Abstract
Nucleic acids containing stretches of tandem guanines can fold into four-stranded structures called G-quadruplexes. The existence of such sequences in genomic DNA suggests the occurrence of these motifs in cells, with potential implications in a number of biological processes relevant to cancer. Small molecules have proven to be valuable tools to dissect cell circuitry. Here, we describe a synthetic small molecule derived from an N,N’-bis(2-quinolinyl)pyridine-2,6-dicarboxamide, which is designed to mediate the selective isolation of G-quadruplex nucleic acids. The methodology was successfully applied to a range of DNA and RNA G-quadruplexes in vitro. We demonstrate the general applicability of the method by isolating telomeric DNA-containing G-quadruplex motifs from cells. We show that telomeres are targets for the probe, providing further evidence of the formation of G-quadruplexes in human cells.
Natural products and related small-molecule affinity-matrices have been used extensively to isolate proteins from cells for the purpose of target discovery1. In pioneering work, Dervan and colleagues created synthetic small molecules with the ability to recognize particular DNA sequences in a programmed fashion with high selectivity2. To the best of our knowledge, there is no report on the use of small organic molecules to recognize and isolate a nucleic acid element in a structure-dependent manner. Several articles have appeared in the literature, suggesting that G-quadruplex nucleic acids may be involved in gene expression3,4, telomere maintenance5 and replication stalling6. Computational analyses have suggested that putative G-quadruplex-forming sequences are prevalent in the human genome7. Although the existence of such structures in vitro is clear, their formation in human cells remains elusive. The development of a small-molecule probe designed to mediate the isolation of G-quadruplex motifs from genomic DNA is needed to establish a protocol conceptually similar to chromatin immunoprecipitation-sequencing8,9. Massively parallel sequencing10 of isolated DNA fragments has the potential to reveal G-quadruplexes that physically fold within a cellular context and inevitably provides additional insights into G-quadruplex biological functions.
We previously described pyridostatin (1)11 (Fig. 1; also known as RR8212), which induces telomere dysfunction and triggers the activation of a DNA damage response13, an effect that we had linked to the stabilization of G-quadruplexes. Here, we describe a novel small molecule derived from 1 that enables the selective isolation of G-quadruplex motifs from human cells. Several structural and functional features were considered for the design of such a probe, including (1) the presence of an affinity-tag to pull down DNA fragments, (2) the ability to recognize various G-quadruplex structures with a high level of specificity over double-stranded DNA (ds-DNA), (3) the capacity to release intact DNA for subsequent analysis by specific polymerase chain reaction (PCR) or sequencing, and (4) a detectable biological activity that can be related to the cellular target of interest (for example, G-quadruplexes).
Figure 1. Molecular structures of 1 and 2.
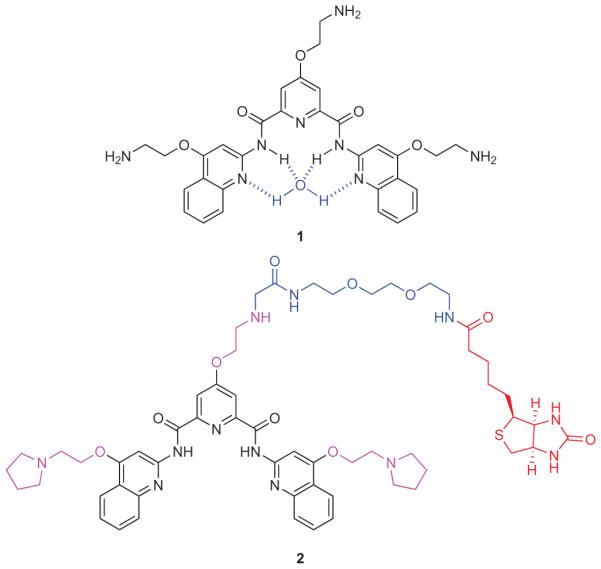
Pyridostatin 1 (black) coordinated with a molecule of water (blue). Small-molecule probe 2 containing an electron-rich aromatic surface (black), water-soluble alkoxy-side appendages (purple), a biotin affinity-tag (red) and a hydrophilic linker (blue).
The successful design of pyridostatin prompted us to synthesize a probe based on an N,N’-bis(2-quinolinyl)pyridine-2,6-dicarboxamide scaffold containing an affinity-tag. To obtain optimal stacking interactions14 with G-quartets, we implemented structural features shared by other potent G-quadruplex-binding small molecules, with particular emphasis on (1) an electron-rich aromatic surface provided by alkoxy-side appendages and (2) the potential to adopt a functional planar conformation15 locked by the internal hydrogen-bonding network16, and a molecule of water potentially coordinated in the centre with available nitrogen lone pairs (Fig. 1)17.
