Abstract
The green sulfur bacterium Chlorobaculum tepidum is proposed to oxidize sulfide and elemental sulfur via sulfite as an obligate intermediate. The sulfite pool is predicted to be contained in the cytoplasm and be oxidized by the concerted action of ApsBA, which directly oxidizes sulfite, and QmoABC, which transfers electrons from ApsBA to the quinone pool. Like other green sulfur bacteria, C. tepidum was unable to use exogenously provided sulfite as the sole electron donor. However, exogenous sulfite significantly stimulated the growth yield of sulfide limited batch cultures. The growth of C. tepidum mutant strains, CT0867/qmoB::TnOGm and CT0868/qmoC::TnOGm, was not increased by sulfite. Furthermore, these strains accumulated sulfite and displayed a growth yield decrease when grown on sulfide as the sole electron donor. These results support an obligate, cytoplasmic sulfite intermediate as part of the canonical sulfur oxidation pathway in C. tepidum that requires the Qmo complex for oxidation.
Keywords: Chlorobi, sulfite, Qmo complex
Introduction
The Chlorobi are obligate phototrophic bacteria that utilize reduced sulfur compounds as electron donors to a photosynthetic electron transport chain that provides energy and reduced ferredoxin to drive carbon fixation, biosynthesis, and cell growth (Bryant and Frigaard, 2006; Overmann and Garcia-Pichel, 2006; Frigaard and Dahl, 2009). These organisms inhabit anoxic environments including sediments and euxinic waters. In these environments, they contribute to the oxidative branch of the sulfur cycle by converting reduced sulfur compounds to sulfate that can subsequently be utilized as an electron acceptor for the degradation of organic compounds by sulfate reducing bacteria. Relative to other phototrophic bacteria, the Chlorobi are generally adapted to lower light intensities and are often found as the deepest layer of phototrophic microbes in stratified sediments and waters (Overmann and Garcia-Pichel, 2006). Consequently, these organisms are often exposed to higher fluxes and concentrations of reduced sulfur compounds than more high light adapted phototrophs. Thus, deep in illuminated anoxic sediments and waters, the Chlorobi facilitate the tight coupling of sulfur and carbon cycling and likely serve to decrease fluxes of reduced sulfur compounds into overlying oxic ecosystems in much that has been observed for chemolithotrophic sulfur oxidizers in marine oxygen minimum zones (Lavik et al., 2009).
The Chlorobi have been intensively studied as models for the reductive TCA pathway of CO2 fixation (Evans et al., 1966) and their highly ordered and efficient light harvesting antenna, the chlorosome (Oostergetel et al., 2010). There currently exist 11 complete and 1 draft genome sequences for members of this group (data from the DOE Joint Genome Institute1). Comparative genomic approaches (Frigaard and Dahl, 2009; Sakurai et al., 2010) have led to the identification of candidate genes encoding sulfur oxidizing activities. Genetic and biochemical analysis of the putative sulfur oxidation pathway outlined by these studies will lead to a detailed picture of the biochemistry of sulfur oxidation in the Chlorobi.
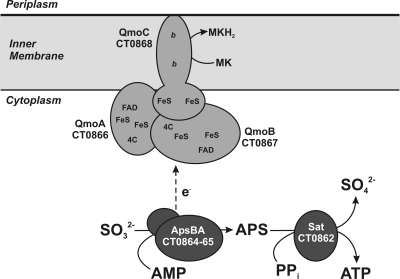
Sulfite is predicted to be an obligate intermediate in sulfur oxidation by the Chlorobi (Frigaard and Dahl, 2009; Sakurai et al., 2010). In general, Chlorobi do not utilize sulfite as the sole electron donor for photosynthesis (Overmann and Garcia-Pichel, 2006). Current models predict that sulfite is generated in the cytoplasm and oxidized by a reversible adenylylphosphosulfate reductase activity (APS reductase, ApsBA) that conjugates AMP and sulfite to produce APS (Figure 1). The electrons liberated during this reaction are thought to be passed to the quinone pool by a quinone interacting membrane bound oxidoreductase (Qmo) complex, which was first described in the sulfate reducing bacterium Desulfovibrio desulfuricans (Pires et al., 2003). The APS is acted upon by a reversible ATP sulfurylase that releases sulfate as the terminal oxidation product with the production of one molecule of ATP. Sulfate is subsequently proposed to exit the cytoplasm by an unknown transporter system. This system is essentially reversal of the initial steps of dissimilatory sulfate reduction and homologous Qmo complexes are encoded by the genomes of some Chlorobi, nearly all sulfate reducing bacteria, and sulfate reducing archaea of the genus Archaeoglobus (Figure 2). In all organisms shown except for the Archaea, the Qmo complex genes are immediately downstream of the genes encoding ApsBA.
Figure 1.
