Abstract
Dissimilatory sulfate-reducing prokaryotes (SRB) are a very diverse group of anaerobic bacteria that are omnipresent in nature and play an imperative role in the global cycling of carbon and sulfur. In anoxic marine sediments sulfate reduction accounts for up to 50% of the entire organic mineralization in coastal and shelf ecosystems where sulfate diffuses several meters deep into the sediment. As a consequence, SRB would be expected in the sulfate-containing upper sediment layers, whereas methanogenic archaea would be expected to succeed in the deeper sulfate-depleted layers of the sediment. Where sediments are high in organic matter, sulfate is depleted at shallow sediment depths, and biogenic methane production will occur. In the absence of sulfate, many SRB ferment organic acids and alcohols, producing hydrogen, acetate, and carbon dioxide, and may even rely on hydrogen- and acetate-scavenging methanogens to convert organic compounds to methane. SRB can establish two different life styles, and these can be termed as sulfidogenic and acetogenic, hydrogenogenic metabolism. The advantage of having different metabolic capabilities is that it raises the chance of survival in environments when electron acceptors become depleted. In marine sediments, SRB and methanogens do not compete but rather complement each other in the degradation of organic matter. Also in freshwater ecosystems with sulfate concentrations of only 10–200 μM, sulfate is consumed efficiently within the top several cm of the sediments. Here, many of the δ-Proteobacteria present have the genetic machinery to perform dissimilatory sulfate reduction, yet they have an acetogenic, hydrogenogenic way of life. In this review we evaluate the physiology and metabolic mode of SRB in relation with their environment.
Keywords: sulfate-reducing bacteria, metabolic flexibility, syntrophy, metabolic interactions
Dissimilatory sulfate-reducing prokaryotes (SRB) are a diverse group of anaerobic bacteria that are widespread in nature and play an essential role in the global cycling of carbon and sulfur. The SRB mainly use sulfate, the most oxidized form of sulfur, as the terminal electron acceptor in the oxidation of hydrogen and various organic compounds (Widdel and Hansen, 1991; Rabus et al., 2006; Muyzer and Stams, 2008). Some SRB can use nitrate as electron acceptor, and their possible microaerophilic nature has also been discussed (Cypionka, 2000).
The anaerobic food chain changes largely when sulfate enters the methanogenic zone. In that case sulfate-reducing bacteria will outcompete methanogenic archaea for hydrogen, formate and acetate, and syntrophic methanogenic communities for substrates like propionate and butyrate (Stams, 1994; Muyzer and Stams, 2008). Interestingly, sulfate reducers can also grow without sulfate and in some cases they grow only in syntrophic association with methanogens or other hydrogen-scavengers. Thus, sulfate reducers may compete with methanogens and grow in syntrophy with methanogens depending on the prevailing environmental conditions (Muyzer and Stams, 2008). Already in the 1970s, the metabolic flexibility of SRB was investigated. Bryant et al. (1977) demonstrated growth of Desulfovibrio on lactate in the absence of sulfate but in the presence of a methanogen. They concluded that the sulfate reducer produced H2 which is used by the methanogen, acting as an alternative electron sink in the absence of sulfate. Therefore, in sulfate-depleted environments, it is possible that sulfate reducers may be metabolically active by living in association with methanogenic bacteria instead of reducing sulfate (Bryant et al., 1977; McInerney et al., 1981). This symbiotic process is known as “syntrophy” and is a widespread type of microbial interaction especially in methanogenic environments (Bryant et al., 1967; Schink, 1997; Stams and Plugge, 2009). Two major life styles are thus performed by some SRB: the sulfidogenic and the syntrophic metabolism. The advantage of having different metabolic potentials is that it enhances the chance of survival of communities of SRB in environments when electron acceptors become depleted.
In sulfate-depleted marine sediments, SRB and methanogens do not compete but rather complement each other in the degradation of organic matter. Also in sulfate-rich marine sediments, SRB and methanogens co-exist, but presumably by competing for common substrates, such as H2 (Oremland et al., 1982; Winfrey and Ward, 1983; Kuivila et al., 1990; Holmer and Kristensen, 1994). Recently, it was found that sulfate reducers were still abundant in the methanogenic zones of Aarhus Bay (Leloup et al., 2009).
In the past decades significant progress has been made through extensive studies of pure cultures, in SRB particularly with its model species Desulfovibrio vulgaris. Genomic analysis gave insight how the utilization of H2 and organic acids (formate and lactate) as electron donors is coupled to sulfate reduction, ATP synthesis and growth (Heidelberg et al., 2004). Lactate is oxidized through several enzymatic steps to acetate resulting in ATP synthesis (Heidelberg et al., 2004). Besides substrate-level ATP synthesis, additional ATP is generated from a proton gradient according to a chemiosmotic model (Peck, 1966), in which the protons and electrons produced during lactate oxidation react with cytoplasmic hydrogenases to form H2, which then diffuses across the membrane where it is re-oxidized by periplasmic hydrogenases to form a proton gradient (Odom and Peck, 1981; Heidelberg et al., 2004). The electrons generated during lactate oxidation are channeled to sulfate through a vast network of hemes that is created by various interconnected c-type cytochromes and involving several transmembrane complexes (Aubert et al., 2000; Heidelberg et al., 2004). Nowadays, research efforts with SRB are significantly aided by the availability of over 20 genome sequences of which 12 representatives of the genus Desulfovibrio (http://img.jgi.doe.gov; Integrated Microbial Genomes). Numerous research groups have since then reported global transcriptomic and proteomic analyses of D. vulgaris under various growth or stress conditions (Chhabra et al., 2006; Clark et al., 2006; He et al., 2006; Mukhopadhyay et al., 2006, 2007; Zhang et al., 2006a,b,c; Bender et al., 2007; Tang et al., 2007; Pereira et al., 2008; Walker et al., 2009; Plugge et al., 2010). As a result, there has been a better understanding of the electron transfer and energy conservation mechanisms of D. vulgaris mainly associated with lactate oxidation during sulfidogenic growth. Yet, the energy metabolism of D. vulgaris is very complex and flexible and as such deserves further study (Pereira et al., 2008).
