Abstract
Background
The objective of this research was to identify the impact of genetic variants of P-glycoprotein (ABCB1) and cytochrome P450 (CYP) on nelfinavir pharmacokinetics and response to highly active antiretroviral therapy (HAART) in HIV-1–infected children.
Methods
HIV-1–infected children (n = 152) from Pediatric AIDS Clinical Trial Group 366 or 377 receiving nelfinavir as a component of HAART were evaluated. Genomic DNA was assayed for ABCB1 and CYP genetic variants using real-time polymerase chain reaction Nelfinavir oral clearance (CL/F), M8 to nelfinavir ratios, CD4+ T cells, and HIV-1-RNA were measured during HAART.
Results
Nelfinavir CL/F and M8 to nelfinavir ratios were significantly associated with the CYP2C19-G681A genotypes (P < 0.001). Furthermore, the CYP2C19-G681A genotype was related to virologic responses at week 24 (P = 0.01). A multivariate analysis demonstrated that age (P = 0.03), concomitant protease inhibitor use (P < 0.001), and the CYP2C19-G681A genotype (P < 0.001) remained significant covariates associated with nelfinavir CL/F.
Conclusions
CYP2C19 genotypes altered nelfinavir pharmacokinetics and the virologic response to HAART in HIV-1–infected children. These findings suggest that CYP2C19 genotypes are important determinants of nelfinavir pharmacokinetics and virologic response in HIV-1-infected children.
Keywords: ABCB1, CYP2C19, children, nelfinavir, virologic response
Introduction
Protease inhibitors (PIs), which have been used widely as a component of highly active antiretroviral therapy (HAART) in children and adults are known to be a substrate of P-glycoprotein1 and metabolized by hepatic cytochrome P450 (CYP), mainly by CYP3A4.2 In particular, nelfinavir (NFV) is metabolized into the metabolite hydroxyl-tert-butylamide (M8) by the CYP2C19 enzyme, and subsequently NFV and M8 are metabolized by CYP3A4.3 Several pharmacogenetic studies have shown that single nucleotide polymorphisms (SNPs) in adenosine triphosphate–binding cassette, subfamily B, member 1 ABCB1 (previously called multidrug resistance 1, MDR1)4 and CYP5 can influence the activity and bioavailability of NFV.
We previously reported that genetic variants in ABCB1 gene encoding for P-glycoprotein was responsible for variability in NFV pharmacokinetics (PKs) and virologic responses to HAART in children.6 However, the study was limited because of the small number of subjects. In addition, our earlier results were in contrast to a study previously reported in adults.4 Therefore, to expand the number of subjects, we examined children who received NFV as a component of HAART from 2 pediatric studies, Pediatric AIDS Clinical Trial Group (PACTG) 3667 and 377,8 to investigate the association between NFV PK and SNPs in ABCB1 and CYP and virologic and immunologic responses.
Materials and Methods
Subjects
This was a retrospective study investigating 152 children who received NFV as a component of HAART regimens although participating in PACTG 366 (n = 75, 49%) and PACTG 377 (n = 77, 51%) (Table 1). Informed consent was obtained from study participants. This study followed the human experimentation guidelines of the US Department of Health and Human Services and the University of California, San Diego review board.
Table 1. Baseline Characteristics of the 152 Children in PACTG 366 and 377 With the CYP2C19-G681A Genotypes.
