Abstract
Posttranslational histone modifications play an important role in regulating chromatin based nuclear processes including transcription. Of these modifications, histone ubiquitination is among the least understood. Histone ubiquitination predominately targets histones H2A and H2B. While ubiquitination of H2B is evolutionarily conserved from budding yeast to mammals, ubiquitination of H2A has not been detected in budding yeast, worms, or plants. Until recently, studies of histone ubiquitination lagged far behind the study of other histone modifications, largely because antibodies specific for ubiquitinated histones are difficult to generate. Despite this obstacle, the identification of the enzymatic machineries involved in histone ubiquitination, together with the successful use of a combination of genetic and immunoblot approaches to detect ubiquitinated histones, have helped to reveal important regulatory roles for this modification in transcriptional initiation and elongation, cell cycle progression, and DNA damage response. With the aid of the recently developed ubiquitinated histone-specific antibodies, an intriguing link between histone ubiquitination and cancer development has been established. While the enzymes involved in H2B ubiquitination were identified first in budding yeast and subsequently in higher organisms based on gene homology, the identification of the enzymatic machineries involved in H2A ubiquitination largely depended on a biochemical purification approach. The unbiased search for ubiquitin ligases targeting histones also led to the identification of a H3 and H4 ubiquitin ligase. Here we detail a protocol for the biochemical approach to identify histone ubiquitin ligase(s) from HeLa cells. Similar approaches have been successfully used to identify histone methyltransferases, histone demethylases, chromatin remodeling factors, and general transcription factors. So long as an in vitro enzymatic assay can be established, the approach we describe can be easily adapted to identify other histone and non-histone modifying enzymes.
1. Introduction
The nucleosome is the basic packaging unit of chromatin, the physiological substrate for transcription (1–3). Chromatin is highly dynamic, undergoing significant structural changes during mitosis and ordinary transcription (4, 5). It has recently become apparent that nucleosomes are integrally involved in modulating chromatin structure and gene accessibility (1–5). The canonical nucleosome is composed of four core histones: a histone H3/H4 tetramer and two H2A/H2B dimers. These four histones possess highly charged N-terminal tails which are subject to extensive posttranslational modifications including acetylation, sumoylation, methylation, phosphorylation and ubiquitination (1–4). These modifications can alter the chromatin structure directly or recruit downstream molecules that mediate specific physiological responses (2–4, 6).
Given the fundamental roles of histone modifications in controlling chromatin structure and function, it is not surprising that misregulation of histone modifications is a hallmark of cancer (7). Aberrant histone modifications, alone or in cooperation with other epigenetic regulatory mechanisms, can alter gene expression (8), potentially activating oncogenes or silencing tumor suppressors (9). Histone modifying enzymes have been implicated in maintenance of genomic stability (10) with mutations in genes encoding histone modifying enzymes associated with several types of cancer (11–14). Transcriptional profiles of cancerous cells reveal a genome wide loss of H4 lysine 16 acetylation and lysine 20 trimethylation in lymphomas and colorectal adenocarcinomas (15). Loss of the repressive histone trimethylation modifications at H3 lysine 27 and H4 lysine 20 are associated with derepression of CLDN3 and CLDN4 during ovarian tumorogenesis (16). Polycomb repressive complex component Bmi1 is overexpressed in many human cancers (17). Interestingly, Bmi1 is also a component of a histone H2A ubiquitin ligase complex and plays a critical role in the regulation of H2A ubiquitination both in vitro and in vivo (18–20).
Extensive efforts have been made to study posttranslational histone modifications. These studies have helped reveal the critical roles of posttranslational histone modifications in regulating virtually all nuclear processes and identifying the important function of aberrant histone modification in human disease. Mass Spectrometry has been used to map many modification sites (21). Antibodies against specific modified histones have been generated and used in chromatin immunoprecipitation assays to localize specific histone modifications throughout the genome (22–24). However, neither approach is capable of establishing a causal relationship between a specific histone modification and a certain biological pathway. The identification of histone modifying enzymes provides a solution to this difficulty.
The genetically tractable organism budding yeast has been used to generate a library of strains containing knockouts of all viable genes. Screening these knockout strains with antibodies for specific histone modifications has provided much information about the enzymes responsible for histone modifications (25, 26). Similar strategies, employing RNA interference (RNAi) mechanisms, have been used to identify signaling molecules involved in specific pathways in mammalian cells (27, 28). However, the large number of mammalian genes and the redundancy of many enzymes within a pathway, limits the usefulness of this method. Biochemical purification employing conventional column chromatography offers an alternative approach. Paired with an in vitro activity assay, this approach allows the systematic search for histone modifying enzymes. Biochemical purification is particularly useful when no information is previously known about a particular modification, as when defining novel histone modifying enzymes. The approach has been successfully used to define histone methyltransferases, histone demethylases, histone ubiquitin ligases, deubiquitinases, and chromatin remodeling factors (29–32). Information derived from these studies has greatly advanced our understanding of the role of histone modifications in epigenetic regulation. Enzymes identified from these studies also provide novel targets for epigenetic-based pharmaceutical interventions.
Histone ubiquitination is a unique modification, where an 8 kDa bulky globular protein is attached to the carboxyl terminus of histone H2A or H2B. Due to potential physical interference, antibodies specific for ubiquitinated histones and suitable for chromatin immunoprecipitation assay could not be generated until very recently. Therefore, despite being known for more than thirty years, the functions of histone ubiquitination remained undetermined. The successful use of a combination of genetic and immunoblot approaches to detect ubiquitinated histones as well as the recent identification of the enzymatic machineries involved in histone ubiquitination have revealed important functions for histone ubiquitination in transcriptional regulation, cell cycle progression, and DNA damage response (33–35).
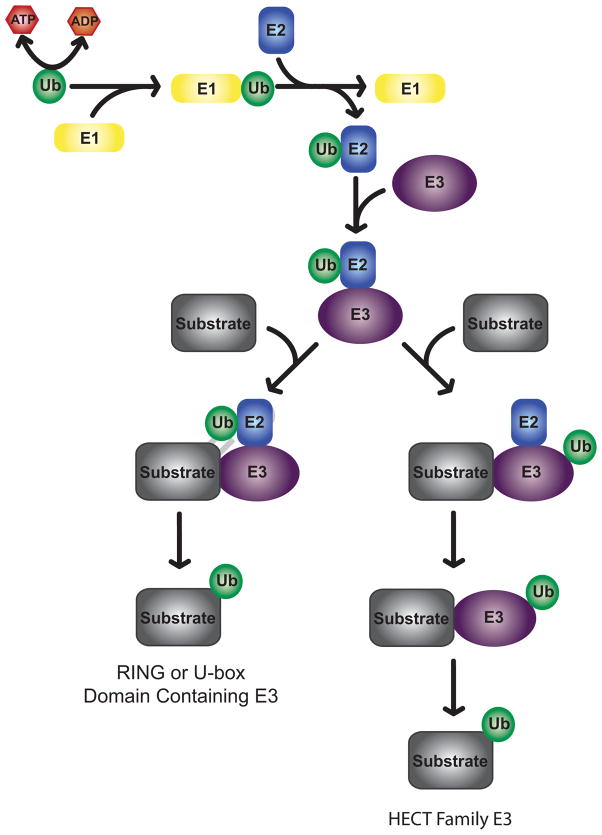
Similar to ubiquitination of other proteins, histone ubiquitination proceeds through a three step process culminating in the attachment of ubiquitin to an acceptor lysine residue via an isopeptide linkage (Figure 1). Ubiquitin is first activated in an ATP dependent manner by an ubiquitin activating enzyme (E1). Next, ubiquitin is transferred to a cysteine residue of an ubiquitin conjugating enzyme (E2) via a thiolester linkage. Finally, ubiquitin is ligated to the target molecule by an E3 ligase. The E3 ubiquitin ligase is responsible for determining substrate specificity. As such, E3 ligases are the most plentiful component of the ubiquitination system with over 600 human E3 ligases identified (36). E3 ligases may be single proteins or exist as a multi-enzyme complex. HECT family E3 ligases receive ubiquitin from an E2 ubiquitin conjugating enzyme and then transfer ubiquitin to target proteins (37). RING or U-box domain containing E3 ligases act as intermediaries, forming a complex that includes the target protein and E2 bound ubiquitin (38). In this case, the E3 ligase stimulates the activity of the interacting E2 and oversees conjugation of E2 bound ubiquitin to the target protein. Histones are primarily subjected to mono-ubiquitination, where only a single ubiquitin molecule is attached to K119 of H2A or K120 of H2B (K123 in budding yeast). While poly-ubiquitination is most often associated with protein degradation, an expanding body of research points to the role of mono-ubiquitination in protein targeting and alteration of protein function and activity (39, 40).
