Abstract
Fusion oncogenes in acute myeloid leukemia (AML) promote self-renewal from committed progenitors, thereby linking transformation and self-renewal pathways. Like most cancers, AML is a genetically and biologically heterogeneous disease, but it is unclear whether transformation results from common or overlapping genetic programs acting downstream of multiple mutations, or by the engagement of unique genetic programs acting cooperatively downstream of individual mutations. This distinction is important, because the involvement of common programs would imply the existence of common molecular targets to treat AML, no matter which fusion oncogenes are involved. Here we demonstrate that the ability to promote self-renewal is a generalized property of leukemia-associated oncogenes. Disparate oncogenes initiated overlapping transformation and self-renewal gene expression programs, the common elements of which were defined in established leukemia stem cells from an animal model as well as from a large cohort of patients with differing AML subtypes, where they strongly predicted pathobiological character. Notably, individual genes commonly activated in these programs could partially phenocopy the self-renewal function of leukemia-associated oncogenes in committed murine progenitors. Further, they could generate AML following expression in murine bone marrow. In summary, our findings reveal the operation of common programs of self-renewal and transformation downstream of leukemia-associated oncogenes, suggesting mechanistically common therapeutic approaches to AML are likely to be possible, regardless of the identity of the driver oncogene involved.
Keywords: Leukemia-associated fusion genes, activation of self-renewal, transcriptional dysregulation, common pathways, therapeutic targets
Introduction
Acute myeloid leukemia (AML) is characterized by a block in terminal myeloid differentiation, accompanied by uncontrolled proliferation of immature myeloid progenitor cells. However, although there are unifying cellular characteristics it remains a very heterogeneous disease, morphologically, molecularly and biologically (1). Hundreds of separate genetic lesions have been described in AML and recent next generation sequencing analysis of AML genomes has demonstrated the occurrence of multiple lesions within individual leukemias (2). Great improvements have been made in the identification of prognostic factors for AML, such as age, white blood cell count (WBC), cytogenetics and mutational analysis of important genes (such as FLT3 and NPM1), however these have reinforced the heterogeneity of AML cases. In addition, gene expression studies have defined signatures downstream of many of these genetic lesions, allowing further molecular characterization (3,4).
Knowledge of the molecular lesions associated with specific subtypes of AML has led to the introduction of specifically targeted therapeutics, such as all trans retinoic acid (ATRA) in PML-RARA positive cases, and selective FLT3 inhibitors in cases which harbor an internal tandem duplication of the FLT3 gene (FLT3-ITD). However, AML remains a significant clinical problem with over 70% of patients succumbing to the disease, accounting for over 9000 deaths per year alone in the US (5), and novel therapeutics with efficacy in the majority of AML patients are urgently required. To this end it is not known whether transformation is mediated by common or overlapping genetic programs downstream of multiple mutations or through the engagement of unique programs downstream of individual mutations. This distinction is important, as the demonstration of common pathways may identify common critical molecular targets for the treatment of AML, in all cases or at least in significant subgroups.
The development of cancer is associated with the acquisition of a number of cellular characteristics, including limitless self-renewal, evasion of apoptosis and self-sufficiency in growth signals (6), and it is possible that these characteristics are governed by a limited number of master regulatory pathways. To use the example of malignant self-renewal, there would appear to be a significant overlap between self-renewal programs in both normal and malignant hematopoietic stem cells. Many genes and pathways implicated in normal stem cell self-renewal, such as the clustered HOX genes, the WNT/β-CATENIN pathway, the PTEN/AKT/FOXO axis and the NOTCH pathway, are also frequently dysregulated in cancer (reviewed in 7,8). However, the finding that these pathways are aberrantly activated or genes in them frequently mutated, along with the demonstration that normal stem cells and cancer stem cells may have different molecular requirements (9), suggests that there are differences between the self-renewal programs in normal and malignant stem cells that may be exploited for therapeutic benefit.
Leukemia-associated fusion genes such as MLL fusions and MOZ-TIF2 generate aberrant transcriptional programs mediated by their ability to modify chromatin, and can confer leukemia stem cell properties when retrovirally overexpressed in committed myeloid progenitors (10-12). We assessed the ability of other AML-associated mutations to alter self-renewal properties and chose two further transcription factor fusion genes, AML1-ETO (RUNX1- RUNX1T1) and NUP98-HOXA9 and the internal tandem duplication (ITD) mutation of the FLT3 receptor tyrosine kinase (RTK) gene (FLT3-ITD). FLT3-ITD mutations are representative of constitutively active RTK signaling, while the AML1-ETO and NUP98-HOXA9 fusion genes, which contain consensus DNA binding sequences, are representative of mutations thought to alter transcriptional programs mainly through transcriptional repression and activation, respectively. In addition, they also represent a prognostic spectrum, with the AML1-ETO rearrangement characteristically associated with a good prognosis (13) and the FLT3-ITD and NUP98-HOXA9 lesions associated with poor prognosis (14,15).
In this report we demonstrate that the ability to alter self-renewal is a more generalized effect of leukemia-associated transcription factor fusion oncogenes. In addition, taking advantage of this restoration of self-renewal properties in vitro, we use this platform to show that three disparate oncogenes initiate early, common and overlapping transformation and self-renewal signatures involved in leukemia induction. Moreover, elements of these signatures can be detected in established leukemia stem cells from an animal model of AML and can also be detected in expression data from a large cohort of patients with differing AML subtypes. Furthermore, these genesets strongly predict disease biology and correlate with existing prognostic factors. Finally, we demonstrate that individual genes from within the signature, such as Sox4 and Bmi1, can at least partially phenocopy the leukemia-associated oncogenes and alter self-renewal in committed murine progenitors and generate AML when expressed in murine bone marrow. This suggests that common transformation and self-renewal pathways downstream of a variety of leukemia-associated oncogenes contribute to the induction and maintenance of AML, a finding with important clinical implications.