| All Subjects* | PACTG 366 | PACTG 377 | CYP2C19-G681A | |||||
|---|---|---|---|---|---|---|---|---|
| n = 152 (%) | n = 75 (49) | n = 77 (51) | P | G/G n = 102 (%) |
G/A n = 42 (%) |
A/A n = 8 (%) |
P | |
| Sex, n (%) | ||||||||
| Male | 79 (52) | 46 (61) | 33 (43) | 0.01 | 53 (67) | 22 (28) | 4 (5) | 0.99 |
| Female | 73 (48) | 29 (39) | 44 (57) | 49 (67) | 20 (27) | 4 (6) | ||
| Race/ethnicity | ||||||||
| Black, non-Hispanic | 92 (61) | 45 (60) | 47 (61) | 0.73 | 58 (63) | 28 (30) | 6 (7) | 0.35 |
| Hispanic | 32 (21) | 15 (20) | 17 (22) | 24 (75) | 8 (25) | 0 (0) | ||
| White, non-Hispanic | 24 (16) | 12 (16) | 12 (16) | 18 (75) | 5 (21) | 1 (4) | ||
| Others | 4 (3) | 3 (4) | 1 (1) | 2 (50) | 1 (25.0) | 1 (25.0) | ||
| Age (yrs) [median, (IQR) ] | 7.1 (3.8-9.6) | 6.9 (3.2-10.6) | 6.8 (4.4-9.2) | 0.42 | 7.1 (3.5-10.1) | 7.1 (3.9-9.4) | 7.9 (5.9-10.7) | 0.82 |
| Concomitant PI or NNRTI | ||||||||
| No | 23 (15) | 0 | 23 (30) | 12 (52) | 9 (39) | 2 (9) | 0.63 | |
| Ritonavir | 40 (26) | 40 (53) | 0 | 29 (73) | 9 (23) | 2 (5) | ||
| Nevirapine | 60 (40) | 6 (8) | 54 (70) | 43 (72) | 14 (23) | 3 (5) | ||
| Ritonavir + nevirapine | 29 (19) | 29 (39) | 0 | 18 (62) | 10 (35) | 1 (3) | ||
| Baseline CD4+ (%) median, (IQR) | 24 (16–32) | 19 (10–28) | 27 (21–35) | <0.001 | 23 (16–32) | 25 (19–31) | 22 (6–40) | 0.85 |
| Mean HIV-1 RNA log10 copies/mL (SD) | 4.56 (0.68) | 4.71 (0.61) | 4.41 (0.72) | 0.003 | 4.54 (0.70) | 4.62 (0.64) | 4.44 (0.74) | 0.65 |
The subjects were selected from the whole study populations if they satisfied the following criteria: (1) received NFV as a component of HAART for >24 weeks with reported excellent compliance to their treatment regimen; (2) virologic and immunologic data were available at baseline, weeks 12 and 24; and (3) PK data for NFV were available at week 4.
NNRTI, nonnucleoside reverse transcriptase inhibitor.
NFV PKs
Among the 152 patients, 106 children (70%) had intensive PK collected over 8 hours at week 4 of treatment and 46 subjects (30%) had sparse PK during HAART These results and detailed methods for the measurement of NFV and calculation of PK parameters have been previously reported from subjects participating in PACTG 377.8,9 The M8 to NFV ratios were calculated based on each M8 and NFV value and averaged in subjects with the intensive PK data. When the M8 level was <50 ng/mL, such numbers were imputed to 50 ng/mL.5
Measurement of Plasma HIV-1 RNA and CD4+ T Cells
Plasma HIV-1 RNA was quantified using the Roche Amplicor HIV-1 Monitor assay (Roche Molecular Systems, Alameda, CA) with a detection limit of 400 copies per milliliter. The numbers and percentages of CD4+ T cells were determined in PACTG certified laboratories by flow cytometry.
Amplification and Detection of Polymorphisms in ABCB1 and CYP Genes by Real-Time Polymerase Chain Reaction
Previously developed fluorescence assays and detection methods were used for analyzing the ABCB1-3435C>T (rs1045642) and CYP3A4-392A>G (rs2740574).6 For the ABCB1-2677G>T (rs2032582),10CYP2C19C*2-681G>A (rs4244285), and CYP2C19*3-636G>A (rs28399504),11 previously reported assays were used. In addition, novel fluorescent assays were developed to detect the ABCB1-1236C>T (rs1128503) and ABCB1-1199G>A (rs2229109) genotypes. The information regarding the sequences of custom designed primers and probes and polymerase chain reaction conditions are available upon request.
Statistical Analysis
Statistical analyses were performed using the SPSS 13.0 software (Chicago, IL). Comparisons among ordered 3 genotypes at each categorical group were performed using the Jonckheere-Terpstra (rank-based trend) test for continuous outcomes and Cochran-Armitage trend test for binomial outcomes. A multivariate analysis for NFV CL/F was performed to evaluate the contribution of covariates. The x2 and Fisher exact tests were used to make pair-wise comparisons. All P values calculated were 2 sided and P value <0.05 was considered to be significant.