Figure 1.
Schematic depiction of histone ubiquitination. Ubiquitin is first activated by an E1 activating enzyme. Ubiquitin is then transferred to an E2 ubiquitin conjugating enzyme. HECT family E3 ligases receive ubiquitin from an E2 and directly conjugate ubiquitin to their target molecules. RING and U-box domain containing E3 ligases mediate the interaction between E2 ubiquitin conjugating enzymes and substrates and oversee the conjugation of ubiquitin to target molecules.
Studies in the biochemically and genetically tractable organism S. cerevisiae identified Rad6 as an ubiquitin conjugating enzyme for H2B (41). Recently a Ring finger-containing protein Bre1 was identified as the E3 ligase for H2B ubiquitination in budding yeast (42, 43). The enzymatic machineries for H2B ubiquitination appear to be evolutionarily conserved, as is the modification itself. Mammalian homologues of Rad6, HR6A and HR6B (44, 45), were recently shown to mediate H2B ubiquitination in higher eukaryotes (46). These two proteins likely play redundant roles (47). Similarly, Bre1 has two homologs in mammals, RNF20/hBre1 and RNF40; however, only RNF20 seems to be required for H2B ubiquitination in vivo (48, 49). These studies, with the aid of the recently developed ubiquitinated histone-specific antibodies, have revealed an intriguing link between histone ubiquitination and cancer development (50, 51). Unlike H2B ubiquitination, H2A ubiquitination has not been detected in budding yeast, Caenorhabditis elegans, or Arabidopsis (41, 52, 53). This prevents the use of genetically tractable lower organisms to study this modification. The H2A ubiquitin ligase was identified as Ring 2 by an unbiased search for E3 ligases that target histones using a biochemical purification approach (18). Interestingly, the H2A ubiquitin ligase identified in this study was coincidently discovered by another group looking at the levels of ubiquitinated H2A on the inactivated X chromosome in a mouse model with knockout of Polycomb protein Ring 2 (52). The identification of Ring2 as the ubiquitin ligase for H2A reveals an intriguing connection between H2A ubiquitination and Polycomb protein mediated gene silencing and resolves the long standing puzzle of this modification (18, 52, 54). During the search for the histone H2A ubiquitin ligase, a histone H3 and H4 ubiquitin ligase, CUL4-DDB-ROC1, was also identified (55). Further analyses revealed a potential link between histone H3 and H4 ubiquitination and the cellular response to DNA damage (55). Here we describe the detailed experimental procedures for the purification and identification of histone ubiquitin ligases from HeLa cells. We also discuss the advantages as well as the limitations of this approach.
2. Methods
Biochemical purification of histone ubiquitin E3 ligases from HeLa cells requires a sensitive and reliable in vitro assay system. We have developed such an assay system using nucleosomes and/or histone octamer as substrates. In the following sections, we detail the experimental procedures for preparation of these substrates, preparation of protein fractions derived from HeLa cell nuclei, establishment of the in vitro histone ubiquitin ligase assay, and outline the strategies used to purify two histone ubiquitin ligases.
2.1 Substrate Preparation
2.1.1 Preparation of histone substrates from large scale cultures of HeLa cells
Histone substrates in different forms should be used in initial establishment of the in vitro ubiquitin ligase assay. Oligonucleosomes, mononucleosomes and histone octamers are prepared from HeLa cells following a published procedure (29). This protocol involves isolation of nuclei, micrococcal nuclease digestion of nuclei, extraction of nucleosomes from digested nuclei, and sucrose gradient purification of crude nucleosomes. To obtain histone octamers, an additional hydroxyapatite chromatography purification step is required. All steps are carried out on ice unless otherwise indicated.
-
Cell culture. HeLa S3 (ATTC CCL-2.2) cells can be purchased from the ATCC for growth in suspension culture.
HeLa S3 cells are grown in media consisting of Joklik Modified MEM powder (Sigma-Aldrich Chemical), 5% Newborn calf serum (Fisher Scientific), 10 ml/L Pen-Strep (Fisher Scientific), 10 ml/L non essential amino acids (Fisher Scientific) and 2 g/L sodium bicarbonate (Fisher Scientific). The Joklik’s basal medium is dissolved in water for irrigation (Hospira) along with all remaining components except the serum. The pH is adjusted to 7.0 with 1M NaOH using a pH meter (Orion). The media is then filtered into sterile bottles using a 0.22μ sterile filter (Millipore) and stored at 4°C until used. Newborn calf serum is thawed and added to the media prior to use. All incubations are done in CO2 incubators (Forma Scientific) at 37°C and 5% CO2 fitted with biological stirrers (Techne) for spinner flasks (Wheaton).
Two 100 mm plates containing 10 ml of media are inoculated with 0.5 ml HeLa cell frozen stock and incubated for 24 hrs. The plates are then washed with 3 ml PBS (Fisher). Two (2) milliliters PBS and 100 μl Trypsin (Fisher) are added to each plate. After a 10 minute incubation 4 ml of media is added and the cells are detached by pipetting across the plate. Each plate is split to 3 plates containing 8 ml of fresh media and incubated for 48 hrs. Cells from these 6 plates are detached as before and transferred to 300 ml of media in a 500 ml spinner flask and incubated at 37°C with 50 rpm. The cells are counted each day using a hemocytometer (Hauser Scientific) and the cell viability is estimated by the trypan blue exclusion method. Once the cell count rises above 1 × 106/ml (with >95% viability) the 300 ml culture is transferred to a 3 L spinner flask containing 2.7 L of fresh media. Once the cell count rises above 1 × 106/ml the culture is expanded to eight 3 L spinner flasks.
The cells are incubated until the cell count is 1–2 × 106/ml (>95% viability) and then harvested. The culture is transferred to sterile 1 L centrifuge bottles (Nalgene) and centrifuged for 10 minutes at 1,500 rpm in an RC-3B centrifuge (Sorvall Instruments) at 4°C. The supernatant is decanted and the cells are gently resuspended in fresh media and pooled. The pooled resuspended cells are then transferred to sterile 250 ml conical bottles (Corning) and centrifuged for 10 minutes at 1,500 rpm. The supernatant is decanted. Cell pellets should be processed as soon as possible after collection. Cells are gently resuspended in PBS supplemented with 1 g MgCl2/L and collected by centrifugation at 3,000 rpm for 10 minutes at 4°C (Fisher Accuspin 3R, rotor #4393). Cells are washed once more in PBS supplemented with 1 g MgCl2/L and collected as before.
-
Isolation of nuclei.
The cell pellet is resuspended in 15 mL of Buffer A (10 mM MES pH 6.5, 5 mM MgCl2, 1 mM CaCl2, 15 mM NaCl, 60 mM KCl, 0.25 M sucrose, 0.5% Triton X-100, 0.5 mM sodium metabisulfite, 0.5 mM benzamidine-HCL, 0.1 mM PMSF, 0.5 mM DDT, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 1 μg/ml aprotinin) per 5 × 108 cells and aliquoted into 3–4 Dounce homogenizers (Wheaton, 357546).
Each aliquot is homogenized 10 times with a type B pestle (loose). The cell suspension is incubated on ice for 15 minutes to allow for the release of nuclei.
Nuclei are collected by centrifugation at 5,000 rpm for 10 minutes (rotor #JA-17, Beckman) and washed once with Buffer A before resuspending in 1.2 ml of Buffer B (10 mM PIPES pH 6.8, 50 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 0.5 mM sodium metabisulfite, 0.5 mM benzamidine-HCl, 0.1 mM PMSF, 0.5 mM DDT, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 1 μg/ml aprotinin) per 5 × 108 cells.
-
Determination of optimal micrococcal digestion conditions.