A schematic of proposed functions for a sulfite oxidation metabolic module including the Qmo complex. The proteins are encoded by basepairs 819,144 to 828,176 of the C. tepidum genome (GenBank accession NC_002932.3). Assignments of putative cofactors in the C. tepidum Qmo complex are from Pires et al. (2003). FAD = FAD nucleotide binding site, FeS = putative 4Fe–4S cluster, b = heme b, 4C = conserved cysteine motif (CXGXRDX6–8CSX2CC), MK = menaquinone.
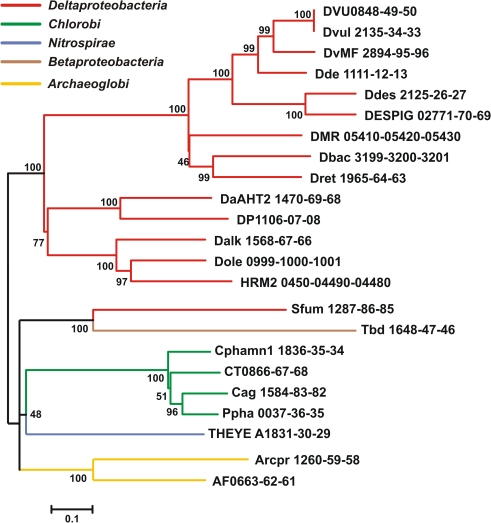
Figure 2.
Maximum likelihood phylogeny inferred from concatenated QmoABC amino acid sequences. The tree was constructed in MEGA5 using the WAG model of amino acid substitution with a gamma distribution = 1.8288 across five rate categories to account for differential substitution rates between sites and an invariable site frequency of 9.82%. A total of 1479 positions were included in the analysis and gaps handled by pairwise deletion. The scale bar represents 0.1 substitutions per site. Sequences are named according to the locus tags found in Table 1 and colored to reflect their phylogenetic affiliation as determined by 16S rRNA sequencing using NCBI taxonomy terms.
In the sulfate reducing bacterium Desulfovibrio vulgaris, a deletion mutant lacking the qmoABC genes was unable to grow with sulfate as the terminal electron acceptor while growth with either sulfite or thiosulfate was normal (Zane et al., 2010). This supports the idea that the QmoABC complex is a critical component for electron flow to the reduction of APS via ApsBA. The Qmo complex is of particular interest as it is homologous to the CoM–CoB heterodisulfide reductase found in the archaea (Hdr, Table 1). Hdr catalyzes the reductive regeneration of two major C1-carrying cofactors in methanogenesis while conserving energy in the form of a proton gradient (Deppenmeier et al., 1999). Recently, this enzyme was proposed to contribute to energy conservation in hydrogenotrophic methanogens by an electron bifurcation mechanism (Thauer et al., 2008). This hypothesis has been supported by protein–protein interaction studies in Methanococcus maripaludis S2 demonstrating that the Hdr forms an apparent complex with formyl-methanofuran dehydrogenase that catalyzes the first step of methanogenesis, and the F420-non-reducing hydrogenase and formate dehydrogenase, both of which provide reducing equivalents for reduction of C1 units to methane (Costa et al., 2010).
Table 1.
Qmo complex sequences used in phylogenetic tree construction, their annotation and properties.
| Qmo complex subunit ortholog annotations | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QmoA | QmoB | QmoC | ||||||||||
| Heterodisulfide reductase subunit A | Heterodisulfide reductase, subunit A/hydrogenase, delta subunit, putative | Heterodisulfide reductase, transmembrane subunit, putative | ||||||||||
| 4Fe–4S ferredoxin, iron–sulfur binding domain protein | Heterodisulfide reductase, subunit A and related polyferredoxins/Coenzyme F420-reducing hydrogenase, delta subunit | Heterodisulfide reductase, subunit E, putative | ||||||||||
| Genome | Locus tag | Length | SigP | TM | Locus tag | Length | SigP | TM | Locus tag | Length | SigP | TM |
| CHLOROBIA | ||||||||||||
| Chlorobaculum tepidum TLS | CT0866 | 413 | Yes | No | CT0867 | 750 | No | No | CT0868 | 391 | No | Yes |
| Chlorobium phaeobacteroides BS1 | Cphamn1_1836 | 413 | Yes | No | Cphamn1_1835 | 750 | No | No | Cphamn1_1834 | 391 | No | Yes |
| Chlorobium chlorochromatii CaD3 | Cag_1584 | 412 | Yes | No | Cag_1583 | 750 | No | No | Cag_1582 | 392 | No | Yes |
| Pelodictyon phaeoclathratiforme BU-1 | Ppha_0037 | 413 | Yes | No | Ppha_0036 | 750 | No | No | Ppha_0035 | 393 | No | Yes |
| DELTAPROTEOBACTERIA | ||||||||||||
| Desulfovibrio vulgaris Hildenborough | DVU0848 | 412 | Yes | No | DVU0849 | 758 | No | No | DVU0850 | 384 | No | Yes |
| D. vulgaris DP4 | Dvul_2135 | 412 | Yes | No | Dvul_2134 | 758 | No | No | Dvul_2133 | 384 | No | Yes |