While the physiology of the symbiotic/syntrophic relationship has been studied for more than 40 years (Bryant et al., 1967, 1977; Stams, 1994; Schink, 1997; Stams and Plugge, 2009), relatively little is known about the metabolic and regulatory networks involved in syntrophic interactions. This may be due to the technical difficulties to establish stable mixed-culture systems and lack of analytical tools for direct large-scale measurement of various biological components (i.e., RNA, proteins or metabolites). However, availability of complete genome sequences and various functional genomics tools in recent years have provided the most needed methodologies to analyze mixed-culture systems. In a recent study, a syntrophic pair, D. vulgaris and a hydrogenotrophic methanogen Methanococcus maripaludis, was cultured syntrophically on lactate in the absence of sulfate (Stolyar et al., 2007). Syntrophic associations were initiated by mixing equal volumes of stationary phase D. vulgaris and M. maripaludis cultures. Experimental measurements with the co-culture were used to test predictions derived from the first multi-species stoichiometric metabolic model involving the two species (Stolyar et al., 2007). The D. vulgaris and M. maripaludis flux-balance models were combined to form one model describing growth and metabolite accumulation when the organisms were growing together. To model the interaction between the two species, a system of three “compartments” was proposed. The first two compartments each contained the metabolite fluxes for one of the single-species models analyzed above. These species’ compartments could each represent the action of single cells, or the combined flux of many cells of the same species. To model the interaction between the two species, a third compartment was added to the model, through which metabolites could be transferred between organisms. Exchange fluxes were added to the model in this compartment. With this modification, species could take up metabolites excreted by the other species. The results of these initial model simulations predicted that D. vulgaris, growing optimally, converts the majority of the carbon contained in the substrate lactate into acetate, and that only 4.8% of the carbon is directed to biomass. The remaining carbon is lost as CO2 or formate. To maintain redox balance in the absence of an external electron acceptor, evolution of a reduced compound, either formate or hydrogen, is predicted. The model shows that when acetate is available, it is the preferred source of biomass carbon for M. maripaludis over the CO dehydrogenase pathway to fix carbon dioxide. This prediction is consistent with published data and the experimental results showed that M. maripaludis consumes acetate and presumably uses it as a carbon source (Stolyar et al., 2007).
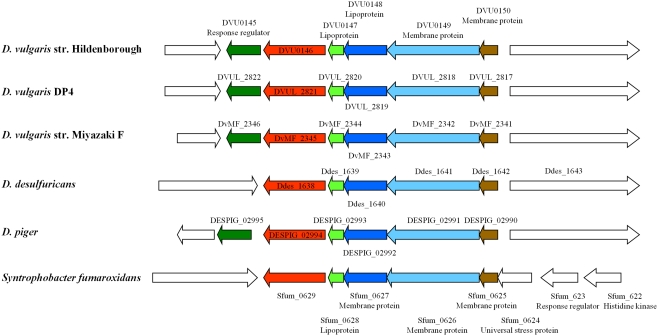
Comparative transcriptomic analysis to investigate syntrophic systems was done recently. In one study, comparative transcriptional analysis of D. vulgaris in two culture conditions was performed: syntrophic co-cultures with M. maripaludis strain S2 (without sulfate) and sulfate-limited monocultures (Walker et al., 2009). During syntrophic growth on lactate with a hydrogenotrophic methanogen, numerous genes involved in electron transfer and energy generation were up-regulated in D. vulgaris compared with their expression in sulfate-limited pure cultures. In another study, whole-genome D. vulgaris microarrays were used to determine relative transcript levels when D. vulgaris shifted its lifestyle from syntroph in a lactate-oxidizing co-culture with Methanosarcina barkeri to a sulfidogenic lifestyle (Plugge et al., 2010). In the study, syntrophic co-cultures were grown in two independent chemostats and perturbation was introduced after six volume changes with the addition of sulfate. Functional analyses revealed that genes involved in cell envelope and energy metabolism were the most regulated when comparing syntrophic and sulfidogenic metabolism. These two studies are similar in many ways, however, there are four major differences in experimental design between the two studies: (i) the methanogenic partner in the study of Walker et al. (2009) was M. maripaludis, whereas it was M. barkeri in the study of Plugge et al. (2010); (ii) the D. vulgaris and M. maripaludis co-culture and the D. vulgaris monoculture were cultivated in parallel in different chemostats by Walker et al. (2009), while a perturbation experiment by adding sulfate was performed to the chemostat co-culture to produce the D. vulgaris sulfidogenic metabolism in Plugge et al. (2010); (iii) the sulfidogenic monoculture of Walker et al. (2009) was sulfate-limited, whereas the D. vulgaris sulfidogenic metabolism was lactate-limited (Plugge et al., 2010); and (iv) the cell ratio (between D. vulgaris and M. maripaludis) during steady-state co-culture growth was higher (4:1) compared with the 1:1 in the D. vulgaris M. barkeri co-culture. Nevertheless, some similar results were obtained. These included the identification of a five-gene cluster encoding several lipo- and membrane-bound proteins which was down-regulated when cells were shifted to a sulfidogenic metabolism (Figure 1). Interestingly, this gene cluster has orthologs found only in the syntrophic bacterium Syntrophobacter fumaroxidans and four recently sequenced Desulfovibrio strains, suggesting that these genes are possible “syntrophic genes.” Both studies demonstrated that syntrophic growth and sulfate respiration use mostly independent energy generation pathways, implying that the molecular mechanism of microbial syntrophic processes cannot be fully interpreted by studying only pure cultures (Walker et al., 2009; Plugge et al., 2010). In addition, the studies also identified the upregulation during the D. vulgaris syntrophic metabolism of the high-molecular-mass cytochrome complex (DVU0533, encoding Hmc protein 4), the DVU0145–0150 cellular membrane gene cluster of unknown function and heterodisulfide reductase (hdrAB), and the downregulation of genes involved in iron transport (feoB and feoA; Walker et al., 2009; Plugge et al., 2010).
Figure 1.
A gene cluster with orthologs only in Syntrophobacter fumaroxidans and four recently sequences Desulfovibrio strains was significantly down-regulated during shift from syntrophic to sulfidogenic metabolism (Printed from Plugge et al., 2010, with permission of SGM).
Among the SRB members of the genus Desulfovibrio are easy to grow and they grow rapidly. Therefore, they have been the subject of the most intensive biochemical and molecular research (Postgate, 1984; Peck, 1993; Voordouw, 1993). However, multiple studies have described physiology and metabolic mode of SRB in relation with their environment. In the following paragraphs we will review a variety of these SRB-containing communities and their role in anaerobic biodegradation.
