Abstract
The Baculoviridae is a large group of insect viruses containing circular double-stranded DNA genomes of 80 to 180 kbp. In this study, genome sequences from 57 baculoviruses were analyzed to reevaluate the number and identity of core genes and to understand the distribution of the remaining coding sequences. Thirty one core genes with orthologs in all genomes were identified along with other 895 genes differing in their degrees of representation among reported genomes. Many of these latter genes are common to well-defined lineages, whereas others are unique to one or a few of the viruses. Phylogenetic analyses based on core gene sequences and the gene composition of the genomes supported the current division of the Baculoviridae into 4 genera: Alphabaculovirus, Betabaculovirus, Gammabaculovirus, and Deltabaculovirus.
1. Background
Baculoviruses are arthropod-specific viruses containing large double-stranded circular DNA genomes of 80,000–180,000 bp. The progeny generation is biphasic, with two different phenotypes during virus infection: budded viruses (BVs), during the initial stage of the multiplication cycle, and occlusion-derived viruses (ODVs), at the final stages of replication [1, 2]. In general, primary infection takes place in the insect midgut cells after ingestion of occlusion bodies (OBs). Following this stage, systemic infection is caused by the initial BV progeny [3, 4]. And finally, OBs are produced during the last stage of the infection. These OBs comprise virions embedded in a protein matrix which protects them from the environment [5, 6].
Baculoviruses have been used extensively in many biological applications such as protein expression systems, models of genetic regulatory networks and genome evolution, putative nonhuman viral vectors for gene delivery, and biological control agents against insect pests [7–17].
The Baculoviridae family is divided into four genera according to common biological and structural characteristics: Alphabaculovirus, which includes lepidopteran-specific baculoviruses and is subdivided into Group I or Group II based on the type of fusogenic protein, Betabaculovirus, comprising lepidopteran-specific granuloviruses, Gammabaculovirus, which includes hymenopteran-specific baculoviruses, and finally Deltabaculovirus which, to date, comprises only CuniNPV and possibly the still undescribed dipteran-specific baculoviruses [1, 18–20].
The comparison between known genome sequences of all baculoviruses has been the source for identifying a common set of genes, the baculovirus core genes. However, there are probably more orthologous sequences that may not be identified due to the accumulation of many mutations throughout evolution. Thus, core genes seem to be a key factor for some of the main biological functions, such as those necessary to transcribe viral late genes, produce virion structure, infect gut cells abrogate host metabolism and establish infections [21–24].
For this report, previous data as well as bioinformatic studies conducted on currently available sets of completely sequenced baculovirus genomes were taken into account and have resulted in a summary of gene content and phylogenetic analyses which validates the classification of this important viral family.
2. Baculovirus Ancestral Genes
There are currently 57 complete baculovirus genomes deposited in GenBank (Table 1). These include 41 Alphabaculoviruses, 12 Betabaculoviruses, 3 Gammabaculoviruses, and 1 Deltabaculovirus.
Table 1.
Baculovirus complete genomes.
| Genus | Name | Abbreviation | Code | Accesion number | Genome (bp) |
Annotated ORFs | GC% | Ref. |
|---|---|---|---|---|---|---|---|---|
| Alphabaculovirus-Group I | Antheraea pernyi NPV-Z | AnpeNPV-Z | APN | NC_008035 | 126629 | 145 | 53.5 | [27] |
| Antheraea pernyi NPV-L2 | AnpeNPV-L2 | AP2 | EF207986 | 126246 | 144 | 53.5 | [28] | |
| Anticarsia gemmatalis MNPV-2D | AgMNPV-2D | AGN | NC_008520 | 132239 | 152 | 44.5 | [29] | |
| Autographa californica MNPV-C6 | AcMNPV-C6 | ACN | NC_001623 | 133894 | 154 | 40.7 | [30] | |
| Bombyx mori NPV | BmNPV | BMN | NC_001962 | 128413 | 137 | 40.4 | [31] | |
| Bombyx mandarina NPV | BomaNPV | BON | NC_012672 | 126770 | 141 | 40.2 | [32] | |
| Choristoneura fumiferana DEF MNPV | CfDEFMNPV | CDN | NC_005137 | 131160 | 149 | 45.8 | [33] | |
| Choristoneura fumiferana MNPV | CfMNPV | CFN | NC_004778 | 129593 | 145 | 50.1 | [34] | |
| Epiphyas postvittana NPV | EppoNPV | EPN | NC_003083 | 118584 | 136 | 40.7 | [35] | |
| Hyphantria cunea NPV | HycuNPV | HCN | NC_007767 | 132959 | 148 | 45.5 | [36] | |
| Maruca vitrata MNPV | MaviMNPV | MVN | NC_008725 | 111953 | 126 | 38.6 | [37] | |
| Orgyia pseudotsugata MNPV | OpMNPV | OPN | NC_001875 | 131995 | 152 | 55.1 | [38] | |
| Plutella xylostella MNPV | PlxyMNPV | PXN | NC_008349 | 134417 | 149 | 40.7 | U | |
| Rachiplusia ou MNPV | RoMNPV | RON | NC_004323 | 131526 | 146 | 39.1 | [39] | |
| Alphabaculovirus-Group II | Adoxophyes honmai NPV | AdhoNPV | AHN | NC_004690 | 113220 | 125 | 35.6 | [40] |
| Adoxophyes orana NPV | AdorNPV | AON | NC_011423 | 111724 | 121 | 35.0 | [41] | |
| Agrotis ipsilon NPV | AgipNPV | AIN | NC_011345 | 155122 | 163 | 48.6 | U | |
| Agrotis segetum NPV | AgseNPV | ASN | NC_007921 | 147544 | 153 | 45.7 | [42] | |
| Apocheima cinerarium NPV | ApciNPV | APO | FJ914221 | 123876 | 118 | 33.4 | U | |
| Chrysodeixis chalcites NPV | ChChNPV | CCN | NC_007151 | 149622 | 151 | 39.0 | [43] | |
| Clanis bilineata NPV | ClbiNPV | CBN | NC_008293 | 135454 | 129 | 37.7 | [44] | |
| Ectropis obliqua NPV | EcobNPV | EON | NC_008586 | 131204 | 126 | 37.6 | [45] | |
| Euproctis pseudoconspersa NPV | EupsNPV | EUN | NC_012639 | 141291 | 139 | 40.4 | [46] | |
| Helicoverpa armigera NPV-C1 | HearNPV-C1 | HA1 | NC_003094 | 130759 | 135 | 38.9 | [47] | |
| Helicoverpa armigera NPV-G4 | HearNPV-G4 | HA4 | NC_002654 | 131405 | 135 | 39.0 | [47] | |
| Helicoverpa armigera MNPV | HearMNPV | HAN | NC_011615 | 154196 | 162 | 40.1 | [48] | |
| Helicoverpa armigera SNPV-NNg1 | HearSNPV-NNg1 | HAS | NC_011354 | 132425 | 143 | 39.2 | [49] | |
| Helicoverpa zea SNPV | HzSNPV | HZN | NC_003349 | 130869 | 139 | 39.1 | U | |
| Leucania separata NPV-AH1 | LeseNPV-AH1 | LSN | NC_008348 | 168041 | 169 | 48.6 | [50] | |
| Lymantria dispar MNPV | LdMNPV | LDN | NC_001973 | 161046 | 163 | 57.5 | [51] | |
| Lymantria xylina MNPV | LyxyMNPV | LXN | NC_013953 | 156344 | 157 | 53.5 | [52] | |
| Mamestra configurata NPV-90-2 | MacoNPV-90-2 | MCN | NC_003529 | 155060 | 169 | 41.7 | [53] | |