Results
Frequencies of ABCB1 and CYP Gene Polymorphisms
The ABCB1-3435-T allelic frequency was 0.35 in the whole cohort; however, the frequencies were less common in black, non-Hispanic (P = 0.006). Similarly, there were significant differences in the frequencies of ABCB1-G2677T (P < 0.001) and ABCB1-C1236T genotypes (P < 0.001) in black, non-Hispanic, compared with others. The CYP2C19-681-A allelic frequency was 0.19 in whole cohort, and the frequencies were similar among race/ethnicity (P = 0.35) (Table 1). All patients had the CYP2C19-636-G/G genotype.
The ABCB1-3435C> T Genotype is Associated With NFV CL/F
NFV CL/F differed significantly among the ABCB1-3435C>T genotype (P < 0.001); children with the ABCB1-3435-C/C genotypes had higher median NFV CL/F [47.2 L/h/m2, interquartile range (IQR): 32.7–68.7 L·h−1·m−2 ] compared with those with the C/T (36.1 L/h/m2, IQR: 28.1– 56.7 L·h−1·m−2) and the T/T genotype (35.4 L/h/m2, IQR: 17.8-61.3 L·h−1·m−2). NFV CL/F did not differ among subjects with the other ABCB1 genotype (P > 0.17). There was no difference in the association when examined by each race/ethnic group (P > 0.11).
The CYP2C19-681G>A Polymorphism is Associated With NFV CL/F
NFV CL/F differed significantly among the CYP2C19-681G>A genotypes (P < 0.001). When the data were analyzed in each race/ethnicity (Fig. 1), a significant difference in NFV CL/F was observed in black, non-Hispanics (P < 0.001). The same trends were also observed for Hispanics (P = 0.12) and white, non-Hispanics (P = 0.25), but the differences did not achieve the level of significance.
Figure 1.
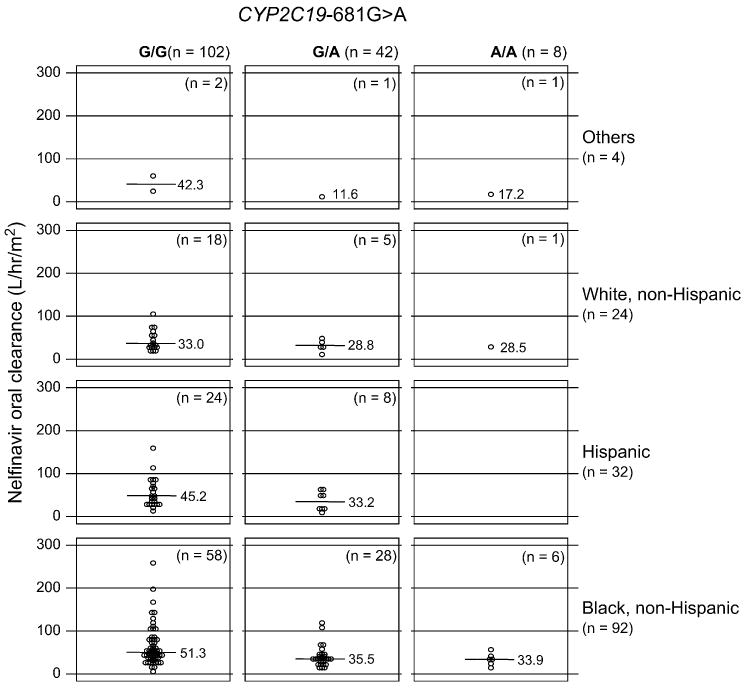
Oral clearance rate (CL/F, L/h/m2) for nelfinavir in children with the CYP2C19-681G>A genotypes in each race/ethnicity. Each circle represents nelfinavir CL/F in each subject with the CYP2C19-681-G/G (left), -G/A (heterozygous, middle), and -A/A (homozygous, right) in each race/ethnicity. The lines in the middle represent the median of CL/F for NFV.
Association Between the CYP2C19-681G>A Genotype and the M8 to NFV Ratio
Overall, the median M8 to NFV ratio was associated with the CYP2C19-681G>A genotype (P < 0.001); the ratio was 0.45 (IQR: 0.22–0.96) in those with the -G/G genotype compared with 0.26 (IQR: 0.13–0.47) in -G/A or 0.02 (IQR: 0.01–0.08) for the -A/A genotype. The association between the CYP2C9-681G>A genotype was particularly strong for the black, non-Hispanic group (P < 0.001), but not for the Hispanic (P = 0.56), and white, non-Hispanic (P = 0.30) groups. No other genotypes were associated with the M8 to NFV ratio (P = 0.29–0.87).