Metabolic Flexibility of Desulfotomaculum Cluster IH
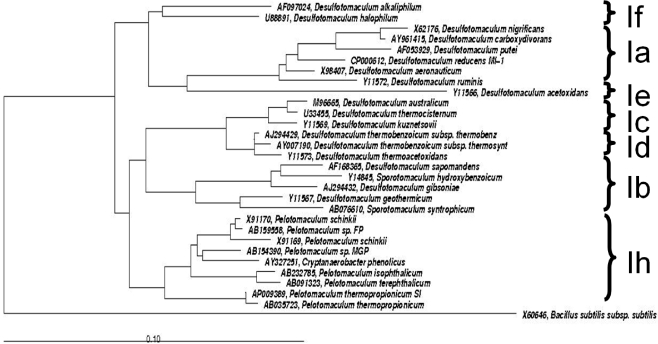
Members of the Gram-positive Desulfotomaculum cluster I are commonly considered as regular sulfate-reducing bacteria. However, in the last decade new representatives have been isolated that lack the ability of sulfate reduction, all phylogenetically grouping in Desulfotomaculum cluster Ih. These representatives are isolated from environments typically low in sulfate and producing methane: anaerobic bioreactors and rice paddy soils. Syntrophic propionate-oxidizing species of the genus Pelotomaculum are Pelotomaculum schinkii, P. thermopropionicum, and P. propionicicum (de Bok et al.,2005; Imachi et al., 2002, 2006, 2007). Additional members of Desulfotomaculum cluster Ih are P. terephthalicum sp. nov., and P. isophthalicum sp. nov. (Qiu et al., 2006) involved in syntrophic degradation of phthalate isomers and also lacking the ability of sulfate reduction. Imachi et al. (2006) demonstrated that none of these species was able to use sulfate, sulfite, or organosulfonates as electron acceptors. A PCR-based screening for dsrAB genes (key genes of the sulfate respiration pathway encoding the alpha and beta subunits of the dissimilatory sulfite reductase) of all cultures tested was negative with the exception of P. propionicicum. Based on these results it was proposed that subcluster Ih bacteria have adapted to anoxic, low-sulfate conditions and thus constitute a significant fraction of the Desulfotomaculum cluster I population in methanogenic microbial communities in a wide variety of methanogenic environments, highlighting its ecological impact in anoxic environments low in sulfate (Imachi et al., 2006; Figure 2). As a consequence of this evolutionary process, they have lost the capability of dissimilatory sulfate reduction and adopted the syntrophic life style, in close proximity to hydrogen- and formate-consuming methanogens. In such a way they can maintain an energetically favorable, low-hydrogen partial pressure that is necessary for the syntrophic oxidation of organic substrates. Given their recognized phenotypes and wide occurrence in low-sulfate, methanogenic environments, descendants of the Desulfotomaculum subcluster Ih branch most likely function as non-sulfate-reducing, syntrophic degraders of organic substrates in situ. This hypothesis received further support from a study that showed, by using rRNA-based stable-isotope probing, that Pelotomaculum species were not only dominant, but also actively involved in syntrophic propionate oxidation in a rice paddy soil (Lueders et al., 2004). Interestingly, the recently sequenced genome of P. thermopropionicum (Kosaka et al., 2008) points out the presence of dsr and aps genes (aps is coding for adenylyl sulfate reductase, another important enzyme in the sulfate respiration pathway that consists of an alpha and beta subunit). The genes are clustered in an operon (PTH_0235 to PTH_0242).
Figure 2.
Phylogenetic tree of Desulfotomaculum cluster I, showing the grouped phylogeny of Pelotomaculum ssp. The tree is based on comparative analyses of 16S rRNA gene sequences. The tree was constructed with sequences greater than 1,000 nucleotides using the latest released version of ARB (ARB 5.2, September 5, 2010). Bacillus subtilis was set as root. The reference bar indicates 10% sequence divergence.
Gene clusters necessary for dissimilatory sulfate reduction, such as those for transmembrane electron transport complexes (Haveman et al., 2004), were not found in the genome of P. thermopropionicum. This is consistent with previous physiological observations showing that this bacterium could not utilize sulfate as an electron acceptor (Imachi et al., 2002). The PCR-based screening for dsrAB genes by Imachi et al. (2006) that tested negative for P. thermopropionicum may have used primer sets that did not amplify the dsrAB genes in P. thermopropionicum. It is also possible that some of these dsrAB-carrying non-sulfate reducers use organosulfonates as electron acceptors for anaerobic respiration instead. e.g., Bilophila wadsworthia degradestaurine to sulfite, which is the actual substrate for its dissimilatory sulfite reductase (Cook et al., 1998). However, a whole range of organosulfonates did not support growth of the thermophilic spore-forming, low-G + C bacteria belonging to the genus Pelotomaculum (Imachi et al., 2006). An alternative explanation is that the real substrate for the dissimilatory sulfite reductase in syntrophic bacteria has not yet been identified.
Desulfotomaculum cluster Ih related species are also involved in the anaerobic mineralization of benzene in the presence of sulfate (Kleinsteuber et al., 2008; Laban et al., 2009; Herrmann et al., 2010). Identification of the benzene-degrading, sulfate-reducing communities was in all three studies based on culture-independent methods. A Pelotomaculum/Cryptanaerobacter-like phylotype represented a syntrophic community responsible for the initiation of the anaerobic benzene degradation in the studies by Kleinsteuber et al. (2008) and Herrmann et al. (2010). Benzene was completely mineralized in the presence of sulfate by a consortium consisting of syntrophs, hydrogenotrophic sulfate reducers and to a minor extent aceticlastic methanogens. Also Laban et al. (2009) showed that organisms phylogenetically related to the Gram-positive genus Pelotomaculum were responsible for benzene degradationcoupled to sulfate reduction. However, the 16S rRNA gene-based sequence similarity to the next cultivated representative of Desulfotomaculum cluster Ih constituted only 95%. These sequences could be clustered between the genera Desulfotomaculum and Pelotomaculum. The similarity of the sequences of the Pelotomaculum-related phylotypes described by Kleinsteuber et al. (2008) to those identified by Laban et al. (2009) range from 88.8 to 95.7% indicating that these phylotypes are indeed different.