| Mamestra configurata NPV-90-4 | MacoNPV-90-4 | MC4 | AF539999 | 153656 | 168 | 41.7 | [54] | |
| Mamestra configurata NPV-B | MacoNPV-B | MCB | NC_004117 | 158482 | 169 | 40.0 | [55] | |
| Orgyia leucostigma NPV | OrleNPV | OLN | NC_010276 | 156179 | 135 | 39.9 | U | |
| Spodoptera exigua MNPV | SeMNPV | SEN | NC_002169 | 135611 | 142 | 43.8 | U | |
| Spodoptera frugiperda MNPV-3AP2 | SfMNPV-3AP2 | SF2 | NC_009011 | 131330 | 143 | 40.2 | [56] | |
| Spodoptera frugiperda MNPV-19 | SfMNPV-19 | SF9 | EU258200 | 132565 | 141 | 40.3 | [57] | |
| Spodoptera litura NPV-II | SpliNPV-II | SLN | NC_011616 | 148634 | 147 | 45.0 | U | |
| Spodoptera litura NPV-G2 | SpliNPV-G2 | SL2 | NC_003102 | 139342 | 141 | 42.8 | [58] | |
| Trichoplusia ni SNPV | TnSNPV | TNN | NC_007383 | 134394 | 144 | 39.0 | [59] | |
| Betabaculovirus | Adoxophyes orana GV | AdorGV | AOG | NC_005038 | 99657 | 119 | 34.5 | [60] |
| Agrotis segetum GV | AgseGV | ASG | NC_005839 | 131680 | 132 | 37.3 | U | |
| Choristoneura occidentalis GV | ChocGV | COG | NC_008168 | 104710 | 116 | 32.7 | [61] | |
| Cryptophlebia leucotreta GV | CrleGV | CLG | NC_005068 | 110907 | 129 | 32.4 | [62] | |
| Cydia pomonella GV | CpGV | CPG | NC_002816 | 123500 | 143 | 45.3 | [63] | |
| Helicoverpa armigera GV | HearGV | HAG | NC_010240 | 169794 | 179 | 40.8 | [64] | |
| Phthorimea operculella GV | PhopGV | POG | NC_004062 | 119217 | 130 | 35.7 | [65] | |
| Plutella xylostella GV | PlxyGV | PXG | NC_002593 | 100999 | 120 | 40.7 | [66] | |
| Pieris rapae GV | PiraGV | PRG | GQ884143 | 108592 | 120 | 33.2 | U | |
| Pseudaletia unipuncta GV-Hawaiin | PsunGV | PUG | EU678671 | 176677 | 183 | 39.8 | U | |
| Spodoptera litura GV-K1 | SpliGV | SLG | NC_009503 | 124121 | 136 | 38.8 | [67] | |
| Xestia c-nigrum GV | XnGV | XCG | NC_002331 | 178733 | 181 | 40.7 | [68] | |
| Gamma | Neodiprion abietis NPV | NeabNPV | NAN | NC_008252 | 84264 | 93 | 33.4 | [69] |
| Neodiprion lecontei NPV | NeleNPV | NLN | NC_005906 | 81755 | 93 | 33.3 | [70, 71] | |
| Neodiprion sertifer NPV | NeseNPV | NSN | NC_005905 | 86462 | 90 | 33.8 | [71, 72] | |
| Delta | Culex nigripalpus NPV | CuniNPV | CNN | NC_003084 | 108252 | 109 | 50.9 | [73] |
This table contains all of baculoviruses used in bioinformatic studies, sorted by genus (and within them by alphabetical order). MNPV is the abbreviation of multicapsid nucleopolyhedrovirus; NPV is the abbreviation of nucleopolyhedrovirus; SNPV is the abbreviation of single nucleopolyhedrovirus; GV is the abbreviation of granulovirus. The accession numbers are from National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) and correspond to the sequences of complete genomes. Code is an acronym used for practicality. U: unpublished.
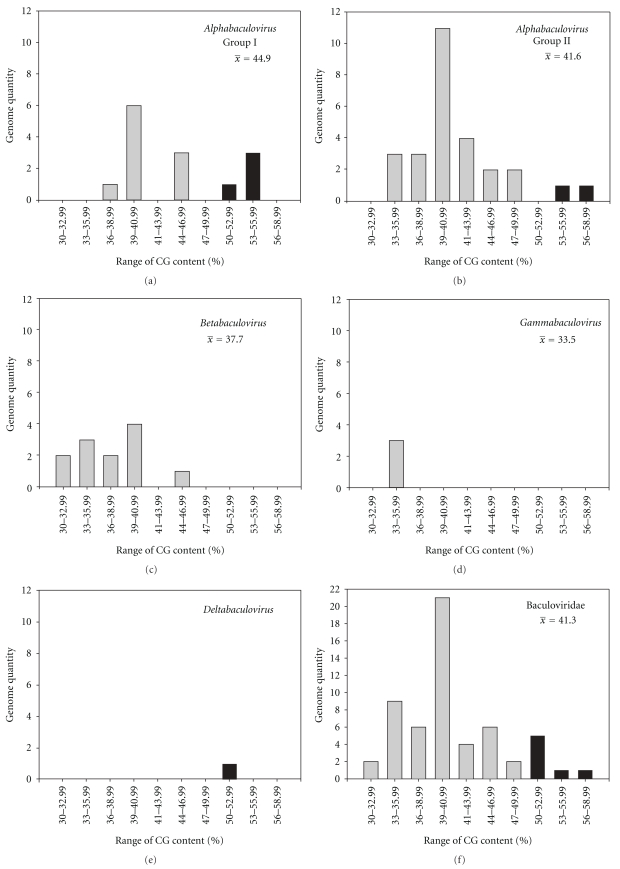
As a first approach to perform a comparative analysis, the GC content of the genomes were calculated (Figure 1). The histogram revealed that many baculoviruses have about 41% of GC content although several of them have significantly higher values (CfMNPV at 50.1%, CuniNPV at 50.9%, AnpeNPV-L2 at 53.5%, AnpeNPV-Z at 53.5%, LyxyNPV at 53.5%, OpMNPV at 55.1%, and LdMNPV at 57.5%). A detailed analysis of DNA content did not show a clear pattern of GC content that could be associated with each genus.
Figure 1.
GC content in baculovirus genomes. The different histograms contain the distribution of baculovirus genomes according to their GC content and their genus classification. Black bars highlight genomes with a GC content higher than 50%.
Further characterization of the patterns of gene content and organization may prove useful for establishing evolutionary relationships among members of Baculoviridae. The high variability observed in the number of coding sequences becomes a key feature of viruses with large DNA genomes that infect eukaryotic cells [18]. Insertions, deletions, duplication events, and/or sequence reorganizations by recombination or transposition processes seem to be the main forces of the macroevolution in this particular kind of biological entities. For example, the loss or gain of genetic material could provide new important abilities for colonization of new hosts, or they could improve performance within established hosts. However, there seems to be a set of core genes whose absence would imply the loss of basic biological functions, and that could be typical of the viral family. In view of this, and considering previous reports [1, 19, 22, 23], the amount and identity of baculovirus common genes were reevaluated (Table 2). As a result, P6.9 and Desmoplakin were recognized in this work, as core proteins by using sequence analysis complementary to the standard ones (see Supplementary files available at doi:10.4061/2011/379424).
Table 2.
Core genes.
| ACN | LDN | CPG | NSN | CNN | |
|---|---|---|---|---|---|
| Replication | |||||
| lef-1 [74] | 14 | 123 | 74 | 68 | 45 |
| lef-2 [74] | 6 | 137 | 41 | 57 | 25 |
| DNA pol [75–78] | 65 | 83 | 111 | 28 | 91 |
| Helicase [79–90] | 95 | 97 | 90 | 61 | 89 |
| Transcription | |||||
| lef-4 [91–95] | 90 | 93 | 95 | 62 | 96 |
| lef-8 [91, 96] | 50 | 51 | 131 | 81 | 26 |
| lef-9 [95, 97] | 62 | 64 | 117 | 40 | 59 |
| p47 [91, 98] | 40 | 48 | 68 | 49 | 73 |
| lef-5 [98–101] | 99 | 100 | 87 | 58 | 88 |
| Packaging, assembly, and release | |||||
| p6.9 [102–104] | 100 | 101 | 86 | 36 | 23 |
| vp39 [105–108] | 89 | 92 | 96 | 89 | 24 |
| vlf-1 [100, 109–113] | 77 | 86 | 106 | 45 | 18 |
| alk-exo [114–116] | 133 | 157 | 125 | 31 | 53 |
| vp1054 [117] | 54 | 57 | 138 | 85 | 8 |
| vp91/p95 [118] | 83 | 91 | 101 | 84 | 35 |
| gp41 [119, 120] | 80 | 88 | 104 | 47 | 33 |
| 38 k [121, 122] | 98 | 99 | 88 | 59 | 87 |
| p33 [123–125] | 92 | 94 | 93 | 24 | 14 |
| odv-ec43 [126–128] | 109 | 107 | 55 | 70 | 69 |
| p49 [129] | 142 | 20 | 15 | 63 | 30 |
| odv-nc42 [130] | 68 | 80 | 114 | 41 | 58 |
| odv-e18 [131] | 143 | 19 | 14 | 65 | 31 |
| desmoplakin [132] | 66 | 82 | 112 | 29 | 92 |
| Cell cycle arrest and/or interaction with host proteins | |||||
| odv-e27 [133, 134] | 144 | 18 | 97 | 66 | 32 |
| ac81 [135] | 81 | 89 | 103 | 48 | 106 |
| Oral infectivity | |||||
| pif-0/p74 [136–141] | 138 | 27 | 60 | 50 | 74 |
| pif-1 [142–144] | 119 | 155 | 75 | 79 | 29 |
| pif-2 [136, 142] | 22 | 119 | 48 | 55 | 38 |
| pif-3 [142] | 115 | 143 | 35 | 69 | 46 |
| pif-4/19k/odv-e28 [145] | 96 | 98 | 89 | 60 | 90 |
| pif-5/odv-e56 [146, 147] | 148 | 14 | 18 | 38 | 102 |
The virus names are indicated in three letter code according to established in Table 1.