We prepared a small library of biotinylated analogues derived from 1, where structural diversity arose from the nature of the biotin-linker, different side-chain lengths and functionalities. Compound 2 (Fig. 1), which contains a hydrophilic polyethylene glycol linked to the central pyridine core, was chosen for its enhanced water solubility properties and tractable synthesis (Supplementary Information). Furthermore, preliminary studies using 2 with synthetic DNA demonstrated the potential to stabilize and isolate G-quadruplex DNA motifs, which has encouraged the investigation described herein.
Results and discussion
We first evaluated the potential of 2 to selectively stabilize a series of structurally distinct DNA G-quadruplexes as compared with ds-DNA by Förster resonance energy transfer melting experiments (Supplementary Fig. S2). Compound 2 was found to significantly increase the thermal stability of the human telomeric G-quadruplex (H-telo)18 and various promoter G-quadruplexes such as C-myc (ref. 4), C-kit1 (ref. 19) and C-kit2 (ref. 20) but not of ds-DNA. For example, 2 induced an increase in the melting temperature (ΔTm) of H-telo by 30.1 K at 1 μM, and by 27.8 K in the presence of 100 mole equivalents of unlabelled ds-DNA competitor. At this concentration, no apparent stabilization of a dual-labelled ds-DNA oligonucleotide was observed. The presence of up to 100 mole equivalents of unlabelled ds-DNA competitor hardly altered the melting profile of the G-quadruplex studied. This is characteristic of a high specificity of 2 towards these motifs, further demonstrating that G-quadruplex structure stabilization is a general feature of 2. In contrast, a ΔTm of 35 K was recorded at 0.2 μM of 1, and remained unaffected by the presence of 100 mole equivalents of ds-DNA competitor11.
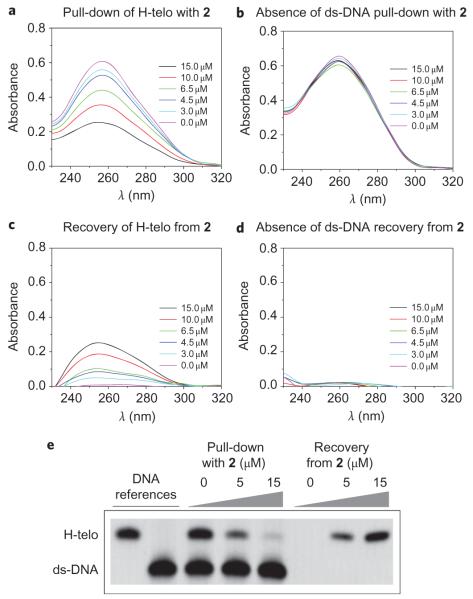
The introduction of a biotin affinity-tag partially altered the potential of 2 to stabilize G-quadruplex motifs compared with pyridostatin, but did not affect its specificity towards G-quadruplexes over ds-DNA. We next explored the aptitude of 2 to selectively pull down G-quadruplex motifs from solution. Nucleic acid solutions containing either H-telo or ds-DNA were incubated with different amounts of 2 and streptavidin-coated magnetic beads. The supernatants were collected and subjected to UV absorbance measurements to measure the amount of residual DNA. Increasing the concentration of 2 resulted in a reduction in absorbance at ~260 nm, indicative of H-telo being pulled down from solution (Fig. 2a). The same protocol was successfully applied to other DNA and RNA G-quadruplexes, including C-myc (ref. 4), C-kit1 (ref. 19), C-kit2 (ref. 20) and N-ras (ref. 21) (Supplementary Fig. S3). In comparison, it was not possible to pull down either a control ds-DNA (Fig. 2b) or a short-hairpin RNA. We then investigated whether it was possible to recover G-quadruplex nucleic acids from beads, a critical step that renders the methodology applicable for future sequencing. After removal of the supernatants, the beads were re-suspended in denaturing buffer and heated. The solutions were separated from the beads and analysed by UV. The amplitudes of recovered H-telo signals were higher for samples obtained from beads with a higher loading of 2 (Fig. 2c). However, no UV signal was observed for ds-DNA, supporting the fact that ds-DNA had not been pulled down (Fig. 2d). Heat and molar concentrations of denaturing agents such as lithium salts or urea were required to unfold and recover the oligonucleotides from beads, characteristic of a strong interaction of 2 with G-quadruplexes.