A 500 μl aliquot of nuclei suspension is removed and adjusted to a final concentration of 5 mM CaCl2 by addition of 1 M CaCl2.
The nuclei are pre-warmed at 37°C in a tabletop water bath (Fisher Isotemp 202) for 5 minutes followed by addition of 4 μl of micrococcal nuclease (MNase) stock (Sigma, 200 U/mL prepared in 5 mM Na2HPO pH 7.0, 2.5 μM CaCl2) 50 μl samples are removed from the suspension at five minute intervals for 45 minutes and transferred immediately to a microcentrifuge tube containing 1 μl 0.5 M EDTA to stop the reaction.
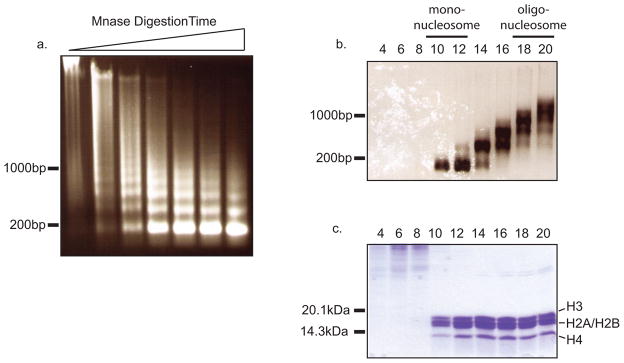
Digestion efficiency is checked by DNA extraction. 75 μl ddH20, 30 μl 10% SDS, 25 μl 4 M NaCl and 200 μl chloroform are sequentially added to each microcentrifuge tube and vortexed after each addition. Each tube is centrifuged for 5 minutes at 132,000 rpm in a tabletop microcentrifuge (Eppendorf Microcentrifuge 5415D) at room temperature. Following centrifugation, 5 μl of aqueous phase is removed from samples taken from each time point and loaded to a 1% agarose gel. Optimal digestion should yield a bulk of fragments approximately 2 kb in size (Figure 2a).
-
Micrococcal digestion of bulk nuclei.
The CaCl2 concentration for the remaining nuclei suspension is adjusted to 5 mM with 1 M CaCl2 and the suspension is pre-warmed to 37°C for 15 minutes in a water bath.
Micrococcal nuclease is added at 8 μl/mL nuclei and the suspension is digested for the optimized time in step 3.
The reaction is stopped by addition of 0.5 M EDTA to a final concentration of 4 mM.
-
Extraction of nucleosomes.
The digested nuclei are centrifuged at 4,500 rpm for 5 minutes (rotor # JA-17, Beckman) and the supernatant is saved as S1.
The pellet is then resuspend in 0.5 mL nucleosome extraction buffer (20 mM Tris HCl pH 7.9, 10 mM EDTA, 0.5 M NaCl) per 5 × 108 cells and incubated on ice for 15 minutes. The digested nuclei are centrifuged at 10,000 rpm for 10 minutes and the supernatant is saved as S2. The extraction should be repeated 2–3 times until no more protein can be extracted.
Supernatants, which are enriched for nucleosomes, are pooled. If H1-depleted nucleosomes are needed, H1 can be disassociated from core nucleosomes by increasing the NaCl concentration to 0.75 M (final). NaCl stock solution should be added dropwise with constant agitation to avoid precipitation. We have previously noted that the presence of H1 may impair the activity of histone ubiquitin ligases.
The concentration of the extracted nucleosomes is determined by adding 10 μl pooled supernatant to 1 mL 1 M NaOH and measuring the absorption at 260 nanometers using a tabletop spectrophotometer (Bio-rad SmartSpec Plus). A reading of 0.1 is designated as 1 unit. The total units are calculated as volume (ml) × A260 ×10.
-
Sucrose gradient purification of nucleosomes.
Depending upon the amount of nucleosomes extracted in step 5, 3–5 35 mL 5–30% non-linear sucrose gradients should be prepared using 40 ml polyallomer tubes (Beckman) with a binary gradient mixing apparatus (Hoefer SG100, Amersham) in buffer NG (10 mM Tris HCl, 1 mM EDTA, 0.5 M NaCl, 0.3 mM PMSF, pH 7.5). Alternatively, the gradient can be poured with the ÄKTA FPLC system.
Approximately 300 units of the extracted nucleosomes are carefully layered on the top of each gradient and centrifuged at 26,000 rpm for 17 hours (SW28 rotor, Beckman).
The gradients are manually fractionated from top to bottom by removing an aliquot (1 mL) of sample each time.
-
Analysis of sucrose gradient fractions.
The length of the DNA fragments contained in each sucrose gradient fraction can be determined using the procedure described in step 3.
Twenty five microliter aliquots from each fraction are TCA precipitated and the protein profiles are analyzed by 18% SDS-PAGE. Briefly, 25 μl of 25% TCA is added to each 25 μl aliquot and the samples are incubated on ice for 20 minutes to precipitate proteins. Samples are then centrifuged at 132,000 rpm for 15 minutes at 4°C. The supernatant is aspirated and the pellets are washed in 0.5 mL cold acetone (−20°C) before being collected by centrifugation for 10 minutes at 132,000 rpm at 4°C. Again, the supernatant is aspirated and the pellets are air dried. Dried pellets are dissolved in 20 μl 1× SDS sample loading buffer and resolved by 18% SDS-PAGE. Protein profiles can be analyzed by staining the SDS-PAGE with Coomassie Blue.
Fractions containing DNA fragments with lengths between 1 and 2.5 Kb (7–15 nucleosomes) and with high purity are pooled (Figure 2b and 2c fractions 18 and 20) and dialyzed against histone storage buffer for 4 hrs (10 mM Hepes-KOH pH 7.5, 1 mM EDTA, 10 mM KCl, 10% glycerol, 0.2 mM PMSF).
Samples are aliquoted and saved as oligonucleosomes. Oligonucleosomes can be stored at 4°C for 2–3 months and more than two years at −80°C. However, repeated freeze-thaw cycles should be avoided.
-
Preparation of mononucleosomes.
Mononucleosomes can be obtained from the upper fractions of the sucrose gradient (Figure 2b and 2c fractions 10 and 12), as determined by protein and DNA length analyses (step 6–7). However, these mononucleosomes are often contaminated with high molecular weight proteins.
As an alternative, high purity mononucleosomes can be obtained by digestion of the oligonucleosomes obtained in step 7. Briefly, oligonucleosomes are dialyzed against Tris-HCl pH 8.0 containing 1 mM PMSF to remove EDTA and sucrose. After dialysis the CaCl2 concentration is adjusted to 5 mM (final) and the oligonucleosomes are digested with micrococcal nuclease to generate mononucleosomes as in steps 3–4. Mononucleosomes are purified by sucrose gradient as in steps 6–7. Fractions with high purity are pooled and dialyzed against histone storage buffer.
Mononucleosomes are less stable than oligonucleosomes, therefore it is recommended that mononucleosomes be stored in small aliquots at −80°C.
-
Preparation of histone octamer.
To obtain histone octamers, the desired amount of oligonucleosome is dialyzed against T50E (50 mM Tris HCl pH 7.9, 1 mM EDTA, 1 mM DTT, and 0.5 mM PMSF) for 3 hours. Protein concentration should be measured as in step 5 and used to prepare a hydroxyapatite column of appropriate volume (Bio-rad, 130-0151; binding capacity: 4 mg protein/1 ml beads and 40 μg DNA/1 ml beads).
The column is equilibrated with buffer NP300 (40 mM Na2HPO4/NaH2PO4 pH 6.8, 0.3 M NaCl, 1mM DTT, and 0.2 mM PMSF) and the dialyzed sample is loaded to the column.
The column is washed with 6–10 column volumes (cv) of buffer NP500 (40 mM Na2HPO4/NaH2PO4 pH 6.8, 0.5 M NaCl, 1 mM DTT, 0.2 mM PMSF) and the histones are eluted with buffer NP2500 (40 mM Na2HPO4/NaH2PO4 pH 6.8, 2.5 M NaCl, 1 mM DTT, 0.2 mM PMSF).
Protein purity is analyzed by SDS-PAGE as in step 7. Samples are dialyzed against histone storage buffer and stored at −80°C in small aliquots.