Numbers in columns indicates the corresponding ORFs of each genome.
The group of conserved sequences found in all baculovirus genomes is consistently estimated at about 30 shared genes, regardless of the increasing number of genomes analyzed [22, 148]. Meanwhile, the role or function assigned to several sequences has been renewed, according to new studies. In particular, it has been identified that 38k (Ac98) gene encodes a protein which is part of the capsid structure [121, 122]; P33 (Ac92) is a sulfhydryl oxidase which could be related to the proper production of virions in the infected cell nucleus [123–125]; ODV-EC43 (Ac109) is a structural component which would be involved in BV and ODV generation [126]; P49 (Ac142) is a capsid protein important in DNA processing, packaging, and capsid morphogenesis [129]; Ac81 interacts with Actin 3 in the cytoplasm but does not appear in BVs or in ODVs [135]; ODV-E18 (Ac143) would mediate BV production [131]; desmoplakin (Ac66) seems to be essential in releasing processes from virogenic stroma to cytoplasm [132]; PIF-4 (Ac96) and PIF-5 (ODV-56, Ac148) are ODV envelope proteins with an essential role in per os infection route [145, 147]; Ac68 may be involved in polyhedron morphogenesis [130].
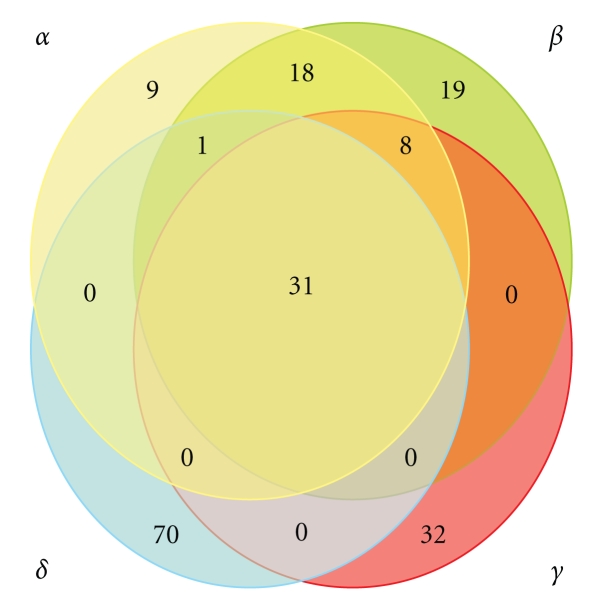
The number and identity of shared orthologous genes in every accepted member of each genus were investigated, and the unique sequences typical of each clade as well as those shared between different phylogenetic groups were identified (Figure 2).
Figure 2.
Baculovirus core genes. The different circles represent the 4 baculovirus genera (in yellow Alphabaculovirus; in green Betabaculovirus; in red Gammabaculovirus; in blue Deltabaculovirus). The numbers contained within the overlapping regions indicate the amount of shared genes between all members of the genera. The numbers within the circles but outside the overlapping regions indicate the amount of genes shared by all members of that genus but with the absence of orthologous sequences in the remaining genera. These estimations were inferred by Blast P algorithm (http://www.ncbi.nlm.nih.gov/) considering E = 0.001 as cutoff value and comparing all reported baculovirus ORFs between them. The identity of common genes is provided in the Supplementary data available at doi:10.4061/2011/379424
This analysis shows that the four accepted baculovirus genera have accumulated a large number of genes during evolution. Probably, many of these sequences have been incorporated into viral genomes prior to diversification processes since they are found in members of different genera. In contrast, other genes are unique to each genus, suggesting that they have been incorporated more recently and after diversification (Table 3). The possibility that nonshared genes found only in one genus which represent baculovirus ancestral sequences deleted in the other lineages should also be considered. In any case, a set of particular genes which could help in an appropriate genus taxonomy of new baculoviruses with partial sequence information were obtained from this analysis.
Table 3.
Shared genes*.
| Core genes |
| lef-2 (ACN6), lef-1 (ACN14), pif-2 (ACN22), p47 (ACN40), lef-8 (ACN50), vp1054 (ACN54), lef-9 (ACN62), DNA polymerase |
| (ACN65), Desmoplakin (ACN66), ACN68, vlf-1 (ACN77), gp41 (ACN80), ACN81, vp91/p95 (ACN83), vp39 (ACN89), lef-4 |
| (ACN90), p33 (ACN92), helicase (ACN95), 19K (ACN96), 38 K (ACN98), lef-5 (ACN99), p6.9 (ACN100), odv-ec43 (ACN109), |
| PIF-3 (ACN115), pif-1 (ACN119), alkaline exonuclease (ACN133), p74 (ACN138), p49 (ACN142), odv-e18 (ACN143), odv-e27 |
| (ACN144), odv-e56 (ACN148) |
| Alpha + Beta + Gamma |
| Polh (ACN8), dbp (ACN25), p48 (ACN103), ACN145, pp34/PEP (ACN131), odv-e25 (ACN94), p40 (ACN101), ACN106/107 |
| Alpha + Beta + Delta |
| F-protein (ACN23) |
| Alpha + Beta |
| pk-1 (ACN10), 38,7 kDa (ACN13), lef-6 (ACN28), pp31/39K (ACN36), ACN38, ACN53, 25K FP (ACN61), LEF-3 (ACN67), ACN75, |
| ACN76, tlp20 (ACN82), p18 (ACN93), P12 (ACN102), ACN108, p24 (ACN129), me53 (ACN139), ACN146, ie-1 (ACN147) |
| Alpha |
| orf1629 capsid (ACN9), ACN19, pkip-1 (ACN24), ACN34, ACN51, iap-2 (ACN58/59), ACN104, p87/vp80 (ACN141), ie-0 (ACN71) |
| Alpha Group I |
| ptp-1/bvp (ACN1), ACN5, odv-e26 (ACN16), iap-1 (ACN27), ACN30, ACN72, ACN73, ACN114, ACN124, gp64 (ACN128), p25 |
| (ACN132), ie-2 (ACN151) |
| Beta |
| CPG4, CPG5, CPG20, CPG23, CPG29, CPG33, CPG39, CPG45, Metalloproteinase (CPG46), CPG62, FGF-1 (CPG76), CPG79, |
| CPG99, CPG100, CPG115, IAP-5 (CPG116), CPG123, CPG135, FGF-3 (CPG140) |
| Gamma |
| NSN3, NSN9, NSN11, NSN12, NSN13, NSN16, NSN18, NSN19, NSN20, NSN26, NSN29, NSN34, NSN37, NSN39, NSN42, NSN43, |
| NSN44, NSN51, NSN52, NSN53, NSN54, NSN56, NSN64, NSN72, NSN74, NSN76, NSN77, NSN79, NSN82, NSN85, NSN86, |
| NSN89 |
| Delta |
| CNN2, CNN3, CNN6, CNN7, CNN9, CNN10, CNN11, CNN12, CNN13, CNN15, CNN16, CNN17, CNN20, CNN21, CNN22, |
| CNN27, CNN28, CNN31, CNN36, CNN37, CNN39, CNN40, CNN41, CNN42, CNN43, CNN44, CNN47, CNN48, CNN49, CNN50, |
| CNN51, CNN52, CNN53, CNN55, CNN56, CNN57, CNN60, CNN61, CNN62, CNN63, CNN64, CNN65, CNN66, CNN67, CNN68, |
| CNN70, CNN71, CNN72, CNN75, CNN76, CNN77, CNN78, CNN79, CNN80, CNN81, CNN82, CNN83, CNN84, CNN85, CNN86, |
| CNN93, CNN94, CNN97, CNN98, CNN99, CNN100, CNN101, CNN103, CNN105, CNN107 |
*Shared genes are indicated only for one selected specie. See supplementary tables for the respective ORF numbers in each specie.