Figure 2. Molecular probe 2 mediates selective G-quadruplex isolation from solution.
a, UV absorbance measurements of remaining DNA in supernatants after pull-down carried out on 5 μM H-telo (5′-(G3TTA)3G3-3′). The decrease in UV absorbance with increasing amounts of 2 shows that the G-quadruplex containing oligonucleotide has been pulled down from solution. b, UV absorbance measurements of remaining DNA in supernatants after pull-down carried out on 5 μM ds-DNA (5′-CAATCGGATCGAATTCGATCCGATTG-3′). The constant amplitude of UV signals demonstrates that 2 is unable to pull down ds-DNA. c, UV absorbance measurements of solutions obtained from 5 μM H-telo containing samples after recovery from beads. The increase in UV absorbance shows that H-telo was loaded on beads after pull-down and was released in solution after urea treatment. d, Residual UV absorbance of solutions obtained from 5 μM ds-DNA containing samples after urea treatment. The absence of signal shows that ds-DNA was not loaded on beads, consistent with the absence of ds-DNA pull-down. e, Gel demonstrating the selective G-quadruplex pull-down and recovery of intact H-telo after urea treatment, and the absence of ds-DNA pull-down. The experiment was performed on a 2.5 μM : 2.5 μM mixture of H-telo : ds-DNA.
To confirm that the protocol was not detrimental to DNA, we performed pull-down experiments on H-telo: ds-DNA mixtures that were monitored by gel electrophoresis. For supernatants obtained after pull-down, the intensity of the band corresponding to H-telo decreased in a dose-dependent manner with increasing amounts of 2, as opposed to the ds-DNA band intensity, which remained unaffected (Fig. 2e). For solutions obtained after DNA recovery, bands corresponding to H-telo were restored with intensities that increased in a dose-dependent manner with increasing amounts of 2, whereas no band was visible for ds-DNA. This shows that recovery of H-telo from beads is achievable without altering DNA integrity, and confirms that 2 is unable to pull down ds-DNA. A similar protocol was applied to other promoter G-quadruplexes, demonstrating that the molecule operates in a structure-specific as opposed to a sequence-specific manner (Supplementary Fig. S4). We successfully applied this methodology to pull down over 100-mer-long oligonucleotides containing a G-quadruplex-forming sequence, indicating that it can be used to pull down longer genomic DNA fragments (Supplementary Table S1 and Figs S5,S6). In addition, a 101-mer containing the C-myc G-quadruplex motif was isolated in the presence of its complementary strand, suggesting that the motif is in dynamic equilibrium that can be driven towards G-quadruplex folding on the addition of 2, as previously observed for other small molecules22. Moreover, the introduction of mutations that prohibit G-quadruplex formation abrogated DNA pull-down, highlighting the structural requirement for pull-down to occur.
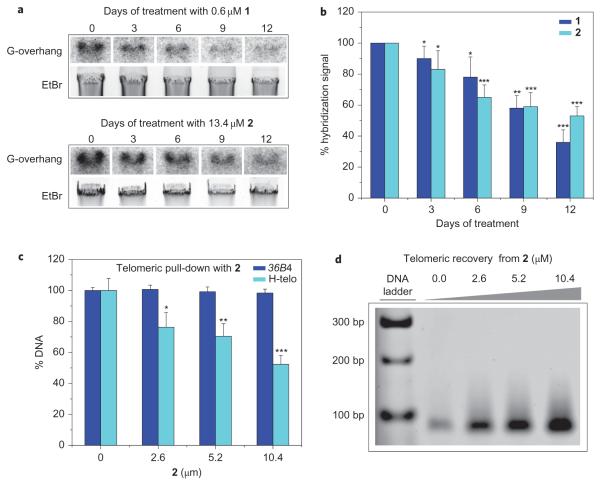
To ascertain that the biotin–linker moiety did not impair the biological properties of 2, we evaluated the growth inhibitory properties of pyridostatin and 2 towards HT1080 human cancer cells. After 72 h of incubation, 1 and 2 induced the inhibition of cell growth with respective IC50 values of 0.6 and 13.4 μM. No cell death occurred at these concentrations. Furthermore, time-dependent G-overhang shortening23 was observed (Fig. 3a,b), accompanied by long-term growth arrest and β-galactosidase activity characteristic of replicative senescence24 (Supplementary Figs S7–S9). The increase in the IC50 value measured for 2 may be the result of a decrease in its G-quadruplex stabilization properties compared with 1 or an alteration of its cellular uptake and metabolism because of the presence of the biotin–linker moiety. Nevertheless, the conservation of growth inhibitory properties and the alteration of telomere length make 2 a suitable molecular probe25 to study G-quadruplexes in cells.