Based on their results with a highly enriched culture, where only one dominant species was present, Laban et al. (2009) proposed that these bacteria with Pelotomaculum-related 16S rRNA gene sequences oxidize benzene directly coupled to sulfate reduction.
Clearly we cannot exclude that members of the Desulfotomaculum subcluster 1 h are capable of sulfate reduction, but apparently they can not be easily adapted to do so. The information presented here is based on our present knowledge.
Metabolic Flexibility by Members of the Order Syntrophobacterales
The Syntrophobacterales are an order of the δ-Proteobacteria, with three families, the Syntrophaceae, Syntrophobacteraceae, and Syntrophorhabdaceae (McInerney et al., 2008). Many of the members of these families are SRB, but also representatives of these families have been isolated that lack the ability for anaerobic sulfate respiration, or can grow as SRB or fermentative organism, depending on the environmental conditions (Wallrabenstein et al., 1994, 1995; Van Kuijk and Stams, 1995; Liu et al., 1999; Chen et al., 2005). With respect to 16S rRNA phylogeny, no predictions can be made on their metabolic flexibility.
Research has focused on propionate-degrading bacteria from the order Syntrophobacterales, specifically members that were enriched and isolated in the absence of sulfate. All species studied (Table 1) were capable of propionate degradation in syntrophic co-culture with a syntrophic partner but also as a pure culture coupled to dissimilatory sulfate reduction.
Table 1.
Growth rates of selected propionate-degrading bacteria with and without sulfate.
| Growth rate (day−1) | References | ||
|---|---|---|---|
| Propionate | Propionate + sulfate | ||
| Syntrophobacter fumaroxidans | 0.17 | 0.024 | Van Kuijk and Stams (1995), Harmsen et al. (1998) |
| Syntrophobacter pfennigii | 0.066 | 0.063 | Wallrabenstein et al. (1995) |
| Syntrophobacter sulfatireducens | n.d.* | 0.12 | Chen et al. (2005) |
| Syntrophobacter wolinii | 6.7** | 0.062 | Wallrabenstein et al. (1994) |
| Smithella propionica | n.d.* | n.d.*** | Liu et al. (1999) |
*Capable of syntrophic growth on propionate, but growth rate not determined.
**Only a doubling time (days) was determined.
***Capable of sulfate reduction, but growth rate not reported.
It remains unknown why some bacteria have actively expressed dsrAB genes, but cannot utilize sulfate, sulfite, and/or organosulfonates for anaerobic respiration. One might speculate that these microbes were formerly active sulfate reducers, but have lost this trait since they have to deal with low-sulfate and/or sulfite levels in methanogenic environments (Imachi et al., 2006). For this reason, the presence of dsrAB in these bacteria, which most often live in close association with hydrogen-consuming microorganisms for the syntrophic oxidation of substrates, would be a genetic remnant and thus indicative of an ancient sulfate/sulfite-respiring potential. The physiological data on syntrophs from the Syntrophobacterales order, which are abundantly present in methanogenic environments (Loy et al., 2004; Lueders et al., 2004; Stams and Plugge, 2009), indicate that all have retained their sulfate-reducing capability (Wallrabenstein et al., 1994; Van Kuijk and Stams, 1995). Syntrophic dsrAB-containing non-SRBs, syntrophic SRBs, and real sulfate reducers are phylogenetically fused, indicating an evolutionary connection between the sulfate-reducing and syntrophic lifestyle.
Non-Existing Metabolic Flexibility by Butyrate Oxidizers
Bacteria that grow on butyrate syntrophically with methanogens can only be found within the Gram-positive genera Syntrophomonas and Syntrophus. None of the members of these genera has the ability to reduce sulfate. The recently sequenced genomes of Syntrophomonas wolfei and Syntrophus aciditrophicus clearly show the lack of dsr and aps genes which explains on a molecular basis the inability of these organisms to do so. On the other hand, Gram-positive and Gram-negative butyrate-degrading sulfate-reducing genera have been described (e.g., Desulfobacterium, Desulfosarcina, and Desulfoarculus). None of the isolated sulfate-reducing species was ever shown to be able to grow in the absence of sulfate in syntrophy with methanogens (Rabus et al., 2006; Muyzer and Stams, 2008). This suggests that butyrate-degrading communities have a different response to changes in sulfate availability than propionate-degrading communities. In granular sludge butyrate-degrading bacteria that grow in syntrophy with methanogens are not easily outcompeted by sulfate-reducing bacteria (Oude Elferink et al., 1994). Instead, hydrogen-consuming methanogens seem to be replaced by hydrogen-consuming sulfate reducers, while propionate-degrading communities are easily outcompeted by typical propionate-degrading sulfate-reducing bacteria. Recently, it was observed that in communities that degrade long-chain fatty acids Syntrophomonas sp. persisted when methanogenic sludge was exposed to increasing levels of sulfate, but the hydrogen-consuming archaea were outcompeted by hydrogen-consuming sulfate-reducing bacteria (Sousa et al., 2009).
Metabolic Flexibility of SRB in Marine Environments
In sulfate-rich marine sediments, sulfate-reducing bacteria typically use all the products of primary fermentations and oxidize them to CO2 coupled to sulfate reduction (Muyzer and Stams, 2008). Therefore, fermentation by syntrophic communities is thought to be unimportant. However, when sulfate becomes limited the organic matter is no longer mineralized coupled to sulfate reduction, but through methanogenesis. Kendall et al. (2006) described marine syntrophic propionate- and butyrate-degrading cultures. This study implied that syntrophic communities contribute to the vast methane reservoirs in marine sediments. Microbial populations identified in shallow methanogenic sediments in the Gulf of Mexico revealed the presence of members of Syntrophobacteriaceae (Lloyd et al., 2006; McInerney et al., 2008). Cultured members of this family include syntrophic propionate oxidizers that can also reduce sulfate and as such have the capability to switch from sulfate-reducing life style to syntrophic life style as was discussed in paragraph “metabolic flexibility by members of the order Syntrophobacterales” (McInerney et al., 2008). Members of the Syntrophobacteriaceae were also found in the sulfate–methane transition zone in the Black Sea (Leloup et al., 2007, 2009). It was speculated that in these sediments, sulfate-reducing bacteria and methanogens do not compete but rather co-exist when mineralizing organic matter. In the absence of sulfate the reducing equivalents produced (hydrogen or formate) by SRB can be shuttled to a methanogen, serving as the syntrophic partner. More recently in Aarhus Bay it was found, that sulfate reducers were still very abundant in the methanogenic zones (Leloup et al., 2009). It is still unclear which type of metabolism sulfate reducers encompass in the methane zone and vice versa. Non-competitive substrates in marine environments (used by methanogens but not by sulfate reducers) methanol and methylamine were indeed converted by methanogens in sulfate-containing sediments from Aarhus Bay. However, sulfate reducers could scavenge the nanomolar hydrogen that leaked from the methanogenic cells (Finke et al., 2007), once more illustrating the complexity of metabolic interactions.
