi: 10.1021/ja0532842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gubbens J, Ruijter E, de Fays LE, Damen JM, et al. Photocrosslinking and click chemistry enable the specific detection of proteins interacting with phospholipids at the membrane interface. Chem Biol. 2009;16(1):3–14. doi: 10.1016/j.chembiol.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125(16):4686–7. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 49.Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126(46):15046–7. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 50.Breinbauer R, Kohn M. Azide-alkyne coupling: a powerful reaction for bioconjugate chemistry. Chembiochem. 2003;4(11):1147–9. doi: 10.1002/cbic.200300705. [DOI] [PubMed] [Google Scholar]
- 51.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. A comparative study of bioorthogonal reactions with azides. ACS Chem Biol. 2006;1(10):644–8. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 52.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, et al. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci U S A. 2007;104(43):16793–7. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saxon E, Armstrong JI, Bertozzi CR. A “traceless” Staudinger ligation for the chemoselective synthesis of amide bonds. Org Lett. 2000;2(14):2141–3. doi: 10.1021/ol006054v. [DOI] [PubMed] [Google Scholar]
- 54.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287(5460):2007–10. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 55.Kohn M, Breinbauer R. The Staudinger ligation-a gift to chemical biology. Angew Chem Int Ed Engl. 2004;43(24):3106–16. doi: 10.1002/anie.200401744. [DOI] [PubMed] [Google Scholar]
- 56.Sprung R, Nandi A, Chen Y, Kim SC, et al. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J Proteome Res. 2005;4(3):950–7. doi: 10.1021/pr050033j. [DOI] [PubMed] [Google Scholar]
- 57.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci U S A. 2002;99(1):19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luchansky SJ, Argade S, Hayes BK, Bertozzi CR. Metabolic functionalization of recombinant glycoproteins. Biochemistry. 2004;43(38):12358–66. doi: 10.1021/bi049274f. [DOI] [PubMed] [Google Scholar]
- 59.Vila A, Tallman KA, Jacobs AT, Liebler DC, Porter NA, Marnett LJ. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem Res Toxicol. 2008;21(2):432–44. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reddie KG, Seo YH, Muse Iii WB, Leonard SE, Carroll KS. A chemical approach for detecting sulfenic acid-modified proteins in living cells. Mol Biosyst. 2008;4(6):521–31. doi: 10.1039/b719986d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kostiuk MA, Keller BO, Berthiaume LG. Non-radioactive detection of palmitoylated mitochondrial proteins using an azido-palmitate analogue. Methods Enzymol. 2009;457:149–65. doi: 10.1016/S0076-6879(09)05009-5. [DOI] [PubMed] [Google Scholar]
- 62.Chu F, Baker PR, Burlingame AL, Chalkley RJ. Finding Chimeras: A bioinformatic strategy for identification of cross-linked peptides. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M800555-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Veken P, Dirksen EH, Ruijter E, Elgersma RC, et al. Development of a novel chemical probe for the selective enrichment of phosphorylated serine- and threonine-containing peptides. Chembiochem. 2005;6(12):2271–80. doi: 10.1002/cbic.200500209. [DOI] [PubMed] [Google Scholar]
- 64.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org Lett. 2004;6(17):2853–5. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 65.Pearson KM, Pannell LK, Fales HM. Intramolecular cross-linking experiments on cytochrome c and ribonuclease A using an isotope multiplet method. Rapid Commun Mass Spectrom. 2002;16(3):149–159. doi: 10.1002/rcm.554. [DOI] [PubMed] [Google Scholar]
- 66.Dihazi GH, Sinz A. Mapping low-resolution three-dimensional protein structures using chemical cross-linking and Fourier transform ion-cyclotron resonance mass spectrometry. 2003;17(17):2005–2014. doi: 10.1002/rcm.1144. [DOI] [PubMed] [Google Scholar]