Figure 3. Molecular probe 2 induces in cellulo G-overhang shortening and mediates G-quadruplex-containing telomeric fragment isolation from human cells.
a, Telomeric G-overhang erosion in HT1080 cells on incubation with 1 and 2, as observed by hybridization of a radio-labelled telomeric probe with genomic DNA normalized against ethidium bromide (EtBr) signals. This suggests a direct interaction of the small molecules with telomeric G-quadruplexes. b, Histogram showing the time-course hybridization signal decrease on incubation of HT1080 cells with 1 and 2 (n = 4). c, Percentage of the remaining telomeric sequence and 36B4 control gene in supernatants after pull-down performed on 0.52 μM genomic DNA (n = 5). This shows that 2 interacts selectively with G-quadruplex motifs containing human DNA. d, PCR amplification products of telomeric sequences after DNA recovery from 2, demonstrating that 2 mediates the selective pull-down and release of G-quadruplex-containing DNA. This establishes a direct correlation between G-quadruplex motifs existing at human telomeres and the G-overhang shortening induced by 2. s.d. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.0001.
We next applied the methodology to the isolation of telomeric DNA from HT1080 cells to establish a correlation with the phenotype induced by 2 and to validate telomeric G-quadruplexes as a cellular target for 2. The small molecule was incubated with human genomic DNA obtained from HT1080 cells. Before the incubation with 2 and streptavidin-coated beads, DNA was subjected to shearing, typically producing 100–300 bp fragments suitable for pull-down (Supplementary Fig. S10). Specific quantitative PCR (qPCR) amplification was performed on the supernatant with primers designed for telomeric DNA and for a control gene containing no putative G-quadruplex sequence26. Comparative analysis of the percentage of remaining DNA before and after pull-down showed a decrease in the total amount of telomeric DNA in a dose-dependent manner, whereas the quantity of control gene remained unaffected, in agreement with a selective pull-down of telomeric fragments (Fig. 3c). Specific dose-dependent qPCR amplification performed on the DNA recovered from beads further demonstrated the presence of telomeric DNA in the pull-down extracts (Fig. 3d and Supplementary Fig. S11). It is possible that nucleic acid fragments comprising similar sequences found elsewhere in the genome may have also been pulled down.
These results indicate that 2 is capable of selective recognition and stabilization of G-quadruplex-containing nucleic acids, allowing their isolation from human cells. Specific G-quadruplex antibodies have previously been used to demonstrate the existence of G-quadruplexes at telomeres of Stylonychia lemnae, giving a rationale underlying their putative biological function27. Here, we have shown that human telomeric DNA is a molecular target for 2, providing further evidence of the formation of G-quadruplexes at human telomeres and their implication in telomere maintenance. The small-molecule affinitytag has been used to selectively isolate a nucleic acid element in a structure-dependent manner. The implication of this novel concept may be extended to explore other putative G-quadruplex sequences in the human genome and will be used to provide insights into G-quadruplex functions in a cellular context.
Methods
Protocol for selective pull-down and recovery of G-quadruplex nucleic acids
Oligonucleotide stock solutions of 100 μM were prepared using molecular biology grade RNase-free water. Further dilutions were carried out in annealing buffer containing 10 mM Tris·HCl (pH 7.4) and 60 mM potassium chloride. Samples were annealed by heating at 94 °C for 10 min followed by slow cooling to room temperature at a controlled rate of 0.1 °C min−1.
In a typical pull-down experiment involving synthetic oligonucleotides, 100 μl of oligonucleotide at 5 μM was incubated with a defined quantity of 2 (0, 3.0, 4.5, 6.5, 10.0 and 15.0 μM) for 30 min at room temperature. Similarly, in experiments involving G-quadruplex DNA–ds-DNA mixtures, 50 μl of a 5 μM ds-DNA solution was added to 50 μl of a 5 μM G-quadruplex DNA solution, and the mixture was incubated with a defined quantity of 2 (0, 3.0, 4.5, 6.5, 10.0 and 15.0 μM) for 30 min at room temperature. One vial of streptavidin-coated magnetic beads (Streptavidin MagneSphere paramagnetic particles, Promega), which had been washed twice with 300 μl of annealing buffer, was then incubated with the solution for 30 min at room temperature. The supernatant was separated from the beads by magnetic separation and analysed by UV or gel electrophoresis. In a typical recovery experiment, the beads collected after pull-down were washed twice with 100 μl of 10 mM Tris·HCl (pH 7.4). For experiments involving DNA, 100 μl of an 8 M aqueous solution of lithium chloride (LiCl) was added per vial of beads and incubated for 30 min at room temperature. The beads were heated for 5 min at 60 °C and the solution was separated from the beads by magnetic separation. Before analysis by gel electrophoresis, the supernatant was purified using Illustra MicroSpin G-25 columns to remove LiCl (GE-Healthcare). For experiments involving RNA, 100 μl of an 8 M aqueous urea solution was added to each vial of beads and incubated for 30 min at room temperature. The solution was separated by magnetic separation and analysed by UV without further purification.