Material and Methods
Generation of retroviral constructs
The MOZ-TIF2, NUP98-HOXA9, and FLT3-ITD MSCV vectors were generated as previously described (16-18). The AML1-ETO vectors were a kind gift from Michael Tomasson. The cDNA coding regions of murine Sox4 (NM_009238, Geneservice clone BC052736.1), Tcf4 (Geneservice clone BC043050) and murine Bmi1 (Geneservice clone BC053708) were amplified and subcloned into MSCV-IRES-GFP and MSCV-pgk-Neo and vectors re-verified by sequencing.
Staining and sorting of progenitor populations
Bone marrow mononuclear cells were isolated as previously described (11) and LSK, CMP (Common myeloid Progenitors), and GMP (Granulocyte Monocyte Progenitors) populations stained as described (19) with minor changes (see supplementary methods)
Retroviral transductions, bone marrow, progenitor, and limiting dilution transplant assays
Retroviral supernatants were generated in 293T cells as previously described (16). Bone marrow transplants were performed as described (16) with the exception of culture with 10 ng/ml recombinant stem cell factor (PeproTech, Rocky Hill, U.S.A.) and centrifugation at 2500 rpm. For whole bone marrow experiments, 1 × 106 CD45.2+ cells were injected into lethally irradiated (2 × 550 rads) CD45.2+ recipients (Charles River, Margate, UK). Double FACS sorted CD45.2+ progenitors were supplemented with recombinant murine IL11 (30ng/ml) (R & D Systems, Minneapolis, U.S.A.), recombinant murine SCF (150ng/ml) (PeproTech), recombinant murine or human IL6 (30ng/ml) (PeproTech, London, UK), recombinant murine TPO (60ng/ml) (Peprotech, London, UK), 2% fibronectin solution (StemCell Technologies, Vancouver, Canada), 5% FBS, and 1% penicillin/streptomycin (Invitrogen, Paisley, UK). Cells were spininoculated with retroviral supernatants and 4μg/ml polybrene at 1800 rpm for 60 minutes and incubated overnight at 32°C. The next day, progenitors were washed, resuspended in Hanks balanced salt solution (Invitrogen, Paisley, UK), and injected (transplant range CMP: 3.2-5.8×104, GMP: 4.2-16.5×104, MEP (Megakaryocyte Erythrocyte Progenitors) 3.5-6.8 ×104) into the tail vein of two lethally irradiated (2 × 550 rads) CD45.1+ recipients (Charles River, Margate, UK) per progenitor population along with 5×105 CD45.1+ bone marrow mononuclear cells. Each experiment was repeated three times. In the adoptive transfer of MOZ-TIF2-associated leukemia cells to enrich for leukemic stem cells (LSC), leukemic GMPs, leukemic GFP+, and leukemic GFP+Mac1+Gr1+ were transplanted at limiting dilution doses of 1×106, 5×105, 1×105, 1×104, 5×103, 1×103, 1×102, and 10 cells into three to five sublethally irradiated CD45.2+ recipients (1 × 650 rads) as detailed in the text.
Serial replating and growth in liquid culture
Serial replating assays were performed as previously described (11).
RNA amplification and gene expression analysis
36 hours after spininoculation of oncogene or empty vector transduced GMP, GFP positive cells were homogenized with TRIzol reagent (Invitrogen) according to manufacturer’s instructions. RNA isolated from the above populations along with sorted normal LSK, GMP and MOZ-TIF2 leukemic GMP was quantified with the RiboGreen RNA Quantitation Reagent and Kit (Invitrogen) according to manufacturer’s specifications and used for RNA amplification and hybridization as previously published (20) with the modification of biotinylated CTP and UTP (Enzo Diagnostics, Farmingdale, U.S.A.) in a 2.5:1 proportion to non-biotinylated CTP and UTP. All RNA populations were simultaneously amplified.
Gene expression levels were measured using Affymetrix (San Jose, CA) Mouse Genome 430 2.0 GeneChip arrays (45,101 probe sets) with hybridization and washes as per the manufacturers specifications. The starting point for all analyses was the “.CEL” files from the MAS5 software. Data were analyzed using the R statistical package bioconductor (21). Data quality was assessed using functions in the affy and affyPLM packages and outlier arrays were removed from subsequent analysis (22). The GCRMA algorithm (ver. 2.4.1) was used to obtain normalized expression estimates. Genes were selected for further analysis on the following basis: genes that had probe sets for which the expression value was greater than 60 (which in our study constitute the average background reading of all probe sets) and that had a present flag call in at least two of three samples. To detect significant changes in the expression levels, two-sample Welch t-tests (parametric; assuming unequal variances; Benjamini and Hochberg step-up multiple testing correction at a False Discovery rate <0.05) was applied to the resulting genes. Heat map images were developed using GenePattern 3.0 (23) with row normalized color scheme.
Results
Alteration of self-renewal properties is a common finding with AML-associated transcriptional oncogenes
Similarly to MLL fusions or MOZ-TIF2, AML1-ETO or NUP98-HOXA9 were able to alter properties of self-renewal in committed myeloid progenitor cells (CMP and GMP) in vitro, when assessed by the ability to serially replate in cytokine supplemented methylcellulose and to grow in short-term liquid culture (Figure 1 B and C). In contrast, transduction of committed progenitors with FLT3-ITD did not alter their self-renewal properties in in vitro or in vivo assays (Figure 1A), in keeping with our previous findings with another activated tyrosine kinase, BCR-ABL (11). However, differences were demonstrated between the effects of the transcription factor fusion genes in vivo. NUP98-HOXA9 was able to generate AML when transduced committed progenitors (CMP, GMP, and MEP) were transplanted into recipient mice (Figure 1C). Previous studies have demonstrated that expression of AML1-ETO in unfractionated murine bone marrow does not generate AML without additional mutations but does lead to stem cell expansion (24), however we could not detect the presence of any transplanted cells in recipients of AML1-ETO transduced committed myeloid progenitors after 4 months (Figure 1B). These data demonstrate that alteration of self-renewal potential is a more generalized property of AML-associated transcription factor fusion genes and is not solely associated with mutations harboring intrinsic chromatin modification properties. However, the extent of this property appears to be variable between different fusions with some, such as MLL fusions, MOZ-TIF2 and NUP98-HOXA9 able to confer self-renewal properties in vivo and generate AML and others, such as AML1-ETO, only able to alter self-renewal in vitro.