| D. vulgaris Miyazaki F | DvMF_2894 | 412 | Yes | No | DvMF_2895 | 754 | No | No | DvMF_2896 | 384 | No | No |
| Desulfovibrio desulfuricans G20 | Dde_1111 | 412 | Yes | No | Dde_1112 | 756 | No | No | Dde_1113 | 384 | No | Yes |
| D. desulfuricans ATCC 27774 | Ddes_2127 | 411 | Yes | No | Ddes_2126 | 756 | No | No | Ddes_2125 | 393 | No | Yes |
| Desulfovibrio piger ATCC 29098 | DESPIG_02771 | 411 | Yes | No | DESPIG_02770 | 753 | No | No | DESPIG_02769 | 394 | No | Yes |
| Desulfovibrio magneticus RS-1 | DMR_05410 | 413 | Yes | No | DMR_05420 | 763 | No | No | DMR_05430 | 399 | No | Yes |
| Desulfomicrobium baculatum DSM 4028 | Dbac_3199 | 413 | Yes | No | Dbac_3200 | 763 | No | No | Dbac_3201 | 408 | No | Yes |
| Desulfohalobium retbaense DSM 5692 | Dret_1965 | 411 | Yes | No | Dret_1964 | 766 | No | No | Dret_1963 | 407 | No | Yes |
| Desulfurivibrio alkaliphilus AHT2 | DaAHT2_1470 | 419 | Yes | No | DaAHT2_1469 | 752 | No | No | DaAHT2_1468 | 397 | No | Yes |
| Desulfotalea psychrophila LSv54 | DP1106 | 445 | No | No | DP1107 | 753 | No | No | DP1108 | 404 | No | Yes |
| Desulfatibacillum alkenivorans AK-01 | Dalk_1568 | 424 | Yes | No | Dalk_1567 | 774 | No | No | Dalk_1566 | 413 | No | Yes |
| Desulfococcus oleovorans Hxd3 | Dole_0999 | 423 | Yes | No | Dole_1000 | 778 | No | No | Dole_1001 | 398 | No | Yes |
| Desulfobacterium autotrophicum HRM2 | HRM2_04500 | 424 | Yes | No | HRM2_04490 | 778 | No | No | HRM2_04480 | 391 | No | Yes |
| Syntrophobacter fumaroxidans MPOB | Sfum_1287 | 420 | Yes | No | Sfum_1286 | 738 | No | No | Sfum_1285 | 385 | No | Yes |
| BETAPROTEOBACTERIA | ||||||||||||
| Thiobacillus denitrificans ATCC 25259 | Tbd_1648 | 427 | No | No | Tbd_1647 | 752 | No | No | Tbd_1646 | 210 | No | No |
| NITROSPIRA | ||||||||||||
| Thermodesulfovibrio yellowstonii | THEYE_A1831 | 415 | No | No | THEYE_A1830 | 744 | No | No | THEYE_A1829 | 382 | No | No |
| ARCHAEOGLOBUS | ||||||||||||
| Archaeoglobus fulgidus DSM 4304 | AF0663 | 404 | No | No | AF0662 | 747 | No | No | AF0661 | 379 | No | Yes |
| Archaeoglobus profundus DSM 5631 | Arcpr_1260 | 420 | Yes | No | Arcpr_1259 | 740 | No | No | Arcpr_1258 | 421 | No | Yes |
Length = amino acid residues in gene product, SigP = signal peptide prediction, TM = transmembrane helix prediction.
To test the proposed role of the Qmo complex in sulfite processing in Chlorobaculum tepidum, mutants in CT0867 (encoding the QmoB homolog, Table 1) and CT0868 (encoding QmoC, Table 1) were analyzed for defects in sulfite metabolism. The results clearly implicated the Qmo complex of C. tepidum in sulfite processing during growth.
Materials and Methods
Sequence analysis and tree construction
Qmo subunit orthologs were identified using the tools of the Integrated Microbial Genomes database2 using the sequences of C. tepidum ORFs CT0866-0868 to identify homologous sequences. To be included in the alignment as a putative Qmo complex, genes encoding the subunits had to be colocalized as a potential operon in the genome and be a bidirectional BLASTP best hit (i.e., the IMG definition of orthology) of the C. tepidum sequence. All sequence manipulations and calculations were performed in MEGA version 5, which is available for download at http://www.megasoftware.net (Tamura et al., 2007; Kumar et al., 2008). To produce the phylogenetic tree (Figure 2), Qmo subunit amino acid sequences were concatenated in the order noted in Table 1 and subsequently aligned by Muscle. The alignment was then analyzed by maximum likelihood techniques, which identified the WAG model with a gamma rate distribution and fraction of invariant sites as the best descriptor of pairwise distances in the alignment. This model and parameters were used to construct and bootstrap a phylogenetic tree by maximum likelihood. The resulting tree was prepared for publication by coloring branches, formatting text, and re-sizing in CorelDraw version 11 without changing the relative branch lengths.