Virologic and Immunologic Responses During HAART in Children With ABCB1 and CYP Genotypes
The percentages in children who reached plasma HIV RNA <400 copies per milliliter at week 12 did not differ by the CYP2C19-681G>A genotypes (P = 0.14–1.00). However, at week 24, the percentage of subjects among the CYP2C19-681G>A genotype who reached plasma HIV RNA <400 copies per milliliter differed significantly (P = 0.01): 46% of subjects with the CYP2C19-681-G/G genotype achieved virologic suppression compared with 69% of those with the -G/A genotype, and 63% of those with the -A/A genotype. When examined by race/ethnicity, these differences were observed for the black, non-Hispanic group (P = 0.02) and the white, non-Hispanic group (P = 0.03), but not for Hispanics (P = 0.84). No differences were observed when the data were analyzed with the ABCB1-3435C>T genotype (P = 0.06) or the CYP3A4-392A>G genotype at week 24 (P = 0.26). Regarding immunologic recovery, changes in CD4+ T-cell percentage from baseline to weeks 12 and 24 were not different among the 3 genotypes in CYP2C19-681G>A (P = 0.50, P = 0.44, respectively) or ABCB1-3435C>T (P = 0.08, P = 0.21, respectively).
Other Factors Contributing to NFV CL/F Concomitant Antiretrovirals
Because nevirapine induces hepatic CYP3A and decreases the levels of PIs12 and ritonavir acts as a potent PK enhancer for CYP substrates,13 we evaluated the association between NFV CL/F and concomitant use of nevirapine or ritonavir. NFV CL/F was not different between subjects who received nevirapine and those who did not receive nevirapine (P = 0.70). In contrast, ritonavir use decreased NFV CL/F significantly (P = 0.002); the median NFV CL/F in patients who received ritonavir (35.8 L/h/m2, IQR: 24.7– 47.5 L·h−1·m−2) was lower compared with those who did not receive ritonavir (47.4 L/h/m2, IQR: 32.5–70.6 L·h−1·m−2).
Association of Race/Ethnicity on NFV CL/F and Clinical Outcomes
Because race/ethnicity is an important determinant of these SNPs, we also analyzed the data based on their race/ethnicity. Black, non-Hispanics (43.4 L·h−1·m−2, IQR: 33.1–66.6 L·h−1·m−2) and Hispanics (45.2 L·h−1·m−2, IQR: 26.2–65.2 L·h−1·m−2) had higher median NFV CL/F compared with white, non-Hispanics (31.7 L·h−1·m−2, IQR: 27.3–53.3 L·h−1·m−2), but it did not reach a statistical significance (P = 0.09). M8 to NFV ratio was not associated with race/ethnicity (P = 0.67). Furthermore, clinical outcomes including percentages in children who reached plasma HIV RNA <400 copies per milliliter at week 12 (P = 0.54) or changes in CD4+ T-cell percentage from baseline to weeks 12 (P = 0.89) was not associated with race/ethnicity.
A Multivariate Analysis for Predicting NFV CL/F
A multivariate analysis showed that the CYP2C19-681G>A variants (P < 0.001), concomitant use of ritonavir (P < 0.001), and age (P = 0.03) were independently associated with NFV CL/F. However, the ABCB1-3435 variants (P = 0.61), CYP3A4-392 homozygous variants (P = 0.42), and race/ethnicity (black, non-Hispanics) (P = 0.07) were no longer statistically significant. Thus, the CYP2C19-681G>A genotype remains an important pharmacogenetic determinant of NFV CL/F even after controlling for other factors.
Discussion
The data presented here demonstrate that CYP2C19-681G>A variants exert the greatest impact on NFV PK and virologic response. Controlling for various factors, only CYP2C19-681G>A genotype and concomitant PI usage continued to demonstrate a highly significant association with NFV CL/F.