Figure 1. Transcription factor fusions associated with AML commonly alter self-renewal properties.
A) Committed progenitors (CMP, GMP and MEP) transduced with FLT3-ITD do not serially replate in cytokine supplemented methycellulose (left panel). The right panel shows a Kaplan-Meier graph for survival of transplant recipients of FLT3-ITD transduced CMP, GMP and MEP. All animals were healthy until the time of sacrifice (up to 350 days) with no evidence of leukemia or reconstitution by the transplanted cells. B) CMP and GMP transduced with AML1-ETO serially replate in cytokine supplemented methylcellulose and grow in liquid culture (left and middle panels). Following transplantation of these populations no reconstitution was detected (right panel). C) Similar analysis is presented for NUP98-HOXA9, demonstrating that CMP, GMP and MEP transduced with NUP98-HOXA9 serially replate in cytokine supplemented methylcellulose and grow in liquid culture (upper left and middle panels). However, and in contrast to AML1-ETO, NUP98-HOXA9 transduced progenitor populations generate acute leukemias as demonstrated in the Kaplan-Meier graph in the right upper panel and in the lower panels, where representative photomicrographs of peripheral blood, bone marrow, liver and spleen demonstrate significant infiltration with leukemic blasts on a background of myeloproliferation.
Common and overlapping pathways promote transformation and alter self-renewal downstream of AML-associated oncogenes
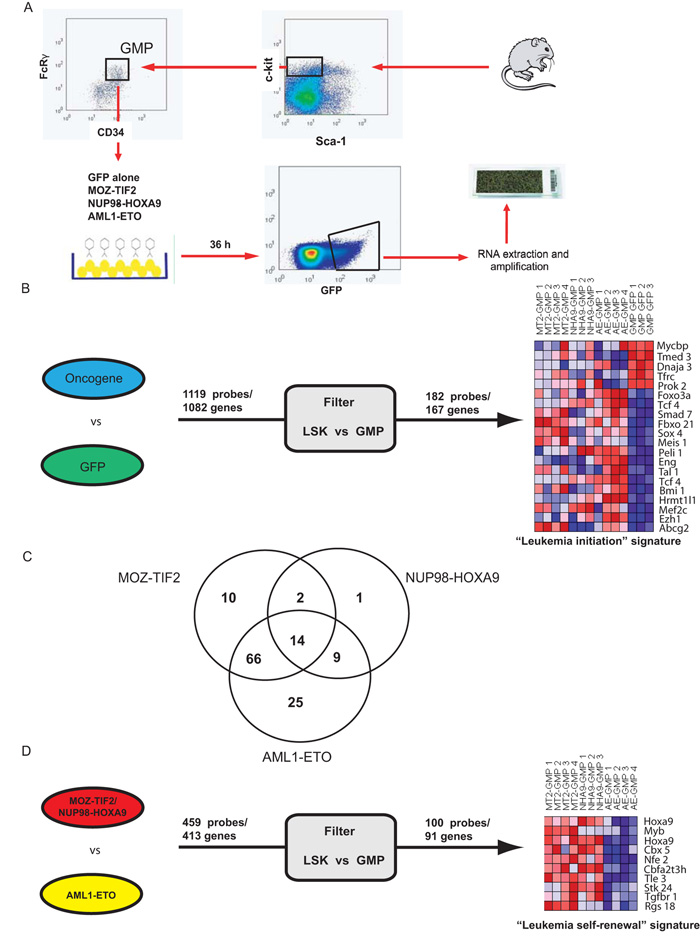
The above findings provided us with a platform to investigate if common, overlapping or separate self-renewal and transformation pathways are activated downstream of individual oncogenes. The MOZ-TIF2, NUP98-HOXA9 and AML1-ETO oncogenes or a control empty vector were retrovirally expressed in the GMP compartment using MSCV-IRES-GFP retroviral vectors. GFP-positive transduced cells were sorted and RNA populations from these cells were simultaneously amplified and then hybridized to Affymetrix 430A 2.0 arrays (Figure 2A).
Figure 2. Identification of immediate leukemia-associated signatures downstream of multiple AML fusion genes.
A) Panel A demonstrates a schema of the experimental strategy used to identify the leukemia-associated signatures. B) Identification and prioritization of genes to generate the 167 gene (182 probe) immediate “leukemia initiation” signature. Initially the expression profiles for multiple replicates of MOZ-TIF2, AML1-ETO and NUP98-HOXA9 transduced GMPs were compared in a single two-class comparison to GFP transduced GMPs. 1119 genes were commonly dysregulated to an FDR significance level of 0.05. A filter comparing expression of the 1119 genes between normal HSC and GMP was then applied, with genes which were significantly and coordinately differentially expressed (p < 0.05) retained. This prioritized 167 genes to generate our signature. Expression patterns of 20 representative genes are presented in the left part of the panel (all genes are listed in Table S1). C) Three single two-class comparisons were also made between each of MOZ-TIF2, AML1-ETO and NUP98-HOXA9 transduced GMPs and GFP transduced GMPs and genes differentially expressed (p<0.05) were overlapped with our leukemia initiation signature. The Venn-diagram details the number of genes dysregulated by each oncogene and demonstrates significant common and overlapping pathways downstream of the three oncogenes. D) Identification and prioritization of genes to generate the 91 gene (100 probe) immediate “leukemia self-renewal” signature. The expression profiles for multiple replicates of MOZ-TIF2 and NUP98-HOXA9 transduced GMPs were compared in a single two-class comparison to AML1-ETO transduced GMPs. 413 genes were commonly dysregulated (p < 0.01). This geneset was then similarly filtered to generate our immediate leukemia self-renewal signature. Expression patterns of 10 representative probes (9 genes) are presented in the left part of the panel (all genes are listed in Table S2).