Microbial strains, growth conditions, and biomass measurements
The wild type strain of C. tepidum used in this study is WT2321, the plating strain derived from the original isolate TLS1. Mutant strains CT0867/qmoB::TnOGm and CT0868/qmoC::TnOGm were produced by in vitro transposition mutagenesis and natural transformation of C. tepidum as previously described (Chan et al., 2008b). The base medium was Pf-7 with sulfide as the sole electron donor prepared as previously described (Chan et al., 2008b, 2009) with the exception that sodium thiosulfate was omitted. Sulfide concentrations in basal medium were typically 1–2 mM. Anoxic, neutralized stock solutions of sulfide and sulfite were used to adjust the concentrations of these electron donors to the concentrations noted in the figures and text after autoclaving. Starter cultures for all experiments were derived from glycerol stocks stored at −70°C and revived in Pf-7 with both sulfide and thiosulfate as electron donors at 42°C. Cultures of strains CT0867/qmoB::TnOGm and CT0868/qmoC::TnOGm were grown in the presence of 4 μg Gm ml−1. Batch cultures for growth yield experiments were performed without antibiotic selection by inoculating cultures to a biomass concentration of ∼4 μg protein ml−1 and incubated in 47°C water baths at a light intensity of ∼20 μmol photons m−2 s−1 from full spectrum incandescent bulbs. Protein concentrations in cultures were measured by Bradford assay as previously described (Mukhopadhyay et al., 1999).
Quantification of sulfur species
Sulfide and sulfite were quantified by bimane derivatization and reversed phase HPLC with fluorescence detection as previously described (Rethmeier et al., 1997). Standard curves were generated from dilution series of concentrated stock solutions diluted in Pf-7 medium base immediately before derivatization. Intracellular and extracellular concentrations of sulfite were separately quantified by capturing cells from 10 to 50 ml of culture on Whatman GF/F filters. A portion of the filtrate was directly added to a bimane derivatization reaction to quantify the extracellular pool. Cells immobilized on the filter were washed with a volume of sulfur free Pf-7 medium followed by three extractions of the filter with the same aliquot of bimane derivatization mix, which consisted of 9.4 mM monobromobimane (Sigma-Aldrich B4380), in 50% v/v acetonitrile:water + 50 mM HEPES + 5 mM EDTA, pH = 8.0. All filtrations and bimane extractions were carried out in a Coy anaerobic chamber using anaerobic solutions.
Chlorobaculum tepidum individual cell size in logarithmic growth was measured by fixing cells with formaldehyde (3.7% w/v final concentration) followed by phase contrast microscopy. Cell counts and samples for protein concentration were taken at the same time. Volume per cell was calculated from cell width (w) and length (l) by V = π w2 (l/4 − w/12) using the assumption that C. tepidum cells are rods composed of two half spheres connected by a cylinder (Sun and Liu, 2003). Based on protein concentration, cell concentration and volume per cell, a conversion factor for protein concentration to total cell volume in logarithmic phase cultures was developed. Intracellular sulfite concentrations were calculated as filter retained sulfite divided by the total cell volume determined from protein concentration measurements made at the same time the sample was taken for sulfite.
Statistics
Data presented are from the means of three independent cultures for each strain and condition. Student's t-tests assuming inequal variance between samples and a two-tailed distribution were performed in Microsoft Excel to assess the significance of differences between means. The resulting P-values are noted in the text where appropriate.
Results
Qmo complex sequence relationships
The sequences of putative Qmo complexes were identified in complete microbial genome sequences. The properties and annotations of the relevant sequences are outlined in Table 1. Putative Qmo complexes have a relatively restricted phylogenetic distribution being found only in multiple members of the Deltaproteobacteria, Chlorobi, and Archaeoglobi and isolated members of the Betaproteobacteria and Nitrospira phyla, although Thermodesulfovibrio yellowstonii is the sole complete genome sequence available from the Nitrospira. The common physiological thread that unites most of the organisms that contain putative Qmo complexes is the utilization of sulfur compounds as either the electron donor (Chlorobi, Thiobacillus denitrificans) or terminal electron acceptor (Deltaproteobacteria, T. yellowstoneii, Archaeoglobi) under anaerobic conditions.