3. Whole Baculovirus Gene Content
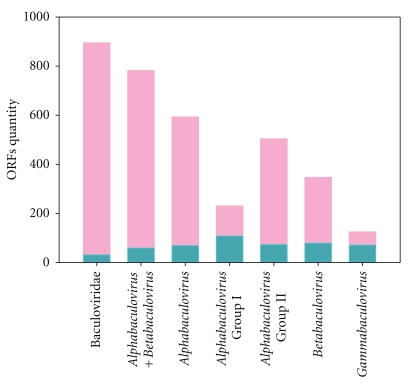
The study of all genes reported in the 57 completely sequenced viral genomes revealed the existence of about 895 different ORFs, a set of sequences that might be called the whole baculovirus gene content. This high number of potential coding sequences contrasts with the range of gene content among the family members, which is between 90–181 genes (Alphabaculovirus: 118–169; Betabaculovirus: 116–181; Gammabaculovirus: 90–93; Deltabaculovirus: 109) as well as with the proportion of core genes which represents only 3%. This curious biological feature supports the hypothesis that highlights the great importance of structural mutations in the macroevolution of viruses with large DNA genomes. From this view, the set of genes shared by all members belonging to each baculovirus genus was compared to those corresponding to the whole genus gene content (Figure 3).
Figure 3.
Whole baculovirus gene content. The histogram shows the amount of different reported genes in each baculovirus genus or recognized lineage (bars in pink color), and the subset of shared genes for all members of the corresponding phylogenetic clade (bars in green color). This bar graph was performed using the information resulting from the comparison of all ORFs reported in the 57 baculovirus with known genomes, analyzing all against all by Blast P algorithm (http://www.ncbi.nlm.nih.gov/) considering E = 0.001 as cutoff value.
The analysis shows that Group I alphabaculoviruses and gammabaculoviruses have a lower diversity of gene content with respect to the rest of lineages. This information, coupled with the significant number of genome sequences obtained from Group I alphabaculoviruses, suggests that this lineage of viruses would constitute the newest clade in baculovirus evolution history [149]. This is based on the assumption that Group I alphabaculoviruses have had less time to incorporate new sequences from different sources (host genomes, other viral genomes, bacterial genomes, etc.) since the appearance of their common ancestor.
4. Baculovirus Core Gene Phylogeny
Traditional attempts to infer relationships between baculoviruses were performed by amino acid or nucleotide sequence analyses of single genes encoding proteins such as polyhedrin/granulin (the major component of OBs), the envelope fusion polypeptides known as F protein and GP64, or DNA polymerase protein, among many other examples [149–152].
Mostly, the evolutionary inferences were in agreement with much stronger subsequent studies based on sequence analyses derived from sets of genes with homologous sequences in all baculoviruses. Thus, these new approaches were based on the construction of common-protein-concatemers which were used to propose evolution patterns for baculoviruses [149].
Then, the fact that a viral family consists of members who share a common pattern of genes and functions and whose proliferation cycle continuously challenges the viral viability turns it essential to take into account their higher or lesser tolerance to the molecular changes. Molecular constraints regarding tolerance to changes in core genes are different from those of other genes. Therefore, core genes should be considered the most ancestral genes which may have diverged in higher or lesser degrees. According to this, a phylogenetic study was performed based on concatemers obtained from multiple alignments of the 31 proteins recognized in this work as core genes for the 57 available baculoviruses with sequenced genomes (Figure 4).
Figure 4.
Baculovirus genome phylogeny. Cladogram based on amino acid sequence of core genes. The 31 identified core genes from Baculoviridae family were independently aligned using MEGA 4 [25] program with gap open penalty = 10, gap extension penalty = 1, and dayhoff matrix [26]. Then, a concatemer was generated and phylogeny inferred using the same software (UPGMA; bootstrap with 1000 replicates; gap/missing data = complete deletion; model = amino (dayhoff matrix); patterns among sites = same (homogeneous); rates among sites = different (gamma distributed); gamma parameter = 2.25). Baculoviruses are identified by the acronyms given in Table 1, and the accepted distribution in lineages and genera are also indicated. Gammabaculovirus and Deltabaculovirus are referenced by Greek letters. The proposed clades of Betabaculoviruses are shown in bold letters.
The obtained cladogram reproduces the current baculovirus classification based on 4 genera. Additionally, this approach consistently separates the alphabaculoviruses into two lineages: Group I and Group II. And the same can be observed when analyzing Group I, where the presence of two different clades can be clearly inferred (clade a and clade b). These groupings result in accordance with previous reports [20, 150]. In Group II alphabaculoviruses, a clear clustering may not be identified and would not allow to suggest a subdivision.
In contrast, in the Betabaculovirus genus, it is possible to propose their separation into two different clades: clade a (XnGV, HearGV, PsunGV, SpliGV, AgseGV, and PlxyGV), and clade b (AdorGV, PhopGV, CpGV, CrleGV, PiraGV, ChocGV).
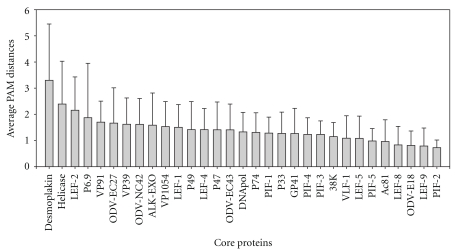
Despite the evolutionary inference based on core genes, there was a remaining question: “is the tolerance to changes in all core genes the same?”. The answer could be reached by an individual core gene variability analysis for which studies of sequence distance for each baculovirus core gene were performed (Figure 5).
Figure 5.
Baculovirus core gene variability. Histograms show the average PAM250 distances for each core gene with their corresponding standard deviations. These values were calculated using MEGA 4 program (UPGMA; bootstrap with 1000 replicates; gap/missing data = complete deletion; model = amino (dayhoff matrix); patterns among sites = same (homogeneous); rates among sites = different (gamma distributed; gamma parameter = 2.25)). PAM (point accepted mutation) matrices refers to the evolutionary distance between pairs of sequences. Given the weak similarity between several core proteins, PAM250 matrix was selected. The divergence considered in this matrix is 250 mutations per 100 amino acid sequence and was calculated to analyze more distantly related sequences. PAM250 is considered a good general matrix for protein similarity search.
The resulting order of core genes shows that pif-2 was the most conserved baculovirus ancestral sequence, whereas desmoplakin was the gene with evidence of greatest variability. This analysis reveals that genomes can be evolutionarily constrained in different ways depending on the proteins they encode.
The gain of access to new hosts might be an important force for gene evolution. During an infection process, the genome variants that appear with mutations introduced by errors in the replication/reparation machinery could be quickly incorporated into the virus population if the nucleotide changes offered a better biological performance when proteins were translated. The DNA helicase gene was considered as an important host range factor being, for this study, the second core sequence showing more variability [87]. However, other sequences like pif-2 gene would not accumulate mutations because the protein encoded might lose vital functions not necessarily associated with the nature of the host.
5. Conclusions
Baculoviridae is a large family of viruses which infect and kill insect species from different orders. The valuable applications of these viruses in several fields of life sciences encourage their constant study with the goal of understanding the molecular mechanisms involved in the generation of progeny in the appropriate cells as well as the processes by which they evolve. The establishment of solid bases to recognize their phylogenetic relationships is necessary to facilitate the generation of new knowledge and the development of better methodologies.
In view of this, many researchers have proposed and used different bioinformatic methodologies to identify genes as well as related baculoviruses. Some of them were based on gene sequences [150], gene content [17], or genome rearrangements [152]. In this work, a combination of core gene sequence and gene content analyses were applied to reevaluate Baculoviridae classification. To our knowledge, the most important fact is that this report is the first work which identifies the whole baculovirus gene content and the shared genes that are unique in different genera and subgenera. All this information should be taken into account to group and classify new virus isolates and to propose molecular methodologies to diagnose baculoviruses based on proper gene targets according to gene variability and gene content.
Supplementary Material
The supplementary text: explains in detail alternative bioinformatic approaches used to validate the recognition of core genes. It also contains a detailed table showing the numbers of ORF homologous within the family Baculoviridae.