The data were analyzed using two complementary methods. Initially, we compared the gene expression profiles of multiple replicates of oncogene transduced GMP together in a single two-class comparison with empty-vector transduced GMP (Figure 2B). In addition, we also compared replicates of each oncogene in three separate two-class comparisons with empty-vector transduced GMP at lower stringency (Figure 2C). In the first analysis we identified 1082 genes/1119 probesets whose expression levels significantly differed (FDR level 0.05) (Figure 2B). There is a loss of self-renewal potential in the transition from the LSK to GMP population and to further enrich for potential self-renewal genes we compared the expression of these probes/genes in sorted normal LSK versus GMP populations. Genes which were not significantly (p<0.05) and similarly differentially expressed between LSK and GMP were excluded, allowing us to prioritize an immediate “leukemia initiation” program of 167 genes (182 probesets, Figure 2B and Table S5). This gene signature included genes associated with either normal self-renewal, such as Foxo3a (25), with malignant transformation, such as Meis1 (26) and Mef2c (12), or with both processes, such as Bmi1 (27).
Our second analysis demonstrated differential expression of 5750, 1161 and 5109 genes, when MOZ-TIF2, NUP98-HOXA9 and AML1-ETO transduced GMP were respectively compared with empty vector transduced GMP(p <0.05). When our “leukemia initiation” signature was compared to the single comparisons, 127/167 (76%) genes overlapped.
Fourteen genes were differentially expressed downstream of all three oncogenes, 77 genes were differentially expressed downstream of two oncogenes and the remaining 36 genes were only differentially expressed downstream of a single oncogene (Figure 2C and Table S5). Taken together these data demonstrates that both common and overlapping genes are transcriptionally dysregulated downstream of disparate AML-associated oncogenes.
As there was a distinction between the effects of MOZ-TIF2, NUP98-HOXA9 and AML1-ETO in their ability to alter self-renewal in vivo and generate AML from committed progenitors, we next assessed differences between the gene expression signatures of MOZ-TIF2 and NUP98-HOXA9 transduced GMP in comparison with AML1-ETO transduced GMP. We detected 413 genes (459 probesets) that were differentially expressed and using a similar filtering strategy we identified an immediate “leukemia self-renewal” program of 91 genes/100 probesets (Figure 2D and Table S6). This gene program was mutually exclusive from the leukemia initiation program but also included genes previously implicated in self-renewal and transformation such as Hoxa9 (28), c-Myb (29) and Cbx5 (Hp1α) (29).
Established leukemia stem cells exhibit induction of similar oncogenic pathways
To assess the requirement for self-renewal and transformation pathways during leukemic evolution, we next wanted to compare our immediate “preleukemic” signatures with the gene expression pattern in established murine leukemia stem cells. We have previously demonstrated that the GMP compartment is the earliest phenotypic population present in MOZ-TIF2 associated leukemias (11). In addition, this same phenotypic population was highly enriched for LSC activity in an MLL fusion model (12). However, a report from another MLL fusion model of AML has suggested that lineage positive leukemia cells also retain significant LSC potential (30). To allow for either possibility, we tested the potential of bulk leukemic cells (EGFP+), a leukemic population with a “mature” surface phenotype (EGFP+, Gr-1+/Mac1+ population), and an “immature” leukemic population with the surface phenotype of GMP (EGFP+/Lin-/c-Kit +/Sca-1 -/CD34+/FcRγ+; L-GMP) to transfer MOZ-TIF2-associated leukemias at limiting dilution (Figure 3). Mice developed phenotypically identical leukemias (data not shown), but at incidences that varied dramatically according to the transplanted population. Our results demonstrated a hierarchy in MOZ-TIF2-associated leukemias, and consistent with normal ontogeny we found the L-GMP population to be at the apex of leukemic differentiation and to be highly enriched for LSC potential (approximately 166 and 3300 fold enrichment over bulk leukemic cells and mature leukemic cells respectively, with an LSC frequency of ~1/300 cells, Figure 3A).
Figure 3. MOZ-TIF2-associated murine leukemias recapitulate an LSC hierarchy and demonstrate the evolutionary nature of LSC transcriptional programs.
A) The experimental schema used to establish an LSC hierarchy for MOZ-TIF2-associated murine leukemias. Briefly, the efficiency with which leukemia was transferred to sublethally irradiated syngeneic recipients was assessed at limiting dilution as is shown for three separate populations. These were defined according to normal myeloid ontogeny and comprised a “mature” population (left panels), a “bulk” leukemia population (middle panels) and an “immature” population with the same surface phenotype as GMP (right panels). The efficiency of transfer and leukemia stem cell frequency as calculated by Poisson statistics for each population is shown below the Kaplan-Meier graphs in the lower row of panels. Greater than 3 log enrichment for leukemia stem cell frequency was demonstrated between the immature “leukemic” GMP (L-GMP) and “mature” (GFP+, Mac+ and Gr1+) populations. B) Gene expression profiles from the L-GMP population, significantly enriched for LSC activity, were then compared with their normal GMP counterpart. Expression data for a representative 20 Probes (19 genes) are shown. This demonstrates that certain genes, such as Bmi1, Meis1, Sox4, Tcf4, Hoxa9 and Smad7 remain significantly upregulated between our immediate signatures and established leukemia stem cells. However, in addition, a number of new genes widely implicated in leukemogenesis become upregulated during leukemic evolution, including Hoxa10, Hoxa7, Runx1, Lmo2 and Ctnna1.