Evolution of Stability and Productivity at the Origin of a Syntrophic Community of an SRB and Methanogen
A syntrophic community was established by combining the D. vulgaris Hildenborough with the methanogenic archaeon M. maripaludis S2 (Hillesland and Stahl, 2010). Both members in the community could be grown in pure culture with appropriate substrates, but under the conditions of the experiments they could grow only through syntrophic cooperation. In the absence of hydrogen and sulfate, the species feed together by cooperating to complete the following energy-yielding reaction:
Twenty-four independent pairings(co-cultures) of the bacterium D. vulgaris and the archaeon M. maripaludis were established and followed for 300 community doublings in two environments, one allowing for the development of a heterogeneous distribution of resources and the other not. Evolved co-cultures grew up to 80% faster and were up to 30% more productive (biomass yield per mole of substrate) than the ancestors. Unfortunately, the authors did not include quantitative data on substrate depletion, product formation, and biomass production in the evolved co-cultures to support their hypothesis.
Unlike eukaryotes that evolve principally through the modification of existing genetic information, bacteria can obtain a significant proportion of their genetic diversity through the acquisition of sequences from distantly related organisms (Scholten et al., 2007). Horizontal gene transfer produces extremely dynamic genomes in which substantial amounts of DNA are introduced into and deleted from the chromosome. These lateral transfers have effectively changed the ecological and pathogenic character of bacterial species (Ochman et al., 2000). Previous studies indicated that the scale of horizontal gene transfer may have previously been underestimated and that horizontal gene transfer may be a major force in the evolution of prokaryotic genomes (Boucher and Doolittle, 2000; Calteau et al., 2005). In a more recent study, a combination of transcriptomics and phylogenomics, codon, and amino acid usage analyses of the syntrophic interaction between D. vulgaris and M. barkeri has led to the discovery of a functionally unknown gene set in the genome of D. vulgaris. This may be the result of ancient horizontal gene transfer from an archaeal methanogen. The gene set consists of three closely located genes on the chromosome, DVU2103 and DVU2104 encoding iron–sulfur cluster-binding/ATPase domain proteins and DVU2108 encoding an MTH1175-like domain family protein (Scholten et al., 2007). Similar lateral gene transfer was also reported between other syntrophic bacteria and methanogenic archaea (Kosaka et al., 2008; Fournier, 2009), suggesting that lateral gene transfer among microorganisms whose niches are closely associated is a common and important evolutionary process (Kato and Watanabe, 2010).
Future Perspectives
The metabolic flexibility of syntrophs and SRBs deserves future attention. The analysis of available and future genome sequences of SRBs, syntrophs and their partners in more detail will provide more insight into horizontal gene transfer, loss of genes (such as dsr and aps), genotypic and phenotypic similarities and its consequences for their ecological specialization. Dedicated physiological studies will unravel the occurrence and significance of less obvious metabolic capabilities of microorganisms in general and of more specifically syntrophic and sulfate-reducing bacteria. Molecular ecological studies such as comparative metagenomics on environments with fluctuating sulfate levels may reveal the in situ metabolism of the microorganisms present.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors were financially supported by the Earth and Life Sciences division (ALW) and Chemical Science division (CW) of the Netherlands Organization for Scientific Research (NWO). The authors thank Tom van den Bogert for beautification of the phylogenetic tree.
References
- Aubert C., Brugna M., Dolla A., Bruschi M., Giudici-Orticoni M. T. (2000). A sequential electron transfer from hydrogenases to cytochromes in sulfate-reducing bacteria. Biochim. Biophys. Acta 1476, 85–92 10.1016/S0167-4838(99)00221-6 [DOI] [PubMed] [Google Scholar]
- Bender K. S., Yen H. C., Hemme C. L., Yang Z., He Z., He Q., Zhou J., Huang K. H., Alm E. J., Hazen T. C., Arkin A. P., Wall J. D. (2007). Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 73, 5389–5400 10.1128/AEM.00276-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher Y., Doolittle W. F. (2000). The role of lateral gene transfer in the evolution of isoprenoid biosynthesis pathways. Mol. Microbiol. 37, 703–716 10.1046/j.1365-2958.2000.02004.x [DOI] [PubMed] [Google Scholar]
- Bryant M. P., Campbell L. L., Reddy C. A., Crabill M. R. (1977). Growth of Desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria. Appl. Environ. Microbiol. 33, 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M. P., Wolin E. A., Wolin M. J., Wolfe R. S. (1967). Methanobacillus omelianskii a symbiotic association of two species of bacteria. Arch. Microbiol. 59, 20–31 10.1007/BF00406313 [DOI] [PubMed] [Google Scholar]
- Calteau A., Gouy M., Perriere G. (2005). Horizontal transfer of two operons coding for hydrogenases between bacteria and archaea. J. Mol. Evol. 60, 557–565 10.1007/s00239-004-0094-8 [DOI] [PubMed] [Google Scholar]
- Chen S. Y., Liu X. L., Dong X. Z. (2005). Syntrophobacter sulfatireducens sp. nov., a novel syntrophic, propionate-oxidizing bacterium isolated from UASB reactors. Int. J. Syst. Evol. Microbiol. 55, 1319–1324 10.1099/ijs.0.63565-0 [DOI] [PubMed] [Google Scholar]