In a typical pull-down experiment involving genomic DNA, 100 μl of sonicated DNA (0.52 μM annealed in annealing buffer) was incubated with a defined quantity of 2 (0, 2.6, 5.2 and 10.4 μM) for 2 h at room temperature. The solution was then incubated with one vial of streptavidin-coated magnetic beads, which had been washed twice with 300 μl of annealing buffer, for 1 h at room temperature. The supernatant was separated from the beads by magnetic separation, purified using the QIAquick Nucleotide Removal Kit (Qiagen), and subjected to specific qPCR amplification of telomere and control gene 36B4. Following pull-down, the collected beads were washed twice with 100 μl of 10 mM Tris·HCl (pH 7.4). Urea solution (100 μl, 8 M aqueous) was added to each vial of beads and the solution was incubated for 30 min at room temperature. Solutions were recovered by magnetic separation and purified using Illustra MicroSpin G-25 (GE-Healthcare) columns. Telomere-specific qPCR was performed on these solutions using the protocol described in the Supplementary Information.
Supplementary Material
Acknowledgements
The authors thank D.A. Sanders, R. Kranaster, J.–F. Riou, D. Gomez, A.R. Venkitaraman, A.J. Bannister and K. Holmes for fruitful discussions and advice. The authors also acknowledge Cancer Research UK for a studentship (S.M.) and programme funding, and the Biotechnology and Biological Sciences Research Council for project funding. R.R. is a Herchel Smith Research Fellow.
Footnotes
Additional information
The authors declare no competing financial interests.
Supplementary information and chemical compound information accompany this paper at www.nature.com/naturechemistry.
Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 2.Kielkopf CL, et al. A structural basis for recognition of A·T and T·A base pairs in the minor groove of B-DNA. Science. 1998;282:111–115. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]
- 3.Schaeffer C, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 6.Rizzo A, et al. Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signalling pathway. Nucleic Acids Res. 2009;37:5353–5364. doi: 10.1093/nar/gkp582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barski A, et al. High-resolution profiling of histone methylation in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 10.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez R, et al. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008;130:15758–15759. doi: 10.1021/ja805615w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buguat A, Rodriguez R, Kumari S, Hsu S-TD, Balasubramanian S. Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org. Biomol. Chem. 2010;8:2771–2776. doi: 10.1039/c002418j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.d’Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 14.Hunter CA, Sanders JKM. The nature of π-π interactions. J. Am. Chem. Soc. 1990;112:5525–5534. [Google Scholar]
- 15.Müller S, Pantoş GD, Rodriguez R, Balasubramanian S. Controlled-folding of a small molecule modulates DNA G-quadruplex recognition. Chem. Commun. 2009:80–82. doi: 10.1039/b816861j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berl B, Huc I, Khoury RG, Krische MJ, Lehn J-M. Interconversion of single and double helices formed from synthetic molecular strands. Nature. 2000;402:720–723. doi: 10.1038/35037545. [DOI] [PubMed] [Google Scholar]
- 17.Jain SL, et al. New pyridine carboxamide ligands and their complexation to copper(ii). X-ray crystal structure of mono-, di, tri- and tetranuclear copper complexes. Dalton Trans. 2004:862–871. doi: 10.1039/b316519a. [DOI] [PubMed] [Google Scholar]
- 18.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 19.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu S-TD, et al. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 2009;131:13399–13409. doi: 10.1021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nature Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez R, Pantoş GD, Gonçalves DPN, Sanders JKM, Balasubramanian S. Ligand-driven G-quadruplex conformational switching by using an unusual mode of interaction. Angew. Chem. Int. Ed. 2007;46:5405–5407. doi: 10.1002/anie.200605075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Temime-Smaali N, et al. The G-quadruplex ligand telomestatin impairs binding of topoisomerase IIIα to G-quadruplex-forming oligonucleotides and uncaps telomeres in ALT cells. PloS One. 2009;4:e6919. doi: 10.1371/journal.pone.0006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nature Rev. Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 25.Frye SV. The art of the chemical probe. Nature Chem. Biol. 2010;6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 26.Cawthon C. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomeres end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nature Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.