To assess the evolutionary nature of transcriptional programs critical for leukemia induction and maintenance, the gene expression profiles of replicate L-GMP were then compared to their normal phenotypic counterpart. 2715 genes were differentially expressed between normal and leukemic GMP (p<0.05). Similarities were seen between our immediate preleukemic signatures and the gene expression profiles in the overt leukemias. The overlap with the leukemia initiation and leukemia self-renewal signatures was highly significant at 59/167 genes (35%) and 29/91 genes (32%), respectively (both p<0.0001). Overlapping genes included Bmi1, Meis1, Sox4, Tcf4, HoxA9 and Smad7. However, new genes, not present in our preleukemic signatures, but widely implicated in leukemogenesis were also significantly upregulated, including Hoxa10, Hoxa7, Runx1, Lmo2 and Ctnna1 (Figure 3B). Therefore, although there are similarities to the genetic programs that establish leukemia stem cells, additional programs, possibly the result of later co-operating mutations, may be required for the further evolution and maintenance of LSC during leukemogenesis.
Preleukemic signatures predict the biology of human AML
Significant differences have been reported in mechanisms of transformation between mice and humans and our signatures have been generated following retroviral overexpression of oncogenes. Therefore, to validate our genetic programs in human AML, we compared our leukemia initiation and leukemia self-renewal genesets across bulk gene expression profiles from 253 unselected cases of AML. These cases represented a wide variety of cytogenetic and genetic mutations and were treated uniformly in the Austrian-German AML studies (Table S7). In comparing the gene expression from this dataset to our own signatures, probes/genes which were not represented on both the human and mouse arrays, or which were not of sufficient quality in the human analyses were excluded. This allowed a direct comparison of 84/167 genes in the leukemia initiation signature and 61/91 genes in the leukemia self-renewal signature. For the leukemia initiation signature 67/84 genes (80%, represented by 109 probesets) were differentially expressed and for the leukemia self-renewal signature 31/61 genes (51%, represented by 37 probesets) were differentially expressed in the human AML cohort.
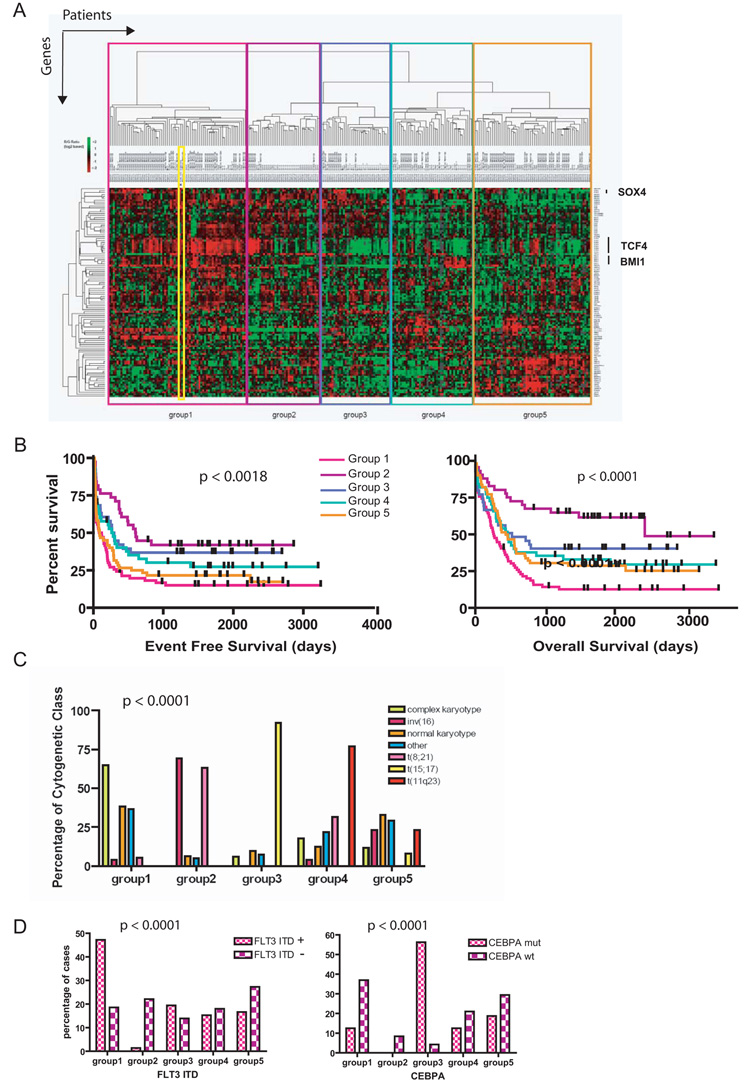
Utilizing the leukemia initiation geneset, we next performed unsupervised hierarchical clustering of the human AML samples. This classified the cohort into five different patient groups (Figure 4A), with this association shown to be stable following iterative consensus clustering. These patient groups differed significantly in known prognostic characteristics (karyotype p < 0.0001, mutational status for molecular lesions such as FLT3-ITD, CEBPA and NPM1c, all p < 0.0001, age, p = 0.005 and white cell count at diagnosis, p = 0.0035, Figure 4C + D and Figure S1) and in survival (both event free p < 0.0018 and overall survival p < 0.0001) (Figure 4B). Although genes from our signature were differentially expressed across all patient groups, the leukemia initiation signature was representative of patient group 1 (containing 71 patients), within which it tightly associated when included in the clustering analysis (Figure 4A). It was noted that group 1 had the worst prognosis both for overall and event free survival (Figure 4B). In this respect, the poor prognosis for group 1 patients may be explained by a direct correlation with known poor prognostic determinants (an increased incidence of complex karyotype and the presence of FLT3-ITD mutations) and an inverse correlation with good prognostic determinants (a decreased incidence of rearrangements of the core-binding factor subunits, retinoic acid receptor alpha and CEBPA mutations) (Figure 4 C and D and Figure S1). In addition, when our geneset was used to classify an independently published dataset of 283 patients (4), similar associations between the leukemia initiation geneset and cytogenetic, molecular genetic and survival findings were demonstrated (Figure S2). Moreover, an association was demonstrated between our geneset and expression of EVI1, a transcriptional repressor whose expression associates with a poor prognosis in AML patients within this dataset (4).
Figure 4. The immediate leukemia initiation signature is present across many human AML samples and predicts for survival and disease biology.