- Chhabra S. R., He Q., Huang K. H., Gaucher S. P., Alm E. J., He Z., Hadi M. Z., Hazen T. C., Wall J. D., Zhou J., Arkin A. P., Singh A. K. (2006). Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188, 1817–1828 10.1128/JB.188.5.1817-1828.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. E., He Q., He Z., Huang K. H., Alm E. J., Wan X. F., Hazen T. C., Arkin A. P., Wall J. D., Zhou J. Z., Fields M. W. (2006). Temporal transcriptomic analysis as Desulfovibrio vulgaris Hildenborough transitions into stationary phase during electron donor depletion. Appl. Environ. Microbiol. 72, 5578–5588 10.1128/AEM.00284-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A. M., Laue H., Junker F. (1998). Microbial desulfonation. FEMS Microbiol. Rev. 22, 399–419 10.1111/j.1574-6976.1998.tb00378.x [DOI] [PubMed] [Google Scholar]
- Cypionka H. (2000). Oxygen respiration in Desulfovibrio species. Ann. Rev. Microbiol. 54, 827–848 10.1146/annurev.micro.54.1.827 [DOI] [PubMed] [Google Scholar]
- de Bok F. A. M., Harmsen H. J. M., Plugge C. M., deVries M. C., Akkermans A. D. L., deVos W. M., Stams A. J. M. (2005). Pelotomaculum schinkii sp. nov., an obligate syntrophic propionate-oxidizing spore-forming bacterium isolated in co-culture with Methanospirillum hungatei. Int. J. Syst. Evol. Microbiol. 55, 1697–1703 10.1099/ijs.0.02880-0 [DOI] [PubMed] [Google Scholar]
- Finke N., Hoehler T. M., Jørgensen B. B. (2007). Hydrogen leakage during methanogenesis from methanol and methylamine: implications for anaerobic carbon degradation pathways in aquatic sediments. Environ. Microbiol. 9, 1060–1071 10.1111/j.1462-2920.2007.01248.x [DOI] [PubMed] [Google Scholar]
- Fournier G. (2009). Horizontal gene transfer and the evolution of methanogenic pathways. Methods Mol. Biol. 532, 163–179 10.1007/978-1-60327-853-9_9 [DOI] [PubMed] [Google Scholar]
- Harmsen H. J. M., van Kuijk B. L. M., Plugge C. M., Akkermans A. D. L., de Vos W. M., Stams A. J. M. (1998). Syntrophobacter fumaroxidans sp. nov., a syntrophic propionate-degrading sulfate-reducing bacterium. Int. J. Syst. Bacteriol. 48, 1383–1387 10.1099/00207713-48-4-1383 [DOI] [PubMed] [Google Scholar]
- Haveman S. A., Greene E. A., Stilwell C. P., Voordouw J. K., Voordouw G. (2004). Physiological and gene expression analysis of inhibition of Desulfovibrio vulgaris Hildenborough by nitrite. J. Bacteriol. 186, 7944–7950 10.1128/JB.186.23.7944-7950.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Huang K. H., He Z., Alm E. J., Fields M. W., Hazen T. C., Arkin A. P., Wall J. D., Zhou J. (2006). Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough inferred from global transcriptional analysis. Appl. Environ. Microbiol. 72, 4370–4381 10.1128/AEM.02609-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg J. F., Seshadri R., Haveman S. A., Hemme C. L., Paulsen I. T., Kolonay J. F., Eisen J. A., Ward N., Methe B., Brinkac L. M., Daugherty S. C., Deboy R. T., Dodson R. J., Durkin A. S., Madupu R., Nelson W. C., Sullivan S. A., Fouts D., Haft D. H., Selengut J., Peterson J. D., Davidsen T. M., Zafar N., Zhou L., Radune D., Dimitrov G., Hance M., Tran K., Khouri H., Gill J., Utterback T. R., Feldblyum T. V., Wall J. D., Voordouw G., Fraser C. M. (2004). The genome sequence of the anaerobic sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22, 554–559 10.1038/nbt959 [DOI] [PubMed] [Google Scholar]
- Herrmann S., Kleinsteuber S., Chatzinotas A., Kuppardt S., Lueders T., Richnow H.-H., Vogt C. (2010). Functional characterization of an anaerobic benzene-degrading enrichment culture by DNA stable isotope probing. Environ. Microbiol. 12, 401–411 10.1111/j.1462-2920.2009.02077.x [DOI] [PubMed] [Google Scholar]
- Hillesland K. L., Stahl D. A. (2010). Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc. Natl. Acad. Sci. U.S.A. 107, 2124–2129 10.1073/pnas.0908456107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmer M., Kristensen E. (1994). Co-existence of sulfate reduction and methane production in an organic-rich sediment. Mar. Ecol. Prog. Ser. 107, 177–184 10.3354/meps107177 [DOI] [Google Scholar]
- Imachi H., Sakai S., Ohashi A., Harada H., Hanada S., Kamagata Y., Sekiguchi Y. (2007). Pelotomaculum propionicicum sp. nov., an anaerobic, mesophilic, obligately syntrophic, propionate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 57, 1487–1492 10.1099/ijs.0.64925-0 [DOI] [PubMed] [Google Scholar]
- Imachi H., Sekiguchi Y., Kamagata Y., Hanada S., Ohashi A., Harada H. (2002). Pelotomaculum thermopropionicum gen. nov., sp. nov., an anaerobic, thermophilic, syntrophic propionate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 52, 1729–1735 10.1099/ijs.0.02212-0 [DOI] [PubMed] [Google Scholar]
- Imachi H., Sekiguchi Y., Kamagata Y., Loy A., Qiu Y.-L., Hugenholtz P., Kimura N., Wagner M., Ohashi A., Harada H. (2006). Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum Cluster I are widely distributed in methanogenic environments. Appl. Environ. Microbiol. 72, 2080–2091 10.1128/AEM.72.3.2080-2091.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S., Watanabe K. (2010). Ecological and evolutionary interactions in syntrophic methanogenic consortia. Microbes Environ. 25, 145–151 10.1264/jsme2.ME10122 [DOI] [PubMed] [Google Scholar]
- Kendall M. M., Liu Y., Boone D. R. (2006). Butyrate- and propionate-degrading syntrophs from permanently cold marine sediments in Skan Bay, Alaska, and description of Algorimarina butyrica gen. nov., sp. nov. FEMS Microbiol. Lett. 262, 107–114 10.1111/j.1574-6968.2006.00380.x [DOI] [PubMed] [Google Scholar]