Figure 2.
Purification of oligonucleosomes and mononucleosomes from HeLa S3 cells. a. MNase test digestion of 500 μl nuclei suspension for increasing lengths of time. b. DNA extraction of from sucrose gradient fractions. Mononucleosomes are enriched in fraction containing a majority of DNA approximately 150bp in length (fractions 10–12). Oligonucleosomes are enriched in fractions 18–20. c. TCA precipitation of different sucrose gradient fractions. Early fractions contain high molecular weight contaminants.
2.1.2 Preparation of histone substrates from small scale cultures of HeLa cells
For laboratories that are not proficient in biochemical or epigenetic techniques, it might be difficult to follow the preceding substrate purification protocol. Therefore, we have developed a convenient small scale protocol for substrate preparation. Substrates can be prepared from a few as 2–3 10 cm cell culture plates making this protocol particularly useful for cell types that are difficult to culture in large scales. We have used this protocol to purify nucleosomes from HeLa stable cell lines and embryonic stem cells. This protocol only requires 2–3 hours and a benchtop microcentrifuge (Eppendorf, Centrifuge 5415D or equivalent). All steps should be carried out on ice unless otherwise indicated.
Culture 2–3 10 cm plates of HeLa cells in DMEM containing 10% FBS and Penicillin and Streptomycin (100 U/mL and 100 μg/mL respectively) until 80% confluent. Cells are harvested by trypsinization and washed twice with PBS containing 1 g MgCl2/L.
Hela cells are resuspended by pipetting in 1.5 mL Buffer A (0.2 5M sucrose, 60 mM KCl, 10 mM NaCl, 15 mM MES pH 6.5, 5 mM MgCl2, 1 mM CaCl2, 0.5% Triton X-100, 1 mM DTT, 0.2 mM PMSF, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 1 μg/ml aprotinin) and incubated on ice for 15 minutes.
Nuclei are collected by centrifugation at 3,600 rpm for 10 minutes and washed once with 1.5 ml Buffer A. Nuclei are centrifuged as before and the pellet is saved.
Pellets are resuspended by pipetting in 1.5 mL TNM Buffer (1 0mM Tris-HCl pH 8.0, 10 mM NaCl, 2 mM MgCl2, 0.3 M Sucrose, 1 mM DTT, 0.2 mM PMSF, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 1 μg/ml aprotinin) containing CaCl2 at 5 mM (final). Nuclei are collected by centrifuging at 3,600 rpm for 10 minutes to equilibrate nuclei with TNM Buffer. Nuclei are recovered and resuspend in 300 μl TNM Buffer containing 5 mM CaCl2. The nuclei should be pre-warmed to 37°C for 5 minutes. After the 5 minute incubation, 5 μl micrococcal nuclease (200 U/μl) is added to nuclei, which are then incubated at 37°C for 20 minutes to give a complete digestion.
After incubation, digested nuclei are collected by centrifuging at 3,600 rpm for 10 minutes and are resuspended in 400 μl nucleosome extraction buffer. Digested nuclei are incubated on ice for 10 minutes and then centrifuged at 132,000 rpm for 10 minutes. The supernatant is highly enriched for mononucleosomes. This extraction is repeated until no more protein is released.
Extracted fractions are pooled and diluted to a protein concentration of 0.2–0.4 μg/mL by dropwise addition of histone storage buffer (with Nonidet-40 concentration of 0.05%) while vortexing.
Samples are dialyzed against histone storage buffer, aliquoted and stored at −80°C.
To obtain oligonucleosomes, the conditions for digestion are optimized as in 2.1.1 step 3. Bulk nuclei are digested to give nucleosomes ranging in size from 1–15 nucleosomes. Mono- and oligo-nucleosomes can be separated on a 5 ml 5–30% linear sucrose gradient, which is prepared by loading layers of increasing concentrations of sucrose (mixing the 5% and 30% sucrose solution at different ratios). Histone octamers can be obtained with a small scale hydroxyapatite column.
2.2 Preparation of nuclear proteins from cultured HeLa cells
Purification of histone ubiquitin ligases using conventional chromatography requires a large amount of starting material. This is to ensure that enough purified sample can be recovered for biochemical assay and mass spectrometry identification. Based on our experience, purification of histone ubiquitin ligases requires a minimum of four grams of nuclear extract or nuclear pellet. Below we describe protocols for the preparation of nuclear extract and nuclear pellet fractions from cultured HeLa cells (56, 57). The nuclear extract contains proteins that are extracted from HeLa nuclei by 500 mM NaCl, while the nuclear pellet contains proteins that remain tightly associated with chromatin under these conditions. These methods can be adapted to other starting materials such as embryonic stem cells and primary tissues.
2.2.1 HeLa S3 cell culture
The HeLa S3 cell line is cultured as in 2.1.1 step 1. In house culturing of HeLa S3 cells may require 3–4 months to produce enough material to start purification. Batches of cells can be processed as they are ready by following the method below and then stored at −80°C until sufficient starting material has been generated.
2.2.2 Fractionation of HeLa cell proteins
The viability (> 97%) and density of HeLa S3 cultures is determined before collecting the cells by centrifugation at 3,000 rpm for 8 minutes (Fisher Accuspin 3R, rotor #4393). The cells are resuspended in one volume of PBS, aliquoted into 50 mL Falcon tubes, centrifuged at 2,500 rpm for 10 minutes and washed once more with PBS. Supernatant is decanted and the volume of the cell pellets is measured.
Five volumes of Buffer A is added (10 mM Tris HCl pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2 mM PMSF) to each cell pellet and the pellets are resuspended completely by pipetting. Complete resuspension of cell pellets is imperative for proper fractionation. Cells are incubated 10 minutes on ice before centrifuging at 2,000 rpm for 10 minutes.
The volume of the swollen cell pellets is measured and 2 volumes of Buffer A are added to each pellet. The cell pellets are resuspended with ten strokes of the type B pestle of a Dounce homogenizer and then centrifuged at 2,500 rpm for 10 minutes.
The supernatant is carefully transferred to a clean graduated cylinder and the pellet is retained for further processing. A 10× stock solution of Buffer B (0.3 M Tris-HCl pH 7.9, 0.03 M MgCl2, 1.4 M KCl, 1 mM DTT, 0.2 mM PMSF, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 1 μg/ml aprotinin) is added to the supernatant to a final concentration of 1× Buffer Band mixed well. The supernatant is centrifuged for 60 minutes at 30,000 rpm (Beckman Coulter Optima XL-100K ultracentrifuge, Type 45Ti rotor). After centrifuging, the supernatant is removed and dialyzed against 4 L of BC50 (20 mM Tris HCl pH 7.9, 50 mM KCl, 0.2 mM EDTA, 10% glycerol, 10 mM DTT, 0.2 mM PMSF) for 4–7 hours. Following dialysis, the supernatant is centrifuged at 12,000 rpm for 20 minutes. The supernatant is recovered as S100 (cytoplasm). Both protein concentration and sample volume are measured, and PMSF is added to a final concentration of 0.2 mM. The S100 is stored at −80°C.
The pellet from step 3 is resuspended in 3 mL Buffer C (20 mM Tris HCl pH 7.9, 0.42 M NaCl, 0.2 mM EDTA, 1.5 mM MgCl2, 25% glycerol, 0.5 mM DTT, 0.5 mM PMSF, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 1 μg/ml aprotinin) per 109 cells with ten strokes of the type B pestle (loose) of a Dounce homogenizer. The pellet is then poured into a small plastic beaker and stirred gently with a magnetic stirring bar for 30 minutes at 4°C. The resuspended pellet is collected and centrifuged at 15,000 rpm for 30 minutes. Both the supernatant and the pellet are recovered.
The supernatant is dialyzed against BC50 for 4–7 hours and then centrifuged at 12,000 rpm for 20 minutes. The recovered supernatant is the nuclear extract. Sample volume and protein concentration should be measured as in step 4. PMSF is added to a final concentration of 0.2 mM, and the nuclear extract is stored at −80°C.
The nuclear pellet from step 5 is resuspended in Buffer E (50 mM Tris HCl pH 7.9, 5 mM MgCl2, 0.5 mM EDTA, 25% glycerol, 5 mM DTT, 0.2 mM PMSF, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 1 μg/ml aprotinin) with the type B pestle of a Dounce homogenizer. The pellet will remain viscous. The nuclear pellet should be stored at 7minus;80°C.