A) Sixty-seven/84 genes (80%) from the immediate leukemia initiation signature were differentially expressed across a cohort of 253 AML patients as shown in the heatmap. The geneset was able to classify this cohort into five groups using unsupervised clustering. The immediate leukemia initiation signature (outlined in the yellow box) is representative of, and tightly segregates with, group 1 when included in this analysis. B) Significant differences in survival for the five groups are demonstrated in this Kaplan-Meier estimation of event-free (p < 0.0018, left panel) and overall survival (p < 0.0001, right panel), with group 1 associated with the poorest prognosis. C) Significant differences in cytogenetic characteristics were demonstrated for individuals in each of the groups (p < 0.0001), with group 1 patients associated with a complex karyotype (poor prognosis) and a lack of rearrangements of core-binding factors and the retinoic acid receptor alpha (RARA) (good prognosis). D) Significant differences in molecular prognostic factors such as mutational status for FLT3-ITD (left panel) and CEBPA (right panel) were also noted (each p < 0.0001). Patients in group 1 demonstrated an increased incidence of the poor prognosis FLT3-ITD mutations and a lower incidence of good prognosis CEBPA mutations (see also Figures S1, 2 and 3 and supplementary Table 3).
Using our leukemia self-renewal geneset to interrogate the initial AML patient cohort, we found similar associations. Using unsupervised clustering, the self-renewal geneset again grouped the cohort into five groups (Figure S3). Similarly, these patient groups differed significantly in survival and known prognostic characteristics. On this occasion our signature tightly clustered with group E (comprising 68 patients). This group again was enriched for poor prognosis patients, and correlated with an increased incidence of poor-risk cytogenetics, although not on this occasion with FLT3-ITD status (Figure S3).
Candidate genes alter self-renewal properties in vitro and generate AML in vivo
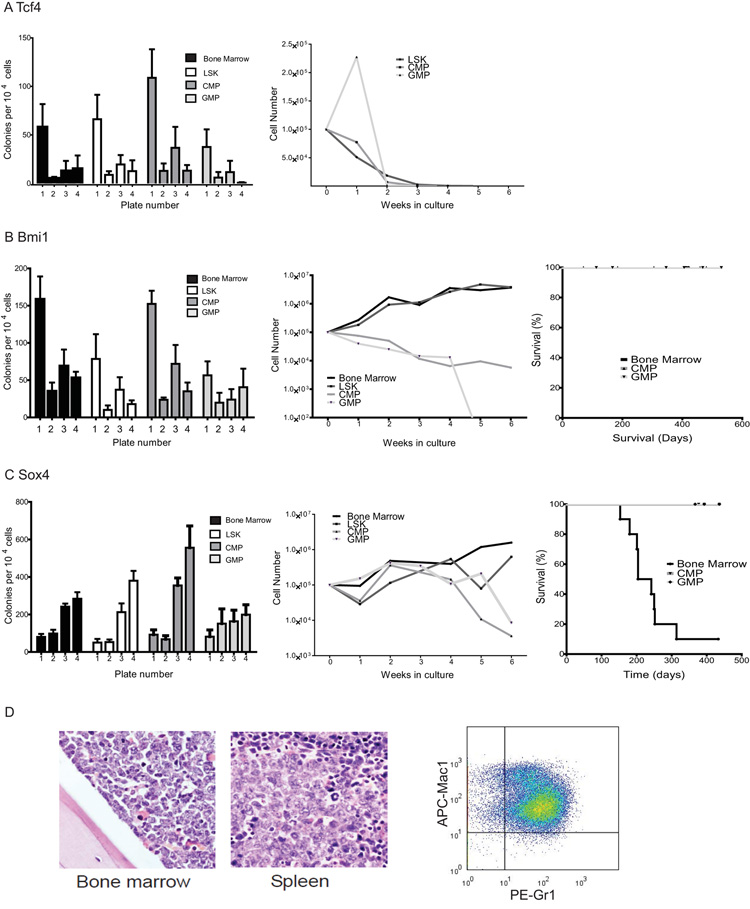
Finally, to provide proof-of-principle that the genes identified in our signatures could alter self-renewal and mediate transformation, three genes from the leukemia initiation signature that were present in mouse and human comparisons were chosen; Tcf4, Bmi1, and Sox4. These genes were assessed for their ability to phenocopy the leukemia-associated fusions in serial replating and transplantation assays. Transcription factor 4, Tcf4, is an E box-binding helix-loop-helix transcription factor associated with the mental retardation Pitt-Hopkins syndrome (31), Bmi1, is a component of the Polycomb PRC1 complex that has been implicated in normal hematopoietic, neural and leukemic stem cell function (27) and Sox4 is an HMG box transcription factor involved in cardiac and lymphoid development and has been described as a retroviral integration site in murine leukemias (32, 33).
No serial replating was seen in LSK cells or whole bone marrow transduced with Tcf4 (Figure 5 A). However, for Bmi1 or Sox4 we demonstrated that retroviral expression could partially phenocopy the effects of fusion oncogene expression and alter the in vitro self-renewal potential of committed progenitors in serial replating assays (Figure 5 B + C). In addition, LSK cells and whole bone marrow transduced with either Sox4 or Bmi1 could grow in cytokine supplemented liquid culture, although CMP and GMP transduced with either gene only grew for a limited period of 4 weeks in culture. Finally, although Bmi1 transduced hematopoietic tissue and Sox4 transduced progenitors did not generate disease, recipient mice transplanted with Sox4 transduced whole bone marrow developed AML and died from AML with a median latency of 28 weeks (Figure 5 D).
Figure 5. Individual genes in the leukemia inititation signature partially phenocopy AML-associated fusion genes.