Hepatic CYP2C19 is the critical enzyme responsible for conversion of NFV to its M8 metabolite.14 Alteration in CYP2C19-681G>A in exon 5, which creates an aberrant splice site resulting in a truncated nonfunctional protein15 is the SNP identified in CYP2C19 most often associated with different clinical responses to pharmacologic agents for treating diseases (eg, treatment of peptic ulcer disease using proton pump inhibitors).16 Previous reports have described the impact of this genotype on the NFV to M8 ratio in HIV-1– infected adult populations.5,17,18 Notably, Haas et al5 reported a trend toward decreased virologic failure associated with the CYP2C19-681G>A genotype. Our current data in children is in agreement with the Haas study and provide further support for an important role of CYP2C19-681G>A variants in NFV PK and virologic response. We cannot rule out that other CYP2C19 SNPs might also alter the PK of NFV or other PIs.19 In addition, we only investigated 7 SNPs which have been reported to be related to NFV PK. Furthermore, when we analyzed the data by each race/ethnicity, significant differences in NFV PK were only observed in black, non-Hispanics and virologic response in black, non-Hispanics and white, non-Hispanics. These apparent differences are likely related, in part, to the lower number of study participants in the white, non-Hispanic and Hispanic cohorts.
NFV was used extensively in years past but is now rarely used in clinical practice for children in developed countries for a few reasons; NFV has been replaced by more potent PIs (eg, lopinavir/ritonavir) with better PK profile and fewer incidences of adverse effects (eg, diarrhea) and recent manufacture problem with a contamination of ethyl methylate.20 However, NFV continues to be used in developing countries. The information learned in this current study may be helpful to improve the clinical outcomes of children who receive NFV.
In conclusion, the CYP2C19-681G>A, age, and concomitant ritonavir are significantly associated with NFV PK in HIV-1–infected children. In addition, favorable virologic response was observed in children with the CYP2C19-681G>A variants associated with lower oral NFV CL/F. These findings suggest that CYP2C19-681G>A is the most important pharmacogenetic determinant for NFV PK and virologic responses in children who receive HAART-containing NFV.
Acknowledgments
Supported by the Pediatric AIDS Clinical Trials Group/International Maternal, Perinatal, Adolescent AIDS Clinical Trials Group, by Grants from the National Institute of Allergy and Infectious Diseases [(U01A141089 and 5K23AI-56931 to AS, AI-41089, AI-39004, AI-27563, AI-33835, AI-41110); AI-36214 (Virology Core University of California, San Diego Center for AIDS Research), AI-32921, AI-68632 and AI-68616] and by Bristol-Myers Squibb.
Footnotes
Presented in part at the: 15th Conference on Retroviruses and Opportunistic Infections, February 2008, Boston, MA. Poster #572.
References
- 1.Lee CG, Gottesman MM, Cardarelli CO, et al. HIV-1 protease inhibitors are substrates for the MDR1 multidrug transporter. Biochemistry. 1998;37:3594–3601. doi: 10.1021/bi972709x. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Chan WK. Transport, metabolism and elimination mechanisms of anti-HIV agents. Adv Drug Deliv Rev. 1999;39:81–103. doi: 10.1016/s0169-409x(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 3.Zhang KE, Wu E, Patick AK, et al. Circulating metabolites of the human immunodeficiency virus protease inhibitor nelfinavir in humans: structural identification, levels in plasma, and antiviral activities. Antimicrob Agents Chemother. 2001;45:1086–1093. doi: 10.1128/AAC.45.4.1086-1093.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fellay J, Marzolini C, Meaden ER, et al. Response to antiretroviral treatment in HIV-1-infected individuals with allelic variants of the multidrug resistance transporter 1: a pharmacogenetics study. Lancet. 2002;359:30–36. doi: 10.1016/S0140-6736(02)07276-8. [DOI] [PubMed] [Google Scholar]