Acknowledgments
This work was supported by research funds from Agencia Nacional de Promoción Científica y Técnica (ANPCyT) and Universidad Nacional de Quilmes. P. D. Ghiringhelli is member of the Research Career of CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas), M. N. Belaich holds a postdoctoral fellowship of CONICET, S. A. B. Miele holds a fellowship of CONICET, and M J Garavaglia holds a fellowship of CIC-PBA (Comisión de Investigaciones Científicas de la Provincia de Buenos Aires). The authors acknowledge to Lic. Javier A. Iserte, Lic. Betina I. Stephan and Lic. Laura Esteban for their helping with the paper. S. A. B. Miele and M. Javier Garavaglia both contributed equally to this work.
References
- 1.Jehle JA, Blissard GW, Bonning BC, et al. On the classification and nomenclature of baculoviruses: a proposal for revision. Archives of Virology. 2006;151(7):1257–1266. doi: 10.1007/s00705-006-0763-6. [DOI] [PubMed] [Google Scholar]
- 2.Blissard GW, Rohrmann GF. Baculovirus diversity and molecular biology. Annual Review of Entomology. 1990;35(1):127–155. doi: 10.1146/annurev.en.35.010190.001015. [DOI] [PubMed] [Google Scholar]
- 3.Kozlov EA, Levitina TL, Gusak NM. The primary structure of baculovirus inclusion body proteins. Evolution and structure-function aspects. Current Topics in Microbiology and Immunology. 1986;131:135–164. doi: 10.1007/978-3-642-71589-1_8. [DOI] [PubMed] [Google Scholar]
- 4.Rohrmann GF. Baculovirus structural proteins. Journal of General Virology. 1992;73(4):749–761. doi: 10.1099/0022-1317-73-4-749. [DOI] [PubMed] [Google Scholar]
- 5.Ohkawa T, Washburn JO, Sitapara R, Sid E, Volkman LE. Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. Journal of Virology. 2005;79(24):15258–15264. doi: 10.1128/JVI.79.24.15258-15264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackes R, Maromorosch E, Sherman K. Stability of Insect Viruses in the Environment. Viral Insecticides for Biological Control. New York, NY, USA: Academic Press; 1985. [Google Scholar]
- 7.Zhang G. Research, development and application of Heliothis viral pesticide in China. Resources and Environment in the Yangtze Valley. 1994;3:1–6. [Google Scholar]
- 8.Possee RD. Baculoviruses as expression vectors. Current Opinion in Biotechnology. 1997;8(5):569–572. doi: 10.1016/s0958-1669(97)80030-4. [DOI] [PubMed] [Google Scholar]
- 9.Moscardi F. Assessment of the application of baculoviruses for control of lepidoptera. Annual Review of Entomology. 1999;44:257–289. doi: 10.1146/annurev.ento.44.1.257. [DOI] [PubMed] [Google Scholar]
- 10.Kost TA, Condreay JP. Recombinant baculoviruses as expression vectors for insect and mammalian cells. Current Opinion in Biotechnology. 1999;10(5):428–433. doi: 10.1016/s0958-1669(99)00005-1. [DOI] [PubMed] [Google Scholar]
- 11.Inceoglu AB, Kamita SG, Hinton AC, et al. Recombinant baculoviruses for insect control. Pest Management Science. 2001;57(10):981–987. doi: 10.1002/ps.393. [DOI] [PubMed] [Google Scholar]
- 12.Kost TA, Condreay JP, Jarvis DL. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nature Biotechnology. 2005;23(5):567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summers MD. Milestones leading to the genetic engineering of baculoviruses as expression vector systems and viral pesticides. Advances in Virus Research. 2006;68:3–73. doi: 10.1016/S0065-3527(06)68001-9. [DOI] [PubMed] [Google Scholar]
- 14.Inceoglu AB, Kamita SG, Hammock BD. Genetically modified baculoviruses: a historical overview and future outlook. Advances in Virus Research. 2006;68:323–360. doi: 10.1016/S0065-3527(06)68009-3. [DOI] [PubMed] [Google Scholar]
- 15.Shi X, Jarvis DL. Protein N-glycosylation in the baculovirus-insect cell system. Current Drug Targets. 2007;8(10):1116–1125. doi: 10.2174/138945007782151360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Condreay JP, Kost TA. Baculovirus expression vectors for insect and mammalian cells. Current Drug Targets. 2007;8(10):1126–1131. doi: 10.2174/138945007782151351. [DOI] [PubMed] [Google Scholar]
- 17.Sun XL, Peng HY. Recent advances in biological control of pest insect by using viruses in China. Virologica Sinica. 2007;22(2):158–162. [Google Scholar]
- 18.Herniou EA, Luque T, Chen X, et al. Use of whole genome sequence data to infer baculovirus phylogeny. Journal of Virology. 2001;75(17):8117–8126. doi: 10.1128/JVI.75.17.8117-8126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herniou EA, Olszewski JA, Cory JS, O’Reilly DR. The genome sequence and evolution of baculoviruses. Annual Review of Entomology. 2003;48:211–234. doi: 10.1146/annurev.ento.48.091801.112756. [DOI] [PubMed] [Google Scholar]
- 20.Jehle JA, Lange M, Wang H, Hu Z, Wang Y, Hauschild R. Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology. 2006;346(1):180–193. doi: 10.1016/j.virol.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 21.van Oers MM, Vlak JM. Baculovirus genomics. Current Drug Targets. 2007;8(10):1051–1068. doi: 10.2174/138945007782151333. [DOI] [PubMed] [Google Scholar]
- 22.Rohrman GF. Baculovirus Molecular Biology. Bethesda, Md, USA: National Library of Medicine (US), NCBI; 2008. [Google Scholar]
- 23.Hayakawa T, Rohrmann GF, Hashimoto Y. Patterns of genome organization and content in lepidopteran baculoviruses. Virology. 2000;278(1):1–12. doi: 10.1006/viro.2000.0668. [DOI] [PubMed] [Google Scholar]
- 24.McCarthy CB, Theilmann DA. AcMNPV ac143 (odv-e18) is essential for mediating budded virus production and is the 30th baculovirus core gene. Virology. 2008;375(1):277–291. doi: 10.1016/j.virol.2008.01.039. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz RM, Dayhoff MO. Matrices for detecting distant relationships. In: Dayhoff MO, editor. Atlas of Protein Sequences. National Biomedical Research Foundation; 1979. pp. 353–358. [Google Scholar]
- 27.Fan Q, Li S, Wang L, et al. The genome sequence of the multinucleocapsid nucleopolyhedrovirus of the Chinese oak silkworm Antheraea pernyi. Virology. 2007;366(2):304–315. doi: 10.1016/j.virol.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 28.Nie ZM, Zhang ZF, Wang D, et al. Complete sequence and organization of Antheraea pernyi nucleopolyhedrovirus, a dr-rich baculovirus. BMC Genomics. 2007;8 doi: 10.1186/1471-2164-8-248. Article ID 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Castro Oliveira JV, Wolff JLC, Garcia-Maruniak A, et al. Genome of the most widely used viral biopesticide: anticarsia gemmatalis multiple nucleopolyhedrovirus. Journal of General Virology. 2006;87(11):3233–3250. doi: 10.1099/vir.0.82161-0. [DOI] [PubMed] [Google Scholar]
- 30.Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202(2):586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 31.Gomi S, Majima K, Maeda S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. Journal of General Virology. 1999;80(5):1323–1337. doi: 10.1099/0022-1317-80-5-1323. [DOI] [PubMed] [Google Scholar]
- 32.Xu YP, Ye ZP, Niu CY, et al. Comparative analysis of the genomes of Bombyx mandarina and Bombyx mori nucleopolyhedroviruses. Journal of Microbiology. 2010;48(1):102–110. doi: 10.1007/s12275-009-0197-4. [DOI] [PubMed] [Google Scholar]
- 33.de Jong JG, Lauzon HAM, Dominy C, et al. Analysis of the Choristoneura fumiferana nucleopolyhedrovirus genome. Journal of General Virology. 2005;86(4):929–943. doi: 10.1099/vir.0.80490-0. [DOI] [PubMed] [Google Scholar]
- 34.Lauzon HAM, Jamieson PB, Krell PJ, Arif BM. Gene organization and sequencing of the Choristoneura fumiferana defective nucleopolyhedrovirus genome. Journal of General Virology. 2005;86(4):945–961. doi: 10.1099/vir.0.80489-0. [DOI] [PubMed] [Google Scholar]
- 35.Hyink O, Dellow RA, Olsen MJ, et al. Whole genome analysis of the Epiphyas postvittana nucleopolyhedrovirus. Journal of General Virology. 2002;83(4):957–971. doi: 10.1099/0022-1317-83-4-957. [DOI] [PubMed] [Google Scholar]