Solublization of nuclear pellet. Nuclear pellet solublization begins with at least 25 batches of nuclear pellet preparation. Nuclear pellets are thawed in cold water and resuspended with pestle B of a Dounce homogenizer by adding Buffer B (50 mM Tris HCl pH 7.9, 5 mM MgCl2, 0.5 mM EDTA, 25% Glycerol, 5 mM DTT, 0.2 mM PMSF, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 1 μg/ml aprotinin) to 36 ml per 1010 cells. The pellet is homogenized 2–3 times to mix well.
The nuclear pellet suspension is transferred to a graduated cylinder and 1/10 volume of 3M (NH4)2SO4 is added by immediately inverting the cylinder several times. The suspension should become thick and gelatinous.
The suspension is transferred to a plastic beaker and sonicated (Fisher 60 Sonic Dismembrator) at an output of approximately 10 watts until it is no longer viscous. Avoid overheating by placing the beaker on dry ice. Sonication may take between 20 and 45 minutes depending upon sample size. The suspension is centrifuged at 26,000 rpm for 60 minutes at 4°C to remove cell debris (Beckman Coulter Optima XL-100K ultracentrifuge, Type 45Ti rotor).
The supernatant is recovered and the (NH4)2SO4 concentration reduced to 0.1 M by addition of 2 volumes of Buffer B. The supernatant should be mixed immediately, spun at 26,000 rpm for 60 minutes at 4°C (Beckman Coulter Optima XL-100K ultracentrifuge, Type 45Ti rotor) and the supernatant is recovered.
Proteins in the supernatant of step 11 are ammonium sulfate precipitated by adding 0.42 g of (NH4)2SO4 per milliliter of supernatant and incubating at 4°C with agitation until all of the ammonium sulfate has dissolved. A mortar and pestle should be used to grind (NH4)2SO4 crystals into smaller sizes as this helps the ammonium sulfate dissolve faster. It is also recommended that the ammonium sulfate be added to the nuclear pellet suspension in three phases, with additional ammonium sulfate added only after all the previous addition has completely dissolved. The mixture should be stirred at 4°C for 30 minutes after the final addition has been dissolved.
Following ammonium sulfate precipitation, the nuclear pellet is centrifuged at 30,000 rpm for 60 minutes at 4°C (Beckman Coulter Optima XL-100K ultracentrifuge, Type 45Ti rotor). The pellet is resuspended with Buffer D (50 mM Tris HCl pH 7.9, 0.1 mM EDTA, 25% glycerol, 2 mM DTT, 0.2 mM PMSF, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, 1 μg/ml aprotinin) at a final concentration of 2 mg/ml. The solublized nuclear pellet should be stored at −80°C until use.
2.2.3 Fractionation of Nuclear Extract and Nuclear Pellet on P11 and DE52
Before being used to screen histone ubiquitin ligases, we first fractionate nuclear extract and nuclear pellet on P11 (Phosphocellulose, Sigma, No. C-2258) and DE52 (Whatman 4057200) columns. These fractionation steps largely remove residual nucleic acids, enrich target proteins in particular fractions, and also significantly reduce sample amounts prior to further purification on an FPLC system.
-
Phosphocellulose P11 Column
P11 resin is prepared according to the manufacturer’s instructions. Briefly, P11 resin is swollen with MilliQ water (200 g of dry beads give approximately 1 L of beads). Fine beads are decanted 3 – 4 times. An equal volume of the bead/water solution is mixed with 1 M NaOH, stirred for 10 minutes and then left to stand for 10 minutes to allow the beads to settle. Beads are washed with MilliQ water until the pH reaches 10. Once the pH is close to 10, equal volumes of beads/water solution and 1 N HCl are stirred for 10 minutes and then left to stand for 10 minutes to allow the beads to settle. Beads are washed with MilliQ water until the pH reaches 4. Once the pH is close to 4, equal volumes of the beads/water solution is mixed with 1 M Tris -HCl (pH 8.0) and stirred for 10 minutes and then left to stand for 10 minutes. Supernatant is removed and the pH checked. The pH should be adjusted back to 8.0 with NaOH and the supernatant mixed with the beads again. This process is repeated until a stable pH of 8.0 is maintained. Subsequently, as much buffer as possible is removed and the beads are suspended in BC100 (20 mM Tris HCl pH 7.9, 100 mM KCl, 0.2 mM EDTA, 10% glycerol, 10 mM DTT, 0.2 mM PMSF).
P11 resin is used to hand pack a column. Column volume is calculated as one liter of beads per 6 g protein. The column is equilibrated with 5–10 column volumes of BC100.
The nuclear extract is diluted to 4 g/L and dialyzed against BC50. The dialysis should be closely monitored. If the extract precipitates, the sample should be further diluted. Dialysis buffer is changed as needed.
Dialyzed sample is loaded to the equilibrated P11 column at a flow rate 2–4 ml/min to allow efficient protein binding. Flowthrough is saved as BC100.
Proteins are stepwise eluted from the column with BC300, BC500 and BC1000.
-
DE-52 Column
DE52 resin is washed with MilliQ water and the fine beads are decanted 3 – 4 times. After washing, DE52 resin is resuspended in BD20 (20 mM Tris HCl pH 7.9, 10% glycerol, 20 mM NH4SO4, 0.2 mM EDTA).
DE52 resin is used to pack a column. Column volume is calculated as one liter of beads per 6 g of protein. The column is equilibrated with 5–10 column volumes of BD20.
The nuclear pellet is diluted to 2 g/L to prevent precipitation and dialyzed against BD20. The dialysis process should be monitored. If any precipitant forms, the sample should be further diluted. The dialysis buffer is changed as needed.
Dialyzed sample is loaded to the equilibrated DE52 column at flow rate 2–4 ml/min to allow efficient protein binding. Flowthrough is saved as the DE52 Ft.
The column is stepwise eluted first with BD350 followed by BD500. The column is eluted with each buffer until no protein is released.
The BD350 fraction is dialyzed against BC100 and further fractionated on a phosphocellulose P11 column as described in steps 3–5 of 2.2.3.
2.3 In vitro histone ubiquitin ligase assay
Substrates including oligonucleosomes, mononucleosome, and histone octamers are used in the initial establishment of the histone ubiquitin ligase assay. HeLa nuclear protein fractions derived from the P11 and DE52 columns are used as enzyme sources. Briefly, nucleosome substrates are incubated with protein fractions in the presence of FLAG-Ubiquitin (F-Ub), ATP, E1 ubiquitin activating enzyme, and certain E2 ubiquitin conjugating enzymes. E1 and E2 are additionally provided, as these enzymes may not elute in the same protein fractions as the E3 ubiquitin ligase(s) of interest. Ubiquitinated histones are detected by western blot assay with anti-FLAG antibody, taking advantage of the fact that ubiquitin conjugation results in a 7–8 kDa super-shift of target protein in SDS-PAGE. Alternatively, antibodies against ubiquitinated histone can be used.
Five micrograms oligonucleosomes, mononucleosomes or histones are coincubated with 20 μl P11 nuclear extract and nuclear pellet fractions in Histone Ubiquitin Ligase reaction buffer [50 mM Tris-HCl pH 7.9, 5 mM MgCl2, 2 mM NaF, 0.6 mM DTT, 2 mM ATP, 10 uM Okadaic acid, 0.1 μg ubiquitin activating enzyme E1 (Calbiochem), 0.6 μg ubiquitin conjugating enzyme Ubc5c, 1 μg FLAG-ubiquitin (Sigma)].
Reactions are incubated in a 37°C water bath for 1 hour. Histone ubiquitin ligase reactions can be stopped by addition of SDS-loading buffer. Samples are resolved on an 8–15% SDS PAGE.
Histone ubiquitination is detected by Western blotting with anti-FLAG antibody. Ubiquitinated histones H3, H4, H2A and H2B can be distinguished based upon size. Alternatively, specific ubiquitin-histone conjugates can be detected using available ubiquitin-histone antibodies.