A) Tcf4 did not alter the properties of bone marrow, LSK or progenitors in methylcellulose (left panel) and Tcf4 cells failed to grow in liquid culture (right panel). B) The left panel demonstrates that whole bone marrow (BM), LSK and myeloid progenitors (CMP and GMP) transduced with Bmi1 serially replate in methylcellose cultures. Both whole BM and LSK transduced with Bmi1 continue to grow in cytokine supplemented liquid culture, while growth of CMP and GMP populations is limited to 4 weeks in culture before their involution (middle panel). The right panel demonstrates that whole bone marrow or progenitors transduced with Bmi1 did not generate AML following transplantation. C) The left and middle panels demonstrate similar findings for stem and progenitor populations transduced with Sox4. In addition, BM transduced with Sox4 generated leukemia with a median latency of 28 weeks (right panel). D) Representative photomicrographs of bone marrow and spleen (left and middle panels respectively) from animals with Sox4-associated AML demonstrate heavy infiltration with immature blasts. Representative flow of the GFP positive cells from these leukemias (right panel) demonstrates a Gr1intermediate/ Mac1intermediate myeloid phenotype.
Discussion
Our data demonstrate an alteration of self-renewal in murine stem and progenitor populations following expression of AML1-ETO and NUP98-HOXA9. A reductionist view of the pathogenesis of AML proposes that its development minimally requires cooperation between type I mutations which alter cellular proliferation and survival (such as FLT3-ITD or c-KIT mutations) and type II mutations which block myeloid differentiation (of which AML1-ETO and NUP98-HOXA9 would be representative examples). A similar ability to alter self-renewal in committed progenitors has been demonstrated by ourselves and others for MLL-fusions, MOZ-TIF2 and CEBP-α mutants (10-12, 34). Taken together, these data strongly link self-renewal and transformation in AML and suggest that an ability to restore self-renewal or to augment existing self-renewal is a more generalized property of these type II mutations. However, our data also demonstrate a differential ability to alter self-renewal in vivo between AML1-ETO and NUP98-HOXA9, with only the latter able to generate AML. As both fusion oncogenes are thought to be initiating events in leukemogenesis (35, 36), our data therefore suggest that the potential cell(s) of origin of the LSCs that propagate the disease may differ according to the properties of the initiating lesion. This may be one explanation for the growing demonstration of heterogeneity with the LSC compartment (37). Furthermore, it also suggests an association between altered self-renewal and aggressive disease, corroborating recent functional findings in murine xenotransplant experiments where levels of engraftment from individual patients correlate with their disease outcome (38).
Global gene expression profiles of bulk blasts from AML patients have greatly informed the study of AML biology, and have been particularly useful in the classification of specific prognostic groups, detailing that many individual genetic lesions are associated with a characteristic gene expression signature (3,4). However, data demonstrating whether the induction of common, overlapping, or unique pathogenetic pathways occurs downstream of multiple mutations in AML are generally lacking. In this report we demonstrate that three disparate but functionally related AML-associated fusion oncogenes cause similar differential expression of common and overlapping gene programs following short-term expression in the murine hematopoietic progenitor compartment. These same pathways were aberrantly regulated in populations enriched for established and functionally validated LSC, although further genes and pathways associated with leukemogenesis were also perturbed. Taken together, these data demonstrate the evolutionary nature of transcriptional programs during leukemogenesis, suggesting that additional programs, possibly the result of later co-operating mutations (39), are required for the further evolution and maintenance of LSC during leukemogenesis.
Comparisons based on differences in the in vitro and in vivo properties conferred by the three oncogenes allowed us to define mutually exclusive leukemia initiation and leukemia self-renewal signatures. Similar signatures defining self-renewal and the maintenance of the LSC have been published for MLL fusion proteins (12, 29). When we compared our leukemia initiation and leukemia self-renewal signatures with those of Krivtsov and Somervaille we found only modest overlap (Figure S4). This corroborates the findings in bulk human AML blasts, where the global gene expression profiles of MLL rearranged and MLL germ line leukemias were seen to segregate (40). However, these small overlaps were greatly enriched for genes demonstrated to alter self-renewal and/or to be critical for AML pathogenesis, and included genes such as Hoxa9 (28), Meis1 (26), Mef2C (12), Myb (41) and Cbx5 (29) (Figure S4 A and B). This is a further demonstration that common pathways are critical effectors of leukemogenesis in subsets of AML patients.
The genesets which comprised our signatures were also able to classify two independent datasets of global gene expression profiles from large cohorts of AML patients. Patients were assigned into groups which significantly differed in existing prognostic characteristics such as patient age, presenting white blood cell count, karyotype and mutation status for critical genes such as FLT3-ITD and CEBPA. Although the majority of genes within our signatures were differentially expressed across all groups, our signatures were most representative of specific groups with a particularly poor prognosis. This probably reflects that patients within these groups demonstrated a decreased incidence of good risk characteristics such as rearrangement of core binding factors and RARA and an increased frequency of known poor prognostic factors such as a complex karyotype, FLT3-ITD and overexpression of EVI1. As such our signatures and genes within them may represent biological effectors downstream of these poor-risk mutations. Furthermore, as long-term survival in patients with poor risk factors may be as low as 10% (13), the identification of downstream pathways and potential targets may inform therapeutic design to improve the dismal current outcomes in this group.
Candidates identified in our common signatures are known to alter self-renewal in hematopoietic cells and to critically mediate leukemogenesis. From our leukemia self-renewal signature Hoxa9 and Myb have been demonstrated to be critical for self-renewal of normal and leukemic stem cells (28,29,41,42), whilst Cbx5 is necessary for MLL fusion LSC maintenance (29). From our leukemia initiation signature, identified genes such as Mef2c, Meis1 and Tal1 have demonstrable roles in leukemia stem cells (12,26). As further proof-of-principle, we validated another two candidates chosen from our leukemia initiation signature to at least partially phenocopy the original leukemia-associated oncogenes. The polycomb group gene Bmi1 has been demonstrated to be requirement for both normal and leukemic stem cell function (27) and was upregulated following overexpression of both MOZ-TIF2 and AML1-ETO (Table S5). Whilst its overexpression as a solitary abnormality was insufficient to generate AML in vivo, it did alter self-renewal properties of hematopoietic stem and progenitor cells in vitro. Sox4 was commonly upregulated downstream of MOZ-TIF2, AML1-ETO and NUP98-HOXA9 (Table S5) and is an HMG box transcription factor involved in cardiac and lymphoid development (32). It has previously been implicated in murine leukemogenesis (33) and is a homologue of Sox2, a critical mediator of the cellular reprogramming of induced pluripotent stem cells (43). Importantly, although it may augment their self-renewal properties upon overexpression (44), Sox4 appears to be dispensable for normal HSC function (32). We demonstrated further that SOX4 expression is altered in human AML, and that overexpression of Sox4 can alter self-renewal properties in hematopoietic stem and progenitor cells in vitro and can generate AML in vivo. Taken as a whole, these cross-species findings validate common and overlapping genes and transcriptional programs downstream of oncogenic transcriptional fusion genes to alter self-renewal properties and contribute to the evolution of AML.