Inspection of subunit length, signal peptide prediction and transmembrane helix prediction (Table 1) suggests that the topology and functional sites of the complex as proposed by Pires et al. (2003) are conserved across these groups (Figure 1). The Betaproteobacterium T. denitrificans is a notable exception, where the integral membrane subunit QmoC is severely truncated. Examination of the coding sequence suggests that this truncation is not a result of misannotation.
Phylogenetic analysis of concatenated Qmo complex subunit amino acid sequences produces a pattern of largely vertical descent of sequences within groups defined by 16S rRNA phylogeny (Figure 2). The exception to this is the strongly supported affiliation of the sequences from the Deltaproteobacterium Syntrophobacter fumaroxidans with the Betaproteobacterium T. denitrificans. This may be consistent with a directional transfer from sulfate reducing Deltaproteobacteria to S. fumaroxidans to T. denitrificans. However, the deeper branches of the phylogeny are not resolved, even when additional sequences, for example those of CoM–CoB heterodisulfide reductase complex subunits from methanogenic archaea, are added as outgroups.
The Qmo complex may have originated in the Archaeoglobi where paralogous genes encoding CoM–CoB heterodisulfide reductases that appear to encode functional products can be found in the same genome as those encoding the Qmo complex. In the Bacteria, the Qmo complex subunits are almost always closely associated with genes encoding the APS reductase, but these are found elsewhere in the A. fulgidus and A. profundicola genomes. The Qmo complex in C. tepidum is part of a large collection of genes colocalized on the genome that are implicated in the oxidation of reduced sulfur compounds. This region encodes one of the two copies of dsrCABL operon (encoding reverse dissimilatory sulfite reductase) that was recently shown to participate in sulfur globule oxidation in C. tepidum (Holkenbrink et al., 2011). In addition this region, sulfur island I (Chan et al., 2008a), contains aprBA, sat (encoding sulfate:adenylyl transferase), the QmoABC homolog encoding genes CT0866-0868 and one of three genes encoding homologs of sulfide:quinone oxidoreductase, CT0876 (Chan et al., 2009; Holkenbrink et al., 2011). The C. tepidum genome also contains a paralog of CT0866 (the QmoA homolog), CT1246, which resides in a potential operon (CT1245-1250) encoding homologs of hydrogenase and sulfur reductase subunits.
Sulfite stimulates the growth of C. tepidum
Genetic experiments have concretely implicated qmo genes in the reduction of sulfite by D. vulgaris (Zane et al., 2010). Therefore, we sought to quantify the effect of sulfite on the growth of C. tepidum as a pre-requisite to examining the physiological role of the putative Qmo complex in this sulfur oxidizing phototroph. In describing the isolation and characterization of the strain, Wahlund et al. (1991) found that adding 2 mM sulfite to medium with approximately 2.5 mM sulfide increased the growth yield of C. tepidum by 14%.
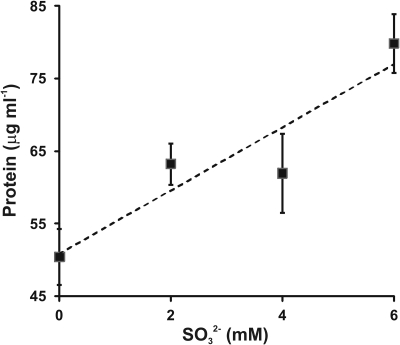
To confirm and extend this observation, varying amounts of sulfite were added to growth medium containing 4 mM sulfide and the protein concentration reached in stationary phase was determined (Figure 3). An increased sulfide concentration was employed based on prior work showing that C. tepidum can tolerate up to 8 mM sulfide without decreases in growth rate or yield (Chan et al., 2009). The increased sulfide concentration allows for a better estimate of growth yield on sulfide alone (i.e., the first point in Figure 3). Clearly, there is a positive correlation between added sulfite and increased biomass. The slope of a linear regression of this data (r2 = 0.86, dashed line in Figure 3) indicates a yield of 4.3 g protein (mol sulfite)−1. In this experiment, the yield from sulfide was 12.6 g protein (mol sulfide)−1, which is consistent with previously published values for sulfide dependent growth yields (9–13 g protein (mol sulfide)−1) (Mukhopadhyay et al., 1999; Chan et al., 2009). No growth was observed when cultures in medium with sulfite as the only electron donor. Thus, as is generally found for other Chlorobi, sulfite appears to not be utilized by C. tepidum as the sole electron donor.
Figure 3.
Sulfite stimulation of C. tepidum growth. Sulfide limited (4 mM initial concentration) batch cultures were amended with the indicated concentrations of sulfite and the protein concentration determined in stationary phase. Data points are the means ± SE of three independent cultures for each concentration. The dashed line is a linear regression of the data that results in the equation [Protein] = 4.3[] + 50.8.