The observed histone ubiquitination must be confirmed to be due to the E3 ligase activity in the protein fractions assayed. In the absence of an E3 ligase (Ring family), E2 enzyme can also ubiquitinate substrates. To determine whether the observed histone ubiquitination is due to a specific ubiquitin E3 ligase within the protein fractions, histone ubiquitin ligase reactions should be performed with omission of E2 and E3 (protein fraction) as well as the substrates from the reaction. Dependence on the E3 ligase (protein fractions) should be established before additional purification.
The choice of E2. During the establishment of the histone ubiquitin reaction, careful choice of E2 ubiquitin conjugating enzymes is critical. In our initial screening, we have used a mixture of E2 enzymes (Ubc1, Ubc2, Ubc3, Ubc7, and Ubc10). When a positive reaction is observed, these E2 enzymes are individually tested for their ability to support the ligase reaction. It should be noted that the E2 (Ubc5c) we have used to purify ubiquitin E3 ligases may not be the bone fide E2 enzyme for histone ubiquitination in vivo.
2.4 Purification of histone ubiquitin ligases from Hela cells
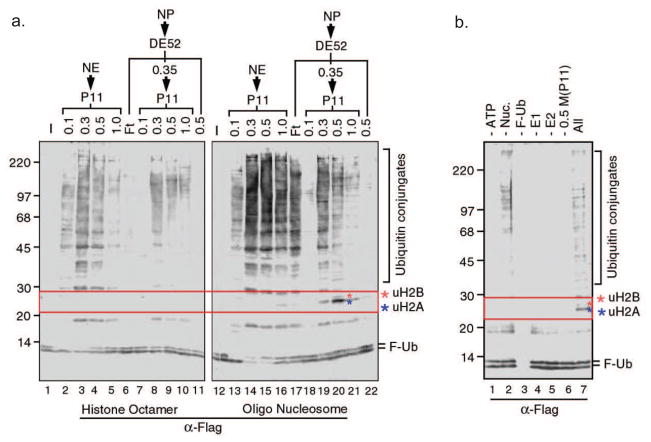
Using the in vitro histone ubiquitin ligase assay established above, we have screened protein fractions derived from HeLa cell nuclei for histone ubiquitin ligase activities. Below we detail the protocols used to purify an H2A and a H3 and H4 ubiquitin ligase in our published studies (18, 55). Using nucleosomes as substrates, Western blot analysis with an antibody against Flag reveals a positive signal around 25 kDa, the size of H2A plus F-Ub in nuclear pellet P11 0.3–0.5 fractions. This activity is nucleosomal histone specific as parallel experiments using core histone octamers fails to detect such an activity (Figure 3a, lanes 8–10). To verify that this activity is due to a bona fide E3 ligase, we have tested the dependency of the activity on each component of the reaction. Results shown in Figure 3b indicate the appearance of the ubiquitinated protein bands around 25 kDa requires the addition of protein fraction as well as nucleosome substrate. To verify the identity of the ubiquitinated 25 kDa histone, the protein band was excised from the gel and subjected to mass spectrometry analysis, revealing the identity as ubiquitinated H2A. With these preliminary studies, we set out to purify the H2A ubiquitin ligase.
Figure 3.
Identification of an H2A ubiquitin ligase activity in HeLa cells. a, Ubiquitin ligase assay using HeLa nuclear proteins fractionated on DE52 and P11 columns. Numbers on top of the panels indicate the salt concentration (M) for step elution. NE and NP represent nuclear extracts and nuclear pellet, respectively. Left and right panels use histone octamer and oligonucleosome substrates, respectively. b, The ligase activity depends on the presence of ATP, E1, E2, ubiquitin, nucleosomal histones, and proteins present in the 0.5M P11 nuclear pellet fraction.
2.4.1 Purification of histone H2A ubiquitin ligase complex
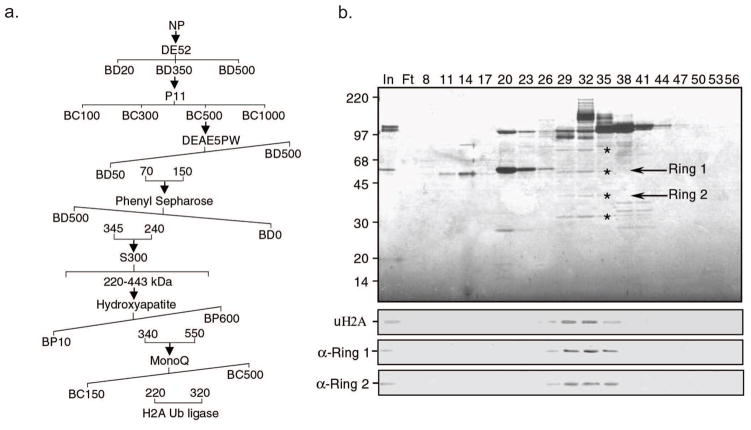
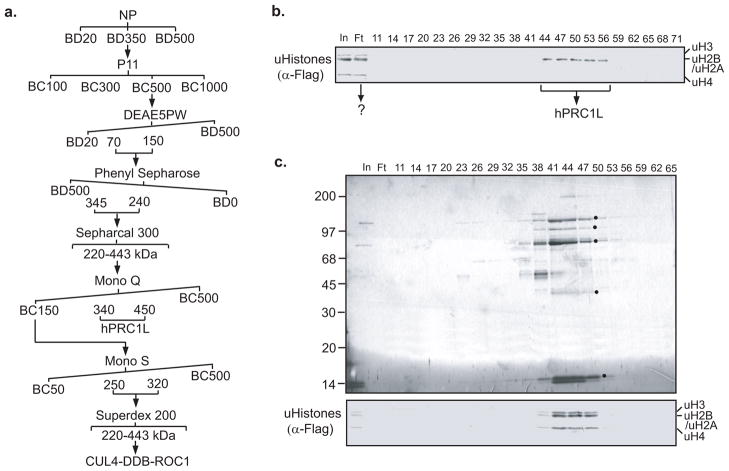
The purification scheme of H2A ubiquitin ligase from HeLa cells is shown in Figure 4a. The key element for successful purification is to choose the right combination of columns (ion exchange, hydrophobic interaction, hydroxyapatite, gel filtration, etc.) and elute the columns under the right conditions. These parameters must be experimentally determined. We have optimized the purification procedures for the H2A ubiquitin ligase and the H3/H4 ubiquitin ligase.
Figure 4.
Purification and identification of the H2A ubiquitination ligase complex. a, Schematic representation of the steps used to purify the H2A ubiquitination ligase complex. Numbers represent the salt concentrations (mM) at which the E3 ligase activity elutes from the columns. b, Silver staining of a polyacrylamide–SDS gel (top panel), H2A ubiquitin ligase activity assay (second panel) and western blot analysis (bottom two panels) of the fractions derived from the MonoQ column. The candidate proteins that co-fractionated with the E3 ligase activity are indicated by *. The positions of the protein size markers on SDS–PAGE are indicated to the left of the panel.
Since the nuclear pellet P11 0.5 M fraction contains the strongest ubiquitin ligase activity for H2A, we used this fraction for further purification. In all subsequent purification steps, aliquots from fractions were analyzed by silver staining and histone ubiquitin ligase activity assay. In certain cases, it was necessary to omit fractions containing a major contaminating protein even if H2A ubiquitin ligase activity was still present in these factions.
After dialyzing against BD20, the nuclear pellet P11 0.5 M fraction was loaded on a DEAE5PW column (TOSOH Bioscience, 45 ml) operated by the ÄKTA FPLC system (GE Healthcare). Proteins bound to the DEAE5PW column were eluted with 12-column volume (cv) linear gradient from 50 mM to 500 mM ammonium sulfate in buffer D [20 mM Tris-HCl (pH 7.9), 0.1 mM EDTA, 2 mM DTT, 0.2 mM PMSF, and 10% glycerol]. Histone ubiquitin ligase assay revealed that the ubiquitin ligase activity was eluted out of the column between 70–150 mM ammonium sulfate.
These fractions were then combined and adjusted to 500 mM ammonium sulfate with saturated ammonium sulfate before loading onto a 22 ml Phenyl Sepharose column (GE). The Phenyl Sepharose column was eluted with 20 cv linear gradient from 500mM to 0 mM ammonium sulfate in buffer D. The H2A ubiquitin ligase activity was eluted between 345–240 mM ammonium sulfate.