- 36.Ikeda M, Shikata M, Shirata N, Chaeychomsri S, Kobayashi M. Gene organization and complete sequence of the Hyphantria cunea nucleopolyhedrovirus genome. Journal of General Virology. 2006;87(9):2549–2562. doi: 10.1099/vir.0.81930-0. [DOI] [PubMed] [Google Scholar]
- 37.Chen YR, Wu CY, Lee ST, et al. Genomic and host range studies of Maruca vitrata nucleopolyhedrovirus. Journal of General Virology. 2008;89(9):2315–2330. doi: 10.1099/vir.0.2008/001412-0. [DOI] [PubMed] [Google Scholar]
- 38.Ahrens CH, Russell RLQ, Funk CJ, Evans JT, Harwood SH, Rohrmann GF. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology. 1997;229(2):381–399. doi: 10.1006/viro.1997.8448. [DOI] [PubMed] [Google Scholar]
- 39.Harrison RL, Bonning BC. The nucleopolyhedroviruses of Rachiplusia ou and Anagrapha falcifera are isolates of the same virus. Journal of General Virology. 1999;80(10):2793–2798. doi: 10.1099/0022-1317-80-10-2793. [DOI] [PubMed] [Google Scholar]
- 40.Nakai M, Goto C, Kang W, Shikata M, Luque T, Kunimi Y. Genome sequence and organization of a nucleopolyhedrovirus isolated from the smaller tea tortrix, Adoxophyes honmai. Virology. 2003;316(1):171–183. doi: 10.1016/j.virol.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Hilton S, Winstanley D. Genomic sequence and biological characterization of a nucleopolyhedrovirus isolated from the summer fruit tortrix, Adoxophyes orana. Journal of General Virology. 2008;89(11):2898–2908. doi: 10.1099/vir.0.2008/002881-0. [DOI] [PubMed] [Google Scholar]
- 42.Jakubowska AK, Peters SA, Ziemnicka J, Vlak JM, van Oers MM. Genome sequence of an enhancin gene-rich nucleopolyhedovirus (NPV) from Agrotis segetum: collinearity with Spodoptera exigua multiple NPV. Journal of General Virology. 2006;87(3):537–551. doi: 10.1099/vir.0.81461-0. [DOI] [PubMed] [Google Scholar]
- 43.van Oers MM, Abma-Henkens MHC, Herniou EA, de Groot JCW, Peters S, Vlak JM. Genome sequence of Chrysodeixis chalcites nucleopolyhedrovirus, a baculovirus with two DNA photolyase genes. Journal of General Virology. 2005;86(7):2069–2080. doi: 10.1099/vir.0.80964-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhu SY, Yi JP, Shen WD, et al. Genomic sequence, organization and characteristics of a new nucleopolyhedrovirus isolated from Clanis bilineata larva. BMC Genomics. 2009;10:9 pages. doi: 10.1186/1471-2164-10-91. Article ID 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma XC, Shang JY, Yang ZN, Bao YY, Xiao Q, Zhang CX. Genome sequence and organization of a nucleopolyhedrovirus that infects the tea looper caterpillar, Ectropis obliqua. Virology. 2007;360(1):235–246. doi: 10.1016/j.virol.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 46.Tang XD, Xiao Q, Ma XC, Zhu ZR, Zhang CX. Morphology and genome of Euproctis pseudoconspersa nucleopolyhedrovirus. Virus Genes. 2009;38(3):495–506. doi: 10.1007/s11262-009-0355-0. [DOI] [PubMed] [Google Scholar]
- 47.Zhang CX, Ma XC, Guo ZJ. Comparison of the complete genome sequence between C1 and G4 isolates of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus. Virology. 2005;333(1):190–199. doi: 10.1016/j.virol.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 48.Ogembo JG, Chaeychomsri S, Kamiya K, et al. Cloning and comparative characterization of nucleopolyhedroviruses isolated from African bollworm, Helicoverpa armigera, (Lepidoptera: Noctudiae) in different geographic regions. Journal of Insect Biotechnology and Sericology. 2007;76(1):39–49. [Google Scholar]
- 49.Chen X, Ijkel WFJ, Dominy C, et al. Identification, sequence analysis and phylogeny of the lef-2 gene of Helicoverpa armigera single-nucleocapsid baculovirus. Virus Research. 1999;65(1):21–32. doi: 10.1016/s0168-1702(99)00097-0. [DOI] [PubMed] [Google Scholar]
- 50.Xiao H, Qi Y. Genome sequence of Leucania seperata nucleopolyhedrovirus. Virus Genes. 2007;35(3):845–856. doi: 10.1007/s11262-007-0106-z. [DOI] [PubMed] [Google Scholar]
- 51.Kuzio J, Pearson MN, Harwood SH, et al. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253(1):17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- 52.Nai YS, Wu CY, Wang TC, et al. Genomic sequencing and analyses of Lymantria xylina multiple nucleopolyhedrovirus. BMC Genomics. 2010;11(1) doi: 10.1186/1471-2164-11-116. Article ID 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li S, Erlandson M, Moody D, Gillott C. A physical map of the Mamestra configurata nucleopolyhedrovirus genome and sequence analysis of the polyhedrin gene. Journal of General Virology. 1997;78(1):265–271. doi: 10.1099/0022-1317-78-1-265. [DOI] [PubMed] [Google Scholar]
- 54.Li L, Li Q, Willis LG, Erlandson M, Theilmann DA, Donly C. Complete comparative genomic analysis of two field isolates of Mamestra configurata nucleopolyhedrovirus-A. Journal of General Virology. 2005;86(1):91–105. doi: 10.1099/vir.0.80488-0. [DOI] [PubMed] [Google Scholar]
- 55.Li L, Donly C, Li Q, et al. Identification and genomic analysis of a second species of nucleopolyhedrovirus isolated from Mamestra configurata. Virology. 2002;297(2):226–244. doi: 10.1006/viro.2002.1411. [DOI] [PubMed] [Google Scholar]
- 56.Harrison RL, Puttler B, Popham HJR. Genomic sequence analysis of a fast-killing isolate of Spodoptera frugiperda multiple nucleopolyhedrovirus. Journal of General Virology. 2008;89(3):775–790. doi: 10.1099/vir.0.83566-0. [DOI] [PubMed] [Google Scholar]
- 57.Wolff JLC, Valicente FH, Martins R, Oliveira JV, Zanotto PM. Analysis of the genome of Spodoptera frugiperda nucleopolyhedrovirus (SfMNPV-19) and of the high genomic heterogeneity in group II nucleopolyhedroviruses. Journal of General Virology. 2008;89(5):1202–1211. doi: 10.1099/vir.0.83581-0. [DOI] [PubMed] [Google Scholar]
- 58.Pang Y, Yu J, Wang L, et al. Sequence analysis of the Spodoptera litura multicapsid nucleopolyhedrovirus genome. Virology. 2001;287(2):391–404. doi: 10.1006/viro.2001.1056. [DOI] [PubMed] [Google Scholar]
- 59.Willis LG, Siepp R, Stewart TM, Erlandson MA, Theilmann DA. Sequence analysis of the complete genome of Trichoplusia ni single nucleopolyhedrovirus and the identification of a baculoviral photolyase gene. Virology. 2005;338(2):209–226. doi: 10.1016/j.virol.2005.04.041. [DOI] [PubMed] [Google Scholar]
- 60.Wormleaton S, Kuzio J, Winstanley D. The complete sequence of the Adoxophyes orana granulovirus genome. Virology. 2003;311(2):350–365. doi: 10.1016/s0042-6822(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 61.Escasa SR, Lauzon HAM, Mathur AC, Krell PJ, Arif BM. Sequence analysis of the Choristoneura occidentalis granulovirus genome. Journal of General Virology. 2006;87(7):1917–1933. doi: 10.1099/vir.0.81792-0. [DOI] [PubMed] [Google Scholar]
- 62.Lange M, Jehle JA. The genome of the Cryptophlebia leucotreta granulovirus. Virology. 2003;317(2):220–236. doi: 10.1016/s0042-6822(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 63.Luque T, Finch R, Crook N, O’Reilly DR, Winstanley D. The complete sequence of the Cydia pomonella granulovirus genome. Journal of General Virology. 2001;82(10):2531–2547. doi: 10.1099/0022-1317-82-10-2531. [DOI] [PubMed] [Google Scholar]
- 64.Harrison RL, Popham HJR. Genomic sequence analysis of a granulovirus isolated from the Old World bollworm, Helicoverpa armigera. Virus Genes. 2008;36(3):565–581. doi: 10.1007/s11262-008-0218-0. [DOI] [PubMed] [Google Scholar]
- 65.Taha A, Nour-el-Din A, Croizier L, López Ferber M, Croizier G. Comparative analysis of the granulin regions of the Phthorimaea operculella and Spodoptera littoralis granuloviruses. Virus Genes. 2000;21(3):147–155. doi: 10.1023/a:1008179228236. [DOI] [PubMed] [Google Scholar]