Mutation of Qmo homolog encoding genes leads to a defect in sulfite growth stimulation
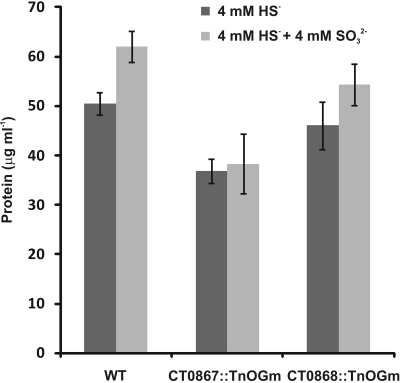
We previously reported that C. tepidum mutant strains carrying transposon insertions in the CT0867/qmoB or CT0868/qmoC genes had no strong phenotype relative to the wild type (Chan et al., 2008b). However, these experiments were carried out in medium with sulfide, thiosulfate or a mixture of both as the electron donor. Given the proposed role of the Qmo complex in sulfite oxidation and the data above that exogenous sulfite stimulates the growth of C. tepidum, these mutants were tested for their ability to benefit from the addition of 4 mM sulfite to medium containing 4 mM sulfide (Figure 4). As expected, the addition of 4 mM sulfite led to a 40% increase in the growth yield of the wild type (P = 0.059). In contrast, the growth yield of strain CT0867/qmoB::TnOGm was not increased by the addition of sulfite (P = 0.430). Strain CT0868/qmoC::TnOGm displayed an 18% increase in growth yield, but this was not statistically significant (P = 0.204). When comparing the growth of each mutant strain to the wild type with sulfide as the sole electron donor, a slight decrease in growth yield was detected: a significant 27% for the CT0867/qmoB::TnOGm (P = 0.007) and a non-significant 9% for the CT0868/qmoC::TnOGm strain (P = 0.291). The growth rates of the mutant and wild type strains in these experiments were not significantly different from one another with a mean doubling time of 2.0 ± 0.2 h, identical to the originally reported maximal growth rate (Wahlund et al., 1991).
Figure 4.
A comparison of sulfite addition to wild type and mutant C. tepidum strains. Strains were grown in batch cultures containing sulfide-only (dark bars) or sulfide + sulfite (light bars) as electron donors for photosynthesis. The protein concentration was determined in stationary phase for three independent cultures for each condition and strain. The data are the means ± SE.
Mutation of Qmo homolog encoding genes results in the accumulation of sulfite
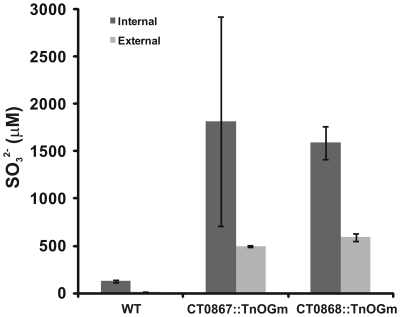
Given that the Qmo complex is predicted to be required for the oxidation of sulfite by C. tepidum, experiments were performed to determine if the mutant strains displayed any defects relative to sulfite metabolism under standard growth conditions (Figure 5). Under these conditions, sulfite should be a pathway intermediate that is predominantly confined to the cytoplasm (Figure 1). Strains were grown with sulfide + thiosulfate as electron donors and the internal and external pools of sulfite determined by bimane derivatization. Low levels of sulfite (<150 μM) were detected in the wild type under these growth conditions, with a 10:1 ratio of internal:external sulfite. In contrast, both mutant strains displayed large increases in both internal (13-fold average) and external (39-fold average) sulfite pools. In the case of the mutant strains, the internal:external ratio was decreased to 3.7- and 2.7-fold in the CT0867/qmoB::TnOGm and CT0868/qmoC::TnOGm strains, respectively.
Figure 5.
A comparison of sulfite pools in wild type and mutant strains of C. tepidums were grown in medium containing sulfide + thiosulfate and harvested during logarithmic growth phase. The values presented are the means of three independent cultures ± SE. Measurement of internal (dark bars) and external (light bars) pools is described in Section “Quantification of Sulfur Species.”
Discussion
The availability of the C. tepidum genome sequence (Eisen et al., 2002) has enabled the production of a detailed model for the oxidation of reduced sulfur compounds and electron transport (Frigaard and Dahl, 2009; Sakurai et al., 2010) that provide the energy for cell growth in this organism. Experimental data are now accumulating that will lead to the refinement of these models over time. The data outlined above provide experimental evidence that a putative Qmo complex in C. tepidum as part of a sulfite oxidation metabolic module (Figure 1).