- Kleinsteuber S., Schleinitz K. M., Breitfeld J., Harms H., Richnow H.-H., Vogt C. (2008). Molecular characterization of bacterial communities mineralizing benzene under sulfate-reducing conditions. FEMS Microbiol. Ecol. 66, 143–157 10.1111/j.1574-6941.2008.00536.x [DOI] [PubMed] [Google Scholar]
- Kosaka T., Kato S., Shimoyama T., Ishii S., Abe T., Watanabe K. (2008). The genome of Pelotomaculum thermopropionicum reveals niche-associated evolution in anaerobic microbiota. Genome Res. 18, 442–448 10.1101/gr.7136508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivila K. M., Murray J. W., Devol A. H. (1990). Methane production in the sulfate depleted sediments of two marine basins. Geochim. Cosmochim. Acta 54, 403–411 10.1016/0016-7037(90)90329-J [DOI] [Google Scholar]
- Laban N. A., Selesi D., Jobelius C., Meckenstock R. U. (2009). Anaerobic benzene degradation by Gram-positive sulfate-reducing bacteria. FEMS Microbiol. Ecol. 68, 300–311 10.1111/j.1574-6941.2009.00672.x [DOI] [PubMed] [Google Scholar]
- Leloup J., Fossing H., Kohls K., Holmkvist L., Jørgensen B. B. (2009). Sulfate-reducing bacteria in marine sediment (Aarhus Bay, Denmark): abundance and diversity related to geochemical zonation. Environ. Microbiol. 11, 1278–1291 10.1111/j.1462-2920.2008.01855.x [DOI] [PubMed] [Google Scholar]
- Leloup J., Loy A., Knab N. J., Borowski C., Wagner M., Jørgensen B. B. (2007). Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ. Microbiol. 9, 131–142 [DOI] [PubMed] [Google Scholar]
- Liu Y., Balkwill D. L., Aldrich H. C., Drake G. R., Boone D. R. (1999). Characterization of the anaerobic propionate-degrading syntrophs Smithella propionica gen. nov., sp. nov. and Syntrophobacter wolinii. Int. J. Syst. Bacteriol. 49, 545–556 10.1099/00207713-49-2-545 [DOI] [PubMed] [Google Scholar]
- Lloyd K. G., Lapham L., Teske A. (2006). An anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Appl. Environ. Microbiol. 72, 7218–7230 10.1128/AEM.00886-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy A., Küsel K., Lehner A., Drake H. L., Wagner M. (2004). Microarray and functional gene analyses of sulfate-reducing prokaryotes in low sulfate, acidic fens reveal co-occurence of recognized genera and novel lineages. Appl. Environ. Microbiol. 70, 6998–7009 10.1128/AEM.70.12.6998-7009.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueders T., Pommerenke B., Friedrich M. W. (2004). Stable-isotope probing of microorganisms thriving at thermodynamic limits: syntrophic propionate oxidation in flooded soil. Appl. Environ. Microbiol. 70, 5778–5786 10.1128/AEM.70.10.5778-5786.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney M. J., Mackie R. I., Bryant M. P. (1981). Syntrophic association of a butyrate-degrading bacterium and Methanosarcina enriched from bovine rumen fluid. Appl. Environ. Microbiol. 41, 826–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney M. J., Struchtemeyer C. G., Sieber J., Mouttaki H., Stams A. J. M., Schink B., Rholin L., Gunsalus R. P. (2008). Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. 1125, 58–72 10.1196/annals.1419.005 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A., He Z., Alm E. J., Arkin A. P., Baidoo E. E., Borglin S. C., Chen W., Hazen T. C., He Q., Holman H. Y., Huang K., Huang R., Joyner D. C., Katz N., Keller M., Oeller P., Redding A., Sun J., Wall J., Wei J., Yang Z., Yen H. C., Zhou J., Keasling J. D. (2006). Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol. 188, 4068–4078 10.1128/JB.01921-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A., Redding A. M., Joachimiak M. P., Arkin A. P., Borglin S. E., Dehal P. S., Chakraborty R., Geller J. T., Hazen T. C., He Q., Joyner D. C., Martin V. J., Wall J. D., Yang Z. K., Zhou J., Keasling J. D. (2007). Cell-wide responses to low-oxygen exposure in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 189, 5996–6010 10.1128/JB.00368-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G., Stams A. J. M. (2008). The ecology and biotechnology of sulphate-reducing bacteria. Nat. Microbiol. Rev. 6, 441–454 [DOI] [PubMed] [Google Scholar]
- Ochman H., Lawrence J. G., Groisman E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- Odom J. M., Peck H. D., Jr. (1981). Hydrogen cycling as a general mechanism for energy coupling in the sulfate-reducing bacteria Desulfovibrio sp. FEMS Microbiol. Lett. 12, 47–50 10.1111/j.1574-6968.1981.tb07609.x [DOI] [Google Scholar]
- Oremland R. S., Marsh L. M., Polcin S. (1982). Methane production and simultaneous sulfate reduction in anoxic saltmarsh sediments. Nature 296, 143–145 10.1038/296143a0 [DOI] [Google Scholar]
- Oude Elferink S. J. W. H., Visser A., Hulshoff Pol L. W., Stams A. J. M. (1994). Sulfate reduction in methanogenic bioreactors. FEMS Microbiol. Rev. 15, 119–136 10.1111/j.1574-6976.1994.tb00130.x [DOI] [Google Scholar]
- Peck H. D., Jr. (1966). Phosphorylation coupled with electron transfer in extracts of the sulfate reducing bacterium Desulfovibrio gigas. Biochem. Biophys. Res. Commun. 22, 112–118 10.1016/0006-291X(66)90611-5 [DOI] [PubMed] [Google Scholar]
- Peck H. D., Jr. (1993). “Bioenergetics strategies of the sulfate-reducing bacteria,” in The Sulfate-Reducing Bacteria: Contemporary Perspectives, eds Odom J. M., Singleton R., Jr (New York: The Springer Verlag; ), 41–76 [Google Scholar]