- 66.Hashimoto Y, Hayakawa T, Ueno Y, Fujita T, Sano Y, Matsumoto T. Sequence analysis of the Plutella xylostella granulovirus genome. Virology. 2000;275(2):358–372. doi: 10.1006/viro.2000.0530. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Choi JY, Roh JY, Woo SD, Jin BR, Je YH. Molecular and phylogenetic characterization of Spodoptera litura granulovirus. Journal of Microbiology. 2008;46(6):704–708. doi: 10.1007/s12275-008-0133-z. [DOI] [PubMed] [Google Scholar]
- 68.Hayakawa T, Ko R, Okano K, Seong SI, Goto C, Maeda S. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology. 1999;262(2):277–297. doi: 10.1006/viro.1999.9894. [DOI] [PubMed] [Google Scholar]
- 69.Duffy SP, Young AM, Morin B, Lucarotti CJ, Koop BF, Levin DB. Sequence analysis and organization of the Neodiprion abietis nucleopolyhedrovirus genome. Journal of Virology. 2006;80(14):6952–6963. doi: 10.1128/JVI.00187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lauzon HAM, Lucarotti CJ, Krell PJ, Feng Q, Retnakaran A, Arif BM. Sequence and organization of the Neodiprion lecontei nucleopolyhedrovirus genome. Journal of Virology. 2004;78(13):7023–7035. doi: 10.1128/JVI.78.13.7023-7035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lauzon HAM, Garcia-Maruniak A, Zanotto PMdeA, et al. Genomic comparison of Neodiprion sertifer and Neodiprion lecontei nucleopolyhedroviruses and identification of potential hymenopteran baculovirus-specific open reading frames. Journal of General Virology. 2006;87(6):1477–1489. doi: 10.1099/vir.0.81727-0. [DOI] [PubMed] [Google Scholar]
- 72.Garcia-Maruniak A, Maruniak JE, Zanotto PMA, et al. Sequence analysis of the genome of the Neodiprion sertifer nucleopolyhedrovirus. Journal of Virology. 2004;78(13):7036–7051. doi: 10.1128/JVI.78.13.7036-7051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Afonso CL, Tulman ER, Lu Z, et al. Genome sequence of a baculovirus pathogenic for Culex nigripalpus. Journal of Virology. 2001;75(22):11157–11165. doi: 10.1128/JVI.75.22.11157-11165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Evans JT, Leisy DJ, Rohrmann GF. Characterization of the interaction between the baculovirus replication factors LEF-1 and LEF-2. Journal of Virology. 1997;71(4):3114–3119. doi: 10.1128/jvi.71.4.3114-3119.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vanarsdall AL, Okano K, Rohrmann GF. Characterization of the replication of a baculovirus mutant lacking the DNA polymerase gene. Virology. 2005;331(1):175–180. doi: 10.1016/j.virol.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 76.Huang J, Levin DB. Expression, purification and characterization of the Spodoptera littoralis nucleopolyhedrovirus (SpliNPV) DNA polymerase and interaction with the SpliNPV non-hr origin of DNA replication. Journal of General Virology. 2001;82(7):1767–1776. doi: 10.1099/0022-1317-82-7-1767. [DOI] [PubMed] [Google Scholar]
- 77.McDougal VV, Guarino LA. Autographa californica nuclear polyhedrosis virus DNA polymerase: measurements of processivity and strand displacement. Journal of Virology. 1999;73(6):4908–4918. doi: 10.1128/jvi.73.6.4908-4918.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hang X, Guarino LA. Purification of Autographa californica nucleopolyhedrovirus DNA polymerase from infected insect cells. Journal of General Virology. 1999;80(9):2519–2526. doi: 10.1099/0022-1317-80-9-2519. [DOI] [PubMed] [Google Scholar]
- 79.Heldens JGM, Liu Y, Zuidema D, Goldbach RW, Vlak JM. Characterization of a putative Spodoptera exigua multicapsid nucleopolyhedrovirus helicase gene. Journal of General Virology. 1997;78(12):3101–3114. doi: 10.1099/0022-1317-78-12-3101. [DOI] [PubMed] [Google Scholar]
- 80.Maeda S, Kamita SG, Kondo A. Host range expansion of Autographa californica nuclear polyhedrosis virus (NPV) following recombination of a 0.6-kilobase-pair DNA fragment originating from Bombyx mori NPV. Journal of Virology. 1993;67(10):6234–6238. doi: 10.1128/jvi.67.10.6234-6238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ito E, Sahri D, Knippers R, Carstens EB. Baculovirus proteins IE-1, LEF-3, and P143 interact with DNA in vivo: a formaldehyde cross-linking study. Virology. 2004;329(2):337–347. doi: 10.1016/j.virol.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 82.Mcdougal VV, Guarino LA. The Autographa californica nuclear polyhedrosis virus p143 gene encodes a DNA helicase. Journal of Virology. 2000;74(11):5273–5279. doi: 10.1128/jvi.74.11.5273-5279.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bideshi DK, Federici BA. DNA-independent ATPase activity of the Trichoplusia ni granulovirus DNA helicase. Journal of General Virology. 2000;81(6):1601–1604. doi: 10.1099/0022-1317-81-6-1601. [DOI] [PubMed] [Google Scholar]
- 84.Liu GE, Carstens EB. Site-directed mutagenesis of the AcMNPV p 143 gene: effects on baculovirus DNA replication. Virology. 1999;253(1):125–136. doi: 10.1006/viro.1998.9485. [DOI] [PubMed] [Google Scholar]
- 85.Bideshi DK, Federici BA. The Trichoplusia ni granulovirus helicase in unable to support replication of Autographa californica multicapsid nucleopolyhedrovirus in cells and larvae of T. ni. Journal of General Virology. 2000;81(6):1593–1599. doi: 10.1099/0022-1317-81-6-1593. [DOI] [PubMed] [Google Scholar]
- 86.Evans JT, Rosenblatt GS, Leisy DJ, Rohrmann GF. Characterization of the interaction between the baculovirus ssDNA-binding protein (LEF 3) and putative helicase (P143) Journal of General Virology. 1999;80(2):493–500. doi: 10.1099/0022-1317-80-2-493. [DOI] [PubMed] [Google Scholar]
- 87.Argaud O, Croizier L, López-Ferber M, Croizier G. Two key mutations in the host-range specificity domain of the p143 gene of Autographa californica nucleopolyhedrovirus are required to kill Bombyx mori larvae. Journal of General Virology. 1998;79(4):931–935. doi: 10.1099/0022-1317-79-4-931. [DOI] [PubMed] [Google Scholar]
- 88.Kamita SG, Maeda S. Abortive infection of the baculovirus Autographa californica nuclear polyhedrosis virus in Sf-9 cells after mutation of the putative DNA helicase gene. Journal of Virology. 1996;70(9):6244–6250. doi: 10.1128/jvi.70.9.6244-6250.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Croizier G, Croizier L, Argaud O, Poudevigne D. Extension of Autographa californica nuclear polyhedrosis virus host range by interspecific replacement of a short DNA sequence in the p143 helicase gene. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(1):48–52. doi: 10.1073/pnas.91.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu G, Carstens EB. Site-directed mutagenesis of the AcMNPV p 143 gene: effects on baculovirus DNA replication. Virology. 1999;253(1):125–136. doi: 10.1006/viro.1998.9485. [DOI] [PubMed] [Google Scholar]
- 91.Guarino LA, Xu B, Jin J, Dong W. A virus-encoded RNA polymerase purified from baculovirus-infected cells. Journal of Virology. 1998;72(10):7985–7991. doi: 10.1128/jvi.72.10.7985-7991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guarino LA, Jin J, Dong W. Guanylyltransferase activity of the LEF-4 subunit of baculovirus RNA polymerase. Journal of Virology. 1998;72(12):10003–10010. doi: 10.1128/jvi.72.12.10003-10010.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gross CH, Shuman S. RNA 5’-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities of baculovirus LEF-4 protein. Journal of Virology. 1998;72(12):10020–10028. doi: 10.1128/jvi.72.12.10020-10028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jin J, Dong W, Guarino LA. The LEF-4 subunit of baculovirus RNA polymerase has RNA 5’- triphosphatase and ATPase activities. Journal of Virology. 1998;72(12):10011–10019. doi: 10.1128/jvi.72.12.10011-10019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gross CH, Shuman S. Characterization of a baculovirus-encoded RNA 5’-triphosphatase. Journal of Virology. 1998;72(9):7057–7063. doi: 10.1128/jvi.72.9.7057-7063.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Titterington JS, Nun TK, Passarelli AL. Functional dissection of the baculovirus late expression factor-8 gene: sequence requirements for late gene promoter activation. Journal of General Virology. 2003;84(7):1817–1826. doi: 10.1099/vir.0.19083-0. [DOI] [PubMed] [Google Scholar]