Active fractions were pooled and concentrated to 5 ml before loading to a 120 ml Sepharcy 300 gel filtration column (GE). The ubiquitin ligase activity eluted out between 220–443 kDa.
Active fractions were combined and dialyzed against buffer P [5 mM HEPES-KOH (pH 7.5), 40 mM KCl, 0.01% Triton X-100, 0.01 mM CaCl2, 0.5 mM PMSF, 1 mM DTT, and 10% glycerol] containing 10 mM potassium phosphate (BP10) and loaded to a 5 ml hydroxyapatite column (Bio-Rad). The bound proteins were eluted with 20 cv linear gradient from BP10 to BP600. The H2A ubiquitin ligase activity eluted out of the column between 340–550 mM potassium phosphate.
Active fractions were combined and loaded to a 1 ml MonoQ column (GE) after dialysis against buffer C [40 mM HEPES-KOH (pH 7.9), 0.1 mM EDTA, 2 mM DTT, 0.2 mM PMSF, and 10% glycerol] containing 150 mM KCl. Bound proteins were eluted with 20cv linear gradient from 150 mM to 500 mM KCl in buffer C. The H2A ubiquitin ligase activity was eluted between 220–320 mM KCl.
At this step, we could correlate the H2A ubiquitin ligase activity to a few polypeptides on SDS-PAGE. Silver staining of an aliquot of fractions derived from this column allowed use to correlate four candidate protein bands, marked by * (Figure 4b, top panel), with the enzymatic activity (Figure 4b, second panel). To identify the proteins that coelute with the H2A ubiquitin ligase activity, active fractions between 29–32 were combined, dialyzed to BC50 and bound to a 200 μl P11 column before being eluted with BC600.
The eluted proteins were resolved in 8–15% gradient SDS-PAGE. After Coomassie blue staining and destaining, candidate polypeptides were excised and subjected to trypsin digestion and a combination of peptide mass fingerprinting using MALDI-TOF MS and MS sequencing using MALDI-TOF/TOF MS/MS (58, 59). Mass spectrometry analysis identified the middle two protein bands as Ring1 and Ring2.
To further identify the complex that is responsible for H2A ubiquitin ligase activity, we employed an affinity purification strategy. Briefly, affinity purified Ring1 antibodies (60) were cross-linked to protein A agarose beads according to the manufacturer’s protocol (IPA300 Repligen) and incubated at 4°C for 4 hrs with an aliquot of hydroxyapatite input dialyzed against BC50. After washing with BC500 3 times and BC50 twice, proteins bound to the anti-Ring1 antibody were eluted out and identified by mass spectrometry. Analysis by mass spectrometry revealed that the H2A ubiquitin E3 ligase complex is composed of four PcG proteins including Ring1, Bmi1, HPH2, and Ring2, which we named hPRC1L (human PRC1 like).
2.4.2 Purification of ubiquitin ligase for H3 and H4
During the purification of the H2A ubiquitin ligase we noticed an ubiquitin ligase activity, which appears to be capable of ubiquitinating all histones, present in the flowthrough from the Mono Q column. This activity was completely separate from the H2A-specific ligase activity (Figure 5b). We further purified this activity through a five column purification scheme as detailed below (Figure 5a).
Figure 5.
Purification of a previously unidentified histone ubiquitin E3 ligase complex. a. Schematic representation of the steps used to purify the histone ubiquitin E3 ligase complex. Numbers represent the salt concentrations (mM) at which the E3 ligase activity elutes from the columns. b. Histone ubiquitin ligase assay of protein fractions derived from a Mono Q column. In addition to the ubiquitin ligase activity specific for histone H2A (hPRC1L), a previously unidentified ubiquitin ligase activity for all the core histones was observed in the flowthrough (Ft). c. Silver staining of a polyacrylamide-SDS gel (top) and histone ubiquitin ligase activity (bottom) of fractions derived from a Mono S column. The protein bands that cofractionated with the histone ubiquitin E3 ligase activity are indicated by an asterisk (*). The protein size marker is indicated on the left side of the panel.
Briefly, the flowthrough of the Mono Q column was loaded onto a Mono S column (GE) after dialysis against buffer C [40 mM HEPES-KOH (pH 7.9), 0.1 mM EDTA, 2 mM DTT, 0.2 mM PMSF, and 10% glycerol] containing 50 mM KCl. The bound proteins were eluted with a 20 cv linear gradient from BC50 to BC500.
The active fractions, which eluted between 250–320 mM KCl, were combined and loaded onto a 25 ml Superdex 200 column (GE). Silver staining and ubiquitin ligase assays of the fractions derived from the Mono S column allowed us to correlate the enzymatic activity with five polypeptides (Figure 5c).
To identify the polypeptides that coelute with the ubiquitin ligase activity, fractions 38–41 of the Superdex 200 column were combined, concentrated and resolved in a 8–15% gradient SDS-PAGE. After Coomassie staining, candidate polypeptides were excised and subjected to trypsin digestion. The proteins were identified by a combination of peptide mass fingerprinting using MALDI-TOF MS and MS sequencing using MALDI-TOF/TOF MS/MS (58, 59). Mass spectrometry analysis revealed that the five polypeptides were DDB1 (damaged DNA-binding protein 1), Cullin 4B, Cullin 4A, DDB2 (damaged DNA-binding protein 2), and ROC1/RBX1. Based on its polypeptide composition, we have named the protein complex CUL4-DDB-ROC1.
During the purification of histone ubiquitin ligase, a weak activity potentially for H2B was also detected (Figure 3a lane 20). However, subsequent purification revealed that the ubiquitinated bands correspond to histone H3.4. The significance of this discovery remains to be elucidated.
3. Discussion
The identification of enzymes responsible for histone modification provides valuable tools for dissecting the functions of a particular histone modification. It also provides pharmaceutical targets for epigenetic-based disease control. Given the critical roles of epigenetics in controlling cell fate and the wide implications of epigenetic mechanism in human disease, including cancer, we envision that epigenetic mechanism represents a promising field for drug development. In this paper we have detailed the experimental procedures for biochemical purification of histone ubiquitin ligases from HeLa cells. Similar biochemical approaches have used to identify enzymes involved in histone methylation, demethylation, deubiquitination, and other processes (29–31). This approach is an unbiased, systemic search for novel histone modifying enzyme activities. The majority of enzymes identified by this approach represent the dominant enzymes in a particular process. However, since this approach is critically dependent on the in vitro histone ubiquitin ligase assay, certain enzymes that have low in vitro activity will be missed by these studies. This was exemplified by our attempt to identify a H2B ubiquitin ligase. Although initial screening revealed a putative ligase activity for H2B, subsequent purification revealed that this activity targeted a histone H3 variant H3.4. By using nucleosomes with mutation at H2A K119, Zhu et al reported a partial purification of H2B ubiquitin ligase using a similar biochemical approach as we have outlined, but the final identification of H2B ubiquitin ligase in mammals ultimately came from genetic studies in budding yeast (48). Finally, this approach may miss enzymes that only function in a particular pathway.