Supplementary Material
Acknowledgements
We would like to thank other members of the authors’ laboratories for helpful discussions, particularly Andre Krivtsov and Zhaohui Feng. Work in the authors’ labs was supported by the MRC (UK), CRUK, the Leukemia and Lymphoma Society of America, the NIHR Cambridge Biomedical Research Centre (BH) and by the Deutsche José Carreras Stiftung e.V. (DJCLS R 08/32f) (LB). BH is supported by an MRC (UK) Senior Clinical Research Fellowship award and the EHA-Jose Carreras Young Investigator prize.
Footnotes
Disclosure of Conflicts of Interest
The authors disclose no conflicts of interest.
References
- 1.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 2.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 4.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg R. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 7.Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5:311–321. doi: 10.1038/nrc1592. [DOI] [PubMed] [Google Scholar]
- 8.Reya T, Morrison S, Clarke M, Weissman I. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 9.Yilmaz OH, Valdez R, Theisen BK, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 10.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huntly BJ, Shigematsu H, Deguchi K, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 13.Grimwade D, Hills RK, Moorman AV, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities amongst 5,876 younger adult patients treated in the UK Medical Research Council trials. Blood. 2010 doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 14.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 15.Chou WC, Chen CY, Hou HA, et al. Acute myeloid leukemia bearing t(7;11)(p15;p15) is a distinct cytogenetic entity with poor outcome and a distinct mutation profile: comparative analysis of 493 adult patients. Leukemia. 2009;23:1303–1310. doi: 10.1038/leu.2009.25. [DOI] [PubMed] [Google Scholar]
- 16.Deguchi K, Ayton PM, Carapeti M, et al. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3:259–271. doi: 10.1016/s1535-6108(03)00051-5. [DOI] [PubMed] [Google Scholar]
- 17.Dash AB, Williams IR, Kutok JL, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci U S A. 2002;99:7622–7627. doi: 10.1073/pnas.102583199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly LM, Liu Q, Kutok JL, Williams IR, Boulton CL, Gilliland DG. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 2002;99:310–318. doi: 10.1182/blood.v99.1.310. [DOI] [PubMed] [Google Scholar]
- 19.Nakorn T, Traver D, Weissman I, Akashi K. Myeloerythroid-restricted progenitors are sufficient to confer radioprotection and provide the majority of day 8 CFU-S. Journal of Clinical Investigation. 2002;109:1579–1585. doi: 10.1172/JCI15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Iwasaki H, Krivtsov A, et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. Embo J. 2005;24:368–381. doi: 10.1038/sj.emboj.7600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 23.Cheng J, Sun S, Tracy A, et al. NetAffx Gene Ontology Mining Tool: a visual approach for microarray data analysis. Bioinformatics. 2004;20:1462–1463. doi: 10.1093/bioinformatics/bth087. [DOI] [PubMed] [Google Scholar]
- 24.Higuchi M, O’Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 25.Tothova Z, Kollipara R, Huntly BJ, et al. FoxOs Are Critical Mediators of Hematopoietic Stem Cell Resistance to Physiologic Oxidative Stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 28.Faber J, Krivtsov AV, Stubbs MC, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somervaille TC, Matheny CJ, Spencer GJ, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Brockschmidt A, Todt U, Ryu S, et al. Severe mental retardation with breathing abnormalities (Pitt-Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Hum Mol Genet. 2007;16:1488–1494. doi: 10.1093/hmg/ddm099. [DOI] [PubMed] [Google Scholar]
- 32.Schilham MW, Oosterwegel MA, Moerer P, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 33.Boyd KE, Xiao YY, Fan K, et al. Sox4 cooperates with Evi1 in AKXD-23 myeloid tumors via transactivation of proviral LTR. Blood. 2006;107:733–741. doi: 10.1182/blood-2003-05-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirstetter P, Schuster MB, Bereshchenko O, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Miyamoto T, Weissman I, Akashi K. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci U S A. 2000;97:7521–7526. doi: 10.1073/pnas.97.13.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore MA, Chung KY, Plasilova M, et al. NUP98 dysregulation in myeloid leukemogenesis. Ann N Y Acad Sci. 2007;1106:114–142. doi: 10.1196/annals.1392.019. [DOI] [PubMed] [Google Scholar]
- 37.Taussig DC, Miraki-Moud F, Anjos-Afonso F, et al. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood. 2008;112:568–575. doi: 10.1182/blood-2007-10-118331. [DOI] [PubMed] [Google Scholar]
- 38.Pearce DJ, Taussig D, Zibara K, et al. AML engraftment in the NOD/SCID assay reflects the outcome of AML: implications for our understanding of the heterogeneity of AML. Blood. 2006;107:1166–1173. doi: 10.1182/blood-2005-06-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilliland DG, Tallman MS. Focus on acute leukemias. Cancer Cell. 2002;1:417–420. doi: 10.1016/s1535-6108(02)00081-8. [DOI] [PubMed] [Google Scholar]
- 40.Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 41.Hess JL, Bittner CB, Zeisig DT, et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence HJ, Christensen J, Fong S, et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Deneault E, Cellot S, Faubert A, et al. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 2009;137:369–379. doi: 10.1016/j.cell.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.