- 97.Iorio C, Vialard JE, McCracken S, Lagacé M, Richardson CD. The late expression factors 8 and 9 and possibly the phosphoprotein p78/83 of Autographa californica multicapsid nucleopolyhedrovirus are components of the virus-induced RNA polymerase. Intervirology. 1998;41(1):35–46. doi: 10.1159/000024913. [DOI] [PubMed] [Google Scholar]
- 98.McLachlin JR, Miller LK. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. Journal of Virology. 1994;68(12):7746–7756. doi: 10.1128/jvi.68.12.7746-7756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu A, Miller LK. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. Journal of Virology. 1995;69(2):975–982. doi: 10.1128/jvi.69.2.975-982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Todd JW, Passarelli AL, Lu A, Miller LK. Factors regulating baculovirus late and very late gene expression in transient-expression assays. Journal of Virology. 1996;70(4):2307–2317. doi: 10.1128/jvi.70.4.2307-2317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Passarelli AL, Miller LK. Identification of genes encoding late expression factors located between 56.0 and 65.4 map units of the Autographa californica nuclear polyhedrosis virus genome. Virology. 1993;197(2):704–714. doi: 10.1006/viro.1993.1646. [DOI] [PubMed] [Google Scholar]
- 102.Wilson ME, Miller LK. Changes in the nucleoprotein complexes of a baculovirus DNA during infection. Virology. 1986;151(2):315–328. doi: 10.1016/0042-6822(86)90052-8. [DOI] [PubMed] [Google Scholar]
- 103.Wilson ME, Mainprize TH, Friesen PD, Miller LK. Location, transcription, and sequence of a baculovirus gene encoding a small arginine-rich polypeptide. Journal of Virology. 1987;61(3):661–666. doi: 10.1128/jvi.61.3.661-666.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang M, Tuladhar E, Shen S, et al. Specificity of baculovirus P6.9 basic DNA-binding proteins and critical role of the C terminus in virion formation. Journal of Virology. 2010;84(17):8821–8828. doi: 10.1128/JVI.00072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thiem SM, Miller LK. Identification, sequence, and transcriptional mapping of the major capsid protein gene of the baculovirus Autographa californica nuclear polyhedrosis virus. Journal of Virology. 1989;63(5):2008–2018. doi: 10.1128/jvi.63.5.2008-2018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pearson MN, Russell RLQ, Rohrmann GF, Beaudreau GS. p39, a major baculovirus structural protein: immunocytochemicalcharacterization and genetic location. Virology. 1988;167(2):407–413. [PubMed] [Google Scholar]
- 107.Blissard GW, Quant-Russell RL, Rohrmann GF, Beaudreau GS. Nucleotide sequence, transcriptional mapping, and temporal expression of the gene encoding p39, a major structural protein of the multicapsid nuclear polyhedrosis virus of Orgyia pseudotsugata. Virology. 1989;168(2):354–362. doi: 10.1016/0042-6822(89)90276-6. [DOI] [PubMed] [Google Scholar]
- 108.Lu S, Ge G, Qi Y. Ha-VP39 binding to actin and the influence of F-actin on assembly of progeny virions. Archives of Virology. 2004;149(11):2187–2198. doi: 10.1007/s00705-004-0361-4. [DOI] [PubMed] [Google Scholar]
- 109.McLachlin JR, Miller LK. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. Journal of Virology. 1994;68(12):7746–7756. doi: 10.1128/jvi.68.12.7746-7756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang S, Miller LK. Control of baculovirus polyhedrin gene expression by very late factor 1. Virology. 1998;248(1):131–138. doi: 10.1006/viro.1998.9272. [DOI] [PubMed] [Google Scholar]
- 111.Yang S, Miller LK. Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology. 1998;245(1):99–109. doi: 10.1006/viro.1998.9152. [DOI] [PubMed] [Google Scholar]
- 112.Mikhailov VS, Rohrmann GF. Binding of the baculovirus very late expression factor 1 (VLF-1) to different DNA structures. BMC Molecular Biology. 2002;3 doi: 10.1186/1471-2199-3-14. Article ID 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vanarsdall AL, Okano K, Rohrmann GF. Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing. Journal of Virology. 2006;80(4):1724–1733. doi: 10.1128/JVI.80.4.1724-1733.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mikhailov VS, Okano K, Rohrmann GF. Baculovirus alkaline nuclease possesses a 5′→3′ exonuclease activity and associates with the DNA-binding protein LEF-3. Journal of Virology. 2003;77(4):2436–2444. doi: 10.1128/JVI.77.4.2436-2444.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mikhailov VS, Okano K, Rohrmann GF. Specificity of the endonuclease activity of the baculovirus alkaline nuclease for single-stranded DNA. Journal of Biological Chemistry. 2004;279(15):14734–14745. doi: 10.1074/jbc.M311658200. [DOI] [PubMed] [Google Scholar]
- 116.Okano K, Vanarsdall AL, Rohrmann GF. Characterization of a baculovirus lacking the alkaline nuclease gene. Journal of Virology. 2004;78(19):10650–10656. doi: 10.1128/JVI.78.19.10650-10656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Olszewski J, Miller LK. Identification and characterization of a baculovirus structural protein, VP1054, required for nucleocapsid formation. Journal of Virology. 1997;71(7):5040–5050. doi: 10.1128/jvi.71.7.5040-5050.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Russell RLQ, Rohrmann GF. Characterization of P91, a protein associated with virions of an Orgyia pseudotsugata baculovirus. Virology. 1997;233(1):210–223. doi: 10.1006/viro.1997.8599. [DOI] [PubMed] [Google Scholar]
- 119.Whitford M, Faulkner P. A structural polypeptide of the baculovirus Autographa californica nuclear polyhedrosis virus contains O-linked N-acetylglucosamine. Journal of Virology. 1992;66(6):3324–3329. doi: 10.1128/jvi.66.6.3324-3329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Olszewski J, Miller LK. A role for baculovirus GP41 in budded virus production. Virology. 1997;233(2):292–301. doi: 10.1006/viro.1997.8612. [DOI] [PubMed] [Google Scholar]
- 121.Wu W, Lin T, Pan L, et al. Autographa californica multiple nucleopolyhedrovirus nucleocapsid assembly is interrupted upon deletion of the 38K gene. Journal of Virology. 2006;80(23):11475–11485. doi: 10.1128/JVI.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu W, Liang H, Kan J, et al. Autographa californica multiple nucleopolyhedrovirus 38K is a novel nucleocapsid protein that interacts with VP1054, VP39, VP80, and itself. Journal of Virology. 2008;82(24):12356–12364. doi: 10.1128/JVI.00948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Long CM, Rohrmann GF, Merrill GF. The conserved baculovirus protein p33 (Ac92) is a flavin adenine dinucleotide-linked sulfhydryl oxidase. Virology. 2009;388(2):231–235. doi: 10.1016/j.virol.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 124.Wu W, Passarelli AL. Autographa californica multiple nucleopolyhedrovirus Ac92 (ORF92, P33) is required for budded virus production and multiply enveloped occlusion-derived virus formation. Journal of Virology. 2010;84(23):12351–12361. doi: 10.1128/JVI.01598-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nie Y, Fang M, Theilmann DA. Autographa californica multiple nucleopolyhedrovirus core gene ac92 (p33) is required for efficient budded virus production. Virology. 2011;409(1):38–45. doi: 10.1016/j.virol.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 126.Fang M, Wang H, Wang H, et al. Open reading frame 94 of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus encodes a novel conserved occlusion-derived virion protein, ODV-EC43. Journal