If sulfite is an obligate intermediate in sulfide oxidation, one would expect a 25% decrease in growth yield if C. tepidum could not oxidize sulfite due to the loss of two out of eight electrons for the complete oxidation of sulfide to sulfate. The Qmo mutant strains displayed an average growth yield decrease of 18% with growth on sulfide as the sole electron donor, consistent with this prediction. Exogenously provided sulfite was found to stimulate the growth of C. tepidum with a yield of 4.3 g protein mol−1, which is lower than the average yield value of 11 g protein mol−1 for sulfide from data in two independent studies (Mukhopadhyay et al., 1999; Chan et al., 2009) in addition to the value of 12.6 g protein mol−1 reported here (Figure 3). However, the growth stimulation was larger than 2.8 g protein mol−1 expected from the electron counting argument. The difference likely reflects the low redox potential of electrons from sulfite (−516 mV) relative to those derived from sulfide (−270 mV), which should lead to greater energy conservation and biomass production per mole of substrate oxidized for sulfite.
If the Qmo complex is indeed essential for sulfite oxidation, then one would expect that the mutant strains should no longer benefit from exogenous sulfite and this was observed. Sulfite produced during standard growth conditions was predominantly found intracellularly, which is consistent with other studies that do not observe significant sulfite concentrations culture supernatants (Chan et al., 2008b; Holkenbrink et al., 2011). This is consistent with the proposal that sulfite is a cytoplasmic intermediate in the sulfur oxidation pathway (Frigaard and Dahl, 2009; Sakurai et al., 2010). This is further supported by the dramatic increases in intracellular sulfite in the mutant strains grown under standard conditions. Together these results suggest there is a transport requirement to deliver exogenous sulfite to AprBA in the cytoplasm. The fact that sulfite does not support the growth of C. tepidum as a sole substrate may indicate a co-solute requirement for sulfite transport or that co-substrates are required to maintain expression of AprBA to facilitate sulfite oxidation.
In general, the CT0867/qmoB::TnOGm mutant strain showed a more severe growth yield and sulfite accumulation defect than the CT0868/qmoC::TnOGm strain. This may occur for several reasons. First and most likely, the transposon insertion in CT0867 is likely polar on CT0868 and may effectively behave as a double qmoB/qmoC mutant. Less likely, but still possible, is that QmoB may be absolutely required to accept electrons from ApsBA while QmoC may be at least partially dispensable for transferring these electrons to the quinone pool. More precise genetic experiments to specifically ablate the heme b sites on QmoC will be valuable to address this question as will protein–protein interaction studies. By analogy to the sulfate reducing Qmo complex (Pires et al., 2003; Zane et al., 2010), the function of the C. tepidum Qmo complex is to transfer electrons from the APS reductase to the quinone pool (Figure 1), which should require at least transient contact between these two complexes. Studies on Hdr proteins homologous to Qmo subunits highlight this point. In methanogenic archaea, HdrA proteins were His-tagged and affinity purified from cell lysates, which allowed the identification of multi-protein interactions with other important enzymes in the methanogenic pathway (Costa et al., 2010). Soluble Hdr subunits have also co-purified with a methyl viologen reducing hydrogenase in Archaeoglobus fulgidus (Mander et al., 2004). As His-tagging of C. tepidum proteins by genetic methods has already been demonstrated with a sulfide:quinone oxidoreductase encoded by CT1087 (Chan et al., 2009), this should be a productive path forward to define the interactions of the C. tepidum Qmo complex with other enzymes. This may show that both Qmo- and Hdr-complexes serve as key organizing points of electron transfer in extremely physiologically diverse bacteria and archaea.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by an NSF CAREER award to Thomas E. Hanson (MCB-0447649), Jesse Rodriguez was supported in part by the University of Delaware McNair Scholars and the EPSCoR REU programs (EPS-0814251). Jennifer Hiras was supported in part by a Preston C. Townsend fellowship.