- 5.Haas DW, Smeaton LM, Shafer RW, et al. Pharmacogenetics of long-term responses to antiretroviral regimens containing efavirenz and/or nelfinavir: an Adult AIDS Clinical Trials Group Study. J Infect Dis. 2005;192:1931–1942. doi: 10.1086/497610. [DOI] [PubMed] [Google Scholar]
- 6.Saitoh A, Singh KK, Powell CA, et al. An MDR1-3435 variant is associated with higher plasma nelfinavir levels and more rapid virologic response in HIV-1 infected children. AIDS. 2005;19:371–380. doi: 10.1097/01.aids.0000161766.13782.2f. [DOI] [PubMed] [Google Scholar]
- 7.Kovacs A, Montepiedra G, Carey V, et al. Immune reconstitution after receipt of highly active antiretroviral therapy in children with advanced or progressive HIV disease and complete or partial viral load response. J Infect Dis. 2005;192:296–302. doi: 10.1086/430922. [DOI] [PubMed] [Google Scholar]
- 8.Floren LC, Wiznia A, Hayashi S, et al. Nelfinavir pharmacokinetics in stable human immunodeficiency virus-positive children: Pediatric AIDS Clinical Trials Group Protocol 377. Pediatrics. 2003;112:e220–e227. doi: 10.1542/peds.112.3.e220. [DOI] [PubMed] [Google Scholar]
- 9.Capparelli EV, Burchett SK, Kovacs A, et al. Characterization of the ritonavir-nelfinavir pharmacokinetic interaction in pediatric patients with advanced HIV disease using a mixed effects modeling approach. Pharmacotherapy. 2003;23:401–402. [Google Scholar]
- 10.Oselin K, Gerloff T, Mrozikiewicz PM, et al. MDR1 polymorphisms G2677T in exon 21 and C3435T in exon 26 fail to affect rhodamine 123 efflux in peripheral blood lymphocytes. Fundam Clin Pharmacol. 2003;17:463–469. doi: 10.1046/j.1472-8206.2003.00163.x. [DOI] [PubMed] [Google Scholar]
- 11.Borlak J, Thum T. Identification of major CYP2C9 and CYP2C19 polymorphisms by fluorescence resonance energy transfer analysis. Clin Chem. 2002;48:1592–1594. [PubMed] [Google Scholar]
- 12.Murphy RL, Sommadossi JP, Lamson M, et al. Antiviral effect and pharmacokinetic interaction between nevirapine and indinavir in persons infected with human immunodeficiency virus type 1. J Infect Dis. 1999;179:1116–1123. doi: 10.1086/314703. [DOI] [PubMed] [Google Scholar]
- 13.Kempf DJ, Marsh KC, Kumar G, et al. Pharmacokinetic enhancement of inhibitors of the human immunodeficiency virus protease by coadministration with ritonavir. Antimicrob Agents Chemother. 1997;41:654–660. doi: 10.1128/aac.41.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirani VN, Raucy JL, Lasker JM. Conversion of the HIV protease inhibitor nelfinavir to a bioactive metabolite by human liver CYP2C19. Drug Metab Dispos. 2004;32:1462–1467. doi: 10.1124/dmd.104.001743. [DOI] [PubMed] [Google Scholar]
- 15.de Morais SM, Wilkinson GR, Blaisdell J, et al. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269:15419–15422. [PubMed] [Google Scholar]
- 16.Padol S, Yuan Y, Thabane M, et al. The effect of CYP2C19 polymorphisms on H. pylori eradication rate in dual and triple first-line PPI therapies: a meta-analysis. Am J Gastroenterol. 2006;101:1467–1475. doi: 10.1111/j.1572-0241.2006.00717.x. [DOI] [PubMed] [Google Scholar]
- 17.Burger DM, Schwietert HR, Colbers EP, et al. The effect of the CYP2C19*2 heterozygote genotype on the pharmacokinetics of nelfinavir. Br J Clin Pharmacol. 2006;62:250–252. doi: 10.1111/j.1365-2125.2006.02635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirt D, Mentre F, Tran A, et al. Effect of CYP2C19 polymorphism on nelfinavir to M8 biotransformation in HIV patients. Br J Clin Pharmacol. 2008;65:548–557. doi: 10.1111/j.1365-2125.2007.03039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibeanu GC, Goldstein JA, Meyer U, et al. Identification of new human CYP2C19 alleles (CYP2C19*6 and CYP2C19*2B) in a Caucasian poor metabolizer of mephenytoin. J Pharmacol Exp Ther. 1998;286:1490–1495. [PubMed] [Google Scholar]
- 20.Wilcox RD. Changes are recommended in use of nelfinavir. HIV Clin. 2008;20:1–4. [PubMed] [Google Scholar]