- 67.Lee YJ, Lackner LL, Nunnari JM, Phinney BS. Shotgun cross-linking analysis for studying quaternary and tertiary protein structures. J Proteome Res. 2007;6(10):3908–17. doi: 10.1021/pr070234i. [DOI] [PubMed] [Google Scholar]
- 68.Guo X, Bandyopadhyay P, Schilling B, Young MM, et al. Partial acetylation of lysine residues improves intraprotein cross-linking. Anal Chem. 2008;80(4):951–60. doi: 10.1021/ac701636w. [DOI] [PubMed] [Google Scholar]
- 69.Mirkin N, Jaconcic J, Stojanoff V, Moreno A. High resolution X-ray crystallographic structure of bovine heart cytochrome c and its application to the design of an electron transfer biosensor. Proteins. 2008;70(1):83–92. doi: 10.1002/prot.21452. [DOI] [PubMed] [Google Scholar]
- 70.Kruppa GH, Schoeniger J, Young MM. A top down approach to protein structural studies using chemical cross-linking and Fourier transform mass spectrometry. Rapid Commun Mass Spectrom. 2003;17(2):155–62. doi: 10.1002/rcm.885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Enrichment of Azide-DSG cross-linked IR-7 peptides by Staudinger ligation. A–C) Cross-linked products of IR-7 prior to enrichment: A) structural illustration of cross-linked peptides, R represents either an additional peptide for inter-linked or a hydroxyl group for a dead-end modified Ac-IR-7 peptides; MS spectra of B) inter-link ([M+3H]3+ 633.66, [M+2H]2+ 949.98) and C) dead-end (MH+ 1073.52) modified Ac-IR-7. D-I) Cross-linked products of Ac-IR-7 after Staudinger ligation (D-F), and affinity purification/TFA cleavage (HI): D) structural illustration of conjugated products after Staudinger ligation; MS spectra of E) inter-linked ([M+3H]3+ 913.77, [M+2H]2+ 1370.15) and F) dead-end ([M+3H]3+ 638.29) products; G) structural illustration of final cross-linked products after affinity purification and TFA cleavage; MS spectra of H) inter-linked ([M+4H]4+ 580.79, [M+3H]3+ 774.05) and I) dead-end ([M+3H]3+ 498.25) modified peptides.
Supplemental Figure 2. MS/MS spectra of the dead-end modified A) Ac-myelin ([M+3H]3+ 648.31); B) Ac-IR-7 ([M+2H]2+ 747.37) peptides. Characteristic fragment ions (i.e. MH+ 447.2, 508.2, 520.2) and corresponding parent ions after these losses were detected.
Supplemental Figure 3. Chemical structures of fragmentation ions of a cross-linked peptide after Staudinger ligation, affinity purification and TFA elution observed in MS/MS. There are three most commonly observed cleavage sites (i.e. I, II, III), resulting in three unique fragment ions, MH+ 447.2, 508.2, 520.2. Structures of the remnant chemical moieties left on the cross-linked peptides are presented.
Supplemental Figure 4. MSn analyses of two inter-linked peptides of cytochrome c after Staudinger ligation and affinity purification/TFA cleavage. MS/MS spectra of inter-linked peptides with A) [M+2H]2+ 762.88 (KATNE interlinked with GKK); B) [M+3H]3+ 573.62 (KATNE interlinked with GGKHK). Characteristic fragment ions (i.e. MH+ 447.2, 508.2, 520.2) were detected in both MS/MS spectra. MS3 spectra of D) a fragment ion (MH+ 1017.49 observed in A), after loss of 508.2 Da from the parent ion ([M+2H]2+ 762.88); E) a fragment ion ([M+2H]2+ 606.54) observed in B), after loss of 508.2 Da from the parent ion ([M+3H]3+ 573.62).
Supplemental Figure 5. MSn analyses of two intra-linked peptides of cytochrome c after Staudinger ligation and affinity purification/TFA cleavage. MS/MS spectra of intra-linked peptides with A) [M+4H]4+ 577.55; B) [M+3H]3+ 561.63. Characteristic fragment ions (i.e. MH+ 447.2, 508.2, 520.2) were detected in both MS/MS spectra. MS3 spectra of D) a fragment ion ([M+3H]3+ 597.08) observed in A), after loss of 520.2 Da from the parent ion ([M+4H]4+ 577.55); E) a fragment ion ([M+2H]2+ 582.28) observed in B), after loss of 520.2 Da from the parent ion ([M+3H]3+ 561.63).
Summary of inter-linked, intra-linked and dead-end modified lysines identified in bovine cytochrome c.
Distances between alpha carbons of the identified cross-linked lysine residues in cytochrome c