Footnotes
References
- Bryant D. A., Frigaard N. U. (2006). Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 14, 488–496 10.1016/j.tim.2006.09.001 [DOI] [PubMed] [Google Scholar]
- Chan L. K., Morgan-Kiss R. M., Hanson T. E. (2008a). “Sulfur oxidation in Chlorobium tepidum (syn. Chlorobaculum tepidum): genetic and proteomic analyses,” in Microbial Sulfur Metabolism, eds Dahl C., Friedrich C. G. (Berlin: Springer-Verlag; ), 117–126 [Google Scholar]
- Chan L. K., Weber T. S., Morgan-Kiss R. M., Hanson T. E. (2008b). A genomic region required for phototrophic thiosulfate oxidation in the green sulfur bacterium Chlorobium tepidum (syn. Chlorobaculum tepidum). Microbiology 154, 818–829 10.1099/mic.0.2007/012583-0 [DOI] [PubMed] [Google Scholar]
- Chan L. K., Morgan-Kiss R. M., Hanson T. E. (2009). Functional analysis of three sulfide:quinone oxidoreductase homologs in Chlorobaculum tepidum. J. Bacteriol. 191, 1026–1034 10.1128/JB.01154-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa K. C., Wong P. M., Wang T., Lie T. J., Dodsworth J. A., Swanson I., Burn J. A., Hackett M., Leigh J. A. (2010). Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc. Natl. Acad. Sci. U.S.A. 107, 11050–11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppenmeier U., Lienard T., Gottschalk G. (1999). Novel reactions involved in energy conservation by methanogenic archaea. FEBS Lett. 457, 291–297 10.1016/S0014-5793(99)01026-1 [DOI] [PubMed] [Google Scholar]
- Eisen J. A., Nelson K. E., Paulsen I. T., Heidelberg J. F., Wu M., Dodson R. J., Deboy R., Gwinn M. L., Nelson W. C., Haft D. H., Hickey E. K., Peterson J. D., Durkin A. S., Kolonay J. L., Yang F., Holt I., Umayam L. A., Mason T., Brenner M., Shea T. P., Parksey D., Nierman W. C., Feldblyum T. V., Hansen C. L., Craven M. B., Radune D., Vamathevan J., Khouri H., White O., Gruber T. M., Ketchum K. A., Venter J. C., Tettelin H., Bryant D. A., Fraser C. M. (2002). The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. U.S.A. 99, 9509–9514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. C. W., Buchanan B. B., Arnon D. I. (1966). A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc. Natl. Acad. Sci. U.S.A. 55, 928–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N. U., Dahl C. (2009). Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54, 103–200 [DOI] [PubMed] [Google Scholar]
- Holkenbrink C., Ocon Barbas S., Mellerup A., Otaki H., Frigaard N. U. (2011). Sulfur globule oxidation in green sulfur bacteria is dependent on the dissimilatory sulfite reductase system. Microbiology. [Epub ahead of print]. 10.1099/mic.0.044669-0. [DOI] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J., Tamura K. (2008). MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9, 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavik G., Stuhrmann T., Bruchert V., Van der Plas A., Mohrholz V., Lam P., Mussmann M., Fuchs B. M., Amann R., Lass U., Kuypers M. M. (2009). Detoxification of sulphidic african shelf waters by blooming chemolithotrophs. Nature 457, 581–854 10.1038/nature07588 [DOI] [PubMed] [Google Scholar]
- Mander G. J., Pierik A. J., Huber H., Hedderich R. (2004). Two distinct heterodisulfide reductase-like enzymes in the sulfate-reducing archaeon Archaeoglobus profundus. Eur. J. Biochem. 271, 1106–1116 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay B., Johnson E. F., Ascano M., Jr. (1999). Conditions for vigorous growth on sulfide and reactor-scale cultivation protocols for the thermophilic green sulfur bacterium Chlorobium tepidum. Appl. Environ. Microbiol. 65, 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostergetel G. T., van Amerongen H., Boekema E. J. (2010). The chlorosome: a prototype for efficient light harvesting in photosynthesis. Photosyn. Res. 104, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmann J., Garcia-Pichel F. (2006). “The phototrophic way of life,” In The Prokaryotes, 3rd Edn, eds Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (New York: Springer; ), 32–85 [Google Scholar]
- Pires R. H., Lourenco A. I., Morais F., Teixeira M., Xavier A. V., Saraiva L. M., Pereira I. A. (2003). A novel membrane-bound respiratory complex from Desulfovibrio desulfuricans ATCC 27774. Biochim. Biophys. Acta 1605, 67–82 [DOI] [PubMed] [Google Scholar]
- Rethmeier J., Rabenstein A., Langer M., Fischer U. (1997). Detection of traces of oxidized and reduced sulfur compounds in small samples by combination of different high-performance liquid chromatography methods. J. Chromatogr. A 760, 295–302 [Google Scholar]
- Sakurai H., Ogawa T., Shiga M., Inoue K. (2010). Inorganic sulfur oxidizing system in green sulfur bacteria. Photosyn. Res. 104, 163–176 [DOI] [PubMed] [Google Scholar]
- Sun J., Liu D. (2003). Geometric models for calculating cell biovolume and surface area for phytoplankton. J. Plankton Res. 25, 1131–1146 10.1093/plankt/fbg096 [DOI] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (mega) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thauer R. K., Kaster A. K., Seedorf H., Buckel W., Hedderich R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591 [DOI] [PubMed] [Google Scholar]
- Wahlund T. M., Woese C. R., Castenholz R. W., Madigan M. T. (1991). A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum Sp–Nov. Arch. Microbiol. 156, 81–90 [Google Scholar]
- Zane G. M., Yen H. C., Wall J. D. (2010). Effect of the deletion of qmoABC and the promoter-distal gene encoding a hypothetical protein on sulfate reduction in Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 76, 5500–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]