- Pereira P. M., He Q., Valente F. M. A., Xavier A. V., Zhou J., Pereira I. A. C., Louro R. O. (2008). Energy metabolism in Desulfovibrio vulgaris Hildenborough: insights from transcriptome analysis. Antonie Van Leeuwenhoek 93, 347–362 10.1007/s10482-007-9212-0 [DOI] [PubMed] [Google Scholar]
- Plugge C. M., Scholten J. C. M., Culley D. E., Nie L., Brockman F. J., Zhang W. (2010). Global transcriptomics analysis of Desulfovibrio vulgaris lifestyle change from syntrophic growth with Methanosarcina barkeri to sulfate reducer. Microbiology 156, 2746–2756 10.1099/mic.0.038539-0 [DOI] [PubMed] [Google Scholar]
- Postgate J. R. (1984). The Sulphate-Reducing Bacteria. Cambridge: Cambridge University Press [Google Scholar]
- Qiu Y.-L., Sekiguchi Y., Hanada S., Imachi H., Tseng I.-C., Cheng S.-S., Ohashi A., Harada H., Kamagata Y. (2006). Pelotomaculum terephthalicum sp. nov. and Pelotomaculum isophthalicum sp. nov.: two anaerobic bacteria that degrade phthalate isomers in syntrophic association with hydrogenotrophic methanogens. Arch. Microbiol. 185, 172–182 10.1007/s00203-005-0081-5 [DOI] [PubMed] [Google Scholar]
- Rabus R., Hansen T. A., Widdel F. (2006). “Dissimilatory sulfate- and sulfur-reducing prokaryotes,” in The Prokaryotes, Vol. 2, eds Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (New York: Springer; ), 659–768 [Google Scholar]
- Schink B. (1997). Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61, 262–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholten J. C., Culley D. E., Brockman F. J., Wu G., Zhang W. (2007). Evolution of the syntrophic interaction between Desulfovibrio vulgaris and Methanosarcina barkeri: Involvement of an ancient horizontal gene transfer. Biochem. Biophys. Res. Commun. 352, 48–54 10.1016/j.bbrc.2006.10.164 [DOI] [PubMed] [Google Scholar]
- Sousa D. Z., Alves J. I., Alves M. M., Smidt H., Stams A. J. M. (2009). Effect of sulfate on methanogenic communities that degrade unsaturated and saturated long chain fatty acids (LCFA). Environ. Microbiol. 11, 68–80 10.1111/j.1462-2920.2008.01740.x [DOI] [PubMed] [Google Scholar]
- Stams A. J. M. (1994). Metabolic interactions between anaerobic bacteria in methanogenic environments. Antonie Van Leeuwenhoek 66, 271–294 10.1007/BF00871644 [DOI] [PubMed] [Google Scholar]
- Stams A. J. M., Plugge C. M. (2009). Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7, 568–577 10.1038/nrmicro2166 [DOI] [PubMed] [Google Scholar]
- Stolyar S., Van Dien S., Hillesland K. L., Pinel N., Lie T. J., Leigh J. A., Stahl D. A. (2007). Metabolic modeling of a mutualistic microbial community. Mol. Syst. Biol. 3, 92. 10.1038/msb4100131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Pingitore F., Mukhopadhyay A., Phan R., Hazen T. C., Keasling J. D. (2007). Pathway confirmation and flux analysis of central metabolic pathways in Desulfovibrio vulgaris Hildenborough using gas chromatography-mass spectrometry and Fourier transform-ion cyclotron resonance mass spectrometry. J. Bacteriol. 189, 940–949 10.1128/JB.00948-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kuijk B. L. M., Stams A. J. M. (1995). Sulfate reduction by a syntrophic propionate- oxidizing bacterium. Antonie Van Leeuwenhoek 68, 293–296 10.1007/BF00874139 [DOI] [PubMed] [Google Scholar]
- Voordouw G. (1993). “Molecular biology of the sulfate-reducing bacteria,” in The Sulfate-Reducing Bacteria: Contemporary Perspectives, eds Odom J. M., Singleton R., Jr (New York: The Springer Verlag; ), 88–130 [Google Scholar]
- Walker C. B., He Z., Yang Z. K., Ringbauer J. A., Jr, He Q., Zhou J., Voordouw G., Wall J. D., Arkin A. P., Hazen T. C., Stolyar S., Stahl D. A. (2009). The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J. Bacteriol. 191, 5793–5801 10.1128/JB.00356-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrabenstein C., Hauschild E., Schink B. (1994). Pure culture and cytological properties of Syntrophobacter wolinii. FEMS Microbiol. Lett. 123, 249–254 10.1111/j.1574-6968.1994.tb07232.x [DOI] [Google Scholar]
- Wallrabenstein C., Hauschild E., Schink B. (1995). Syntrophobacter pfennigii sp. nov., new syntrophically propionate-oxidizing anaerobe growing in pure culture with propionate and sulfate. Arch. Microbiol. 164, 346–352 10.1007/BF02529981 [DOI] [Google Scholar]
- Widdel F., Hansen T. A. (1991). “The sulphate and sulphur-reducing prokaryotes,” in The Prokaryotes, 2nd Edn, Vol. II, eds Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (New York: Springer; ), 583–624 [Google Scholar]
- Winfrey M. R., Ward D. M. (1983). Substrates for sulfate reduction and methane production in intertidal sediments. Appl. Environ. Microbiol. 45, 193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Culley D. E., Scholten J. C., Hogan M., Vitiritti L., Brockman F. J. (2006a). Global transcriptomic analysis of Desulfovibrio vulgaris on different electron donors. Antonie Van Leeuwenhoek 89, 221–237 10.1007/s10482-005-9024-z [DOI] [PubMed] [Google Scholar]
- Zhang W., Culley D. E., Hogan M., Vitiritti L., Brockman F. J. (2006b). Oxidative stress and heat-shock responses in Desulfovibrio vulgaris by genome-wide transcriptomic analysis. Antonie Van Leeuwenhoek 90, 41–55 10.1007/s10482-006-9059-9 [DOI] [PubMed] [Google Scholar]
- Zhang W., Gritsenko M. A., Moore R. J., Culley D. E., Nie L., Petritis K., Strittmatter E., Camp D. G., II, Smith R. D., Brockman F. J. (2006c). A proteomic view of Desulfovibrio vulgaris metabolism as determined by liquid chromatography coupled with tandem mass spectrometry. Proteomics 6, 4286–4299 10.1002/pmic.200500930 [DOI] [PubMed] [Google Scholar]