Population homogeneity, ease of culturing and relatively low production costs have made HeLa cells widely used enzymatic sources for the biochemical purification of histone modifying enzymes. However, the enzymes purified from HeLa cells might only represent enzymatic machineries for general cellular activities. Since HeLa cells are a special cell line, histone modifying enzymes that are active in other cell types or cellular processes will not be identified by this approach. For example, UBC4-testis and LASU1 were reported to be capable of ubiquitinating histones in vitro. However, since these proteins are expressed primarily in testis, biochemical identification of these enzymes required the use of proteins extracted from testis (61, 62). In addition, the 2A-HUB H2A ubiquitin ligase, which is responsible for repression of a specific set of chemokine genes in macrophages, was also identified through alternative approaches (63). Therefore, other cell types or primary tissues, if sufficient amounts of starting material can be obtained, represent additional sources for identifying tissue or cell type-specific histone modifying enzymes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berger SL. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Carey M, Workman JL. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Campos EI, Reinberg D. Annu Rev Genet. 2009;43:559–99. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 5.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Nature. 2009;461:762–7. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger SL. Science. 2001;292:64–5. [PubMed] [Google Scholar]
- 7.Esteller M. Nat Rev Genet. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 8.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, Gold DL, Sekido Y, Huang TH, Issa JP. Nat Genet. 2008;40:741–50. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 9.Jones PA, Baylin SB. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Cell. 2001;107:323–37. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 11.Ropero S, Fraga MF, Ballestar E, Hamelin R, Yamamoto H, Boix-Chornet M, Caballero R, Alaminos M, Setien F, Paz MF, Herranz M, Palacios J, Arango D, Orntoft TF, Aaltonen LA, Schwartz S, Jr, Esteller M. Nat Genet. 2006;38:566–9. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]
- 12.Ozdag H, Teschendorff AE, Ahmed AA, Hyland SJ, Blenkiron C, Bobrow L, Veerakumarasivam A, Burtt G, Subkhankulova T, Arends MJ, Collins VP, Bowtell D, Kouzarides T, Brenton JD, Caldas C. BMC Genomics. 2006;7:90. doi: 10.1186/1471-2164-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, Teague J, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Forbes S, Jia M, Jones D, Knott H, Kok CY, Lau KW, Leroy C, Lin ML, McBride DJ, Maddison M, Maguire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, O’Meara S, Pleasance E, Rajasingham A, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turrell K, Dykema KJ, Khoo SK, Petillo D, Wondergem B, Anema J, Kahnoski RJ, Teh BT, Stratton MR, Futreal PA. Nature. 2010;463:360–3. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O’Meara S, Teague J, Butler A, Hinton J, Latimer C, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Cole J, Forbes S, Jia M, Jones D, Kok CY, Leroy C, Lin ML, McBride DJ, Maddison M, Maquire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, Pleasance E, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Collins VP, Ichimura K, Law S, Wong J, Yuen ST, Leung SY, Tonon G, DePinho RA, Tai YT, Anderson KC, Kahnoski RJ, Massie A, Khoo SK, Teh BT, Stratton MR, Futreal PA. Nat Genet. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 16.Kwon MJ, Kim SS, Choi YL, Jung HS, Balch C, Kim SH, Song YS, Marquez VE, Nephew KP, Shin YK. Carcinogenesis. 2010 doi: 10.1093/carcin/bgp336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Cell. 2004;118:409–18. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 19.Wei J, Zhai L, Xu J, Wang H. J Biol Chem. 2006;281:22537–44. doi: 10.1074/jbc.M600826200. [DOI] [PubMed] [Google Scholar]
- 20.Cao R, Tsukada Y, Zhang Y. Mol Cell. 2005;20:845–54. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Garcia BA, Shabanowitz J, Hunt DF. Curr Opin Chem Biol. 2007;11:66–73. doi: 10.1016/j.cbpa.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 22.Mravinac B, Sullivan LL, Reeves JW, Yan CM, Kopf KS, Farr CJ, Schueler MG, Sullivan BA. PLoS One. 2009;4:e6602. doi: 10.1371/journal.pone.0006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. Dev Cell. 2009;17:425–34. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Nat Cell Biol. 2008;10:483–8. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 25.Jackson J, Shilatifard A. Methods Mol Biol. 2009;548:175–86. doi: 10.1007/978-1-59745-540-4_10. [DOI] [PubMed] [Google Scholar]
- 26.Schneider J, Dover J, Johnston M, Shilatifard A. Methods Enzymol. 2004;377:227–34. doi: 10.1016/S0076-6879(03)77013-X. [DOI] [PubMed] [Google Scholar]
- 27.Cullen LM, Arndt GM. Immunol Cell Biol. 2005;83:217–23. doi: 10.1111/j.1440-1711.2005.01332.x. [DOI] [PubMed] [Google Scholar]
- 28.Lai A, Cairns MJ, Tran N, Zhang HP, Cullen L, Arndt GM. PLoS One. 2009;4:e4758. doi: 10.1371/journal.pone.0004758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang J, Wang H, Zhang Y. Methods Enzymol. 2004;377:213–26. doi: 10.1016/S0076-6879(03)77012-8. [DOI] [PubMed] [Google Scholar]
- 30.Tsukada Y, Zhang Y. Methods. 2006;40:318–26. doi: 10.1016/j.ymeth.2006.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai L, Joo HY, Wang H. Methods Mol Biol. 2009;523:295–309. doi: 10.1007/978-1-59745-190-1_20. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Q, Lieberman PM, Boyer TG, Berk AJ. Genes Dev. 1992;6:1964–74. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]
- 33.Weake VM, Workman JL. Mol Cell. 2008;29:653–63. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 34.Hannah J, Zhou P. DNA Repair (Amst) 2009;8:536–43. doi: 10.1016/j.dnarep.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Game JC, Chernikova SB. DNA Repair (Amst) 2009;8:470–82. doi: 10.1016/j.dnarep.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Deshaies RJ, Joazeiro CA. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 37.Scheffner M, Nuber U, Huibregtse JM. Nature. 1995;373:81–3. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 38.Pickart CM, Fushman D. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson KD. Semin Cell Dev Biol. 2000;11:141–8. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 40.Komander D. Biochemical Society Transactions. 2009;037:937–53. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 41.Robzyk K, Recht J, Osley MA. Science. 2000;287:501–4. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 42.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. Mol Cell. 2003;11:261–6. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 43.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Mol Cell. 2003;11:267–74. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 44.Koken MH, Reynolds P, Jaspers-Dekker I, Prakash L, Prakash S, Bootsma D, Hoeijmakers JH. Proc Natl Acad Sci U S A. 1991;88:8865–9. doi: 10.1073/pnas.88.20.8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koken MH, Hoogerbrugge JW, Jasper-Dekker I, de Wit J, Willemsen R, Roest HP, Grootegoed JA, Hoeijmakers JH. Dev Biol. 1996;173:119–32. doi: 10.1006/dbio.1996.0011. [DOI] [PubMed] [Google Scholar]
- 46.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. Cell. 2009;137:459–71. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roest HP, van Klaveren J, de Wit J, van Gurp CG, Koken MH, Vermey M, van Roijen JH, Hoogerbrugge JW, Vreeburg JT, Baarends WM, Bootsma D, Grootegoed JA, Hoeijmakers JH. Cell. 1996;86:799–810. doi: 10.1016/s0092-8674(00)80154-3. [DOI] [PubMed] [Google Scholar]
- 48.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Mol Cell. 2005;20:601–11. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 49.Kim J, Hake SB, Roeder RG. Mol Cell. 2005;20:759–70. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 50.Espinosa JM. Genes Dev. 2008;22:2743–9. doi: 10.1101/gad.1732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, Vojtesek B, Berger SL, Oren M. Genes Dev. 2008;22:2664–76. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Dev Cell. 2004;7:663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 53.Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, Hasegawa PM, Bressan RA, Zhu JK. Nature. 2007;447:735–8. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 54.Fleming A, Osley MA. Cell. 2004;119:449–51. doi: 10.1016/j.cell.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 55.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Mol Cell. 2006;22:383–94. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 56.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.LeRoy G, Orphanides G, Lane WS, Reinberg D. Science. 1998;282:1900–4. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 58.Devroe E, Erdjument-Bromage H, Tempst P, Silver PA. J Biol Chem. 2004;279:24444–51. doi: 10.1074/jbc.M401999200. [DOI] [PubMed] [Google Scholar]
- 59.Sebastiaan Winkler G, Lacomis L, Philip J, Erdjument-Bromage H, Svejstrup JQ, Tempst P. Methods. 2002;26:260–9. doi: 10.1016/S1046-2023(02)00030-0. [DOI] [PubMed] [Google Scholar]
- 60.Schoorlemmer J, Marcos-Gutierrez C, Were F, Martinez R, Garcia E, Satijn DP, Otte AP, Vidal M. Embo J. 1997;16:5930–42. doi: 10.1093/emboj/16.19.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Z, Oughtred R, Wing SS. Mol Cell Biol. 2005;25:2819–31. doi: 10.1128/MCB.25.7.2819-2831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wing SS, Bedard N, Morales C, Hingamp P, Trasler J. Mol Cell Biol. 1996;16:4064–72. doi: 10.1128/mcb.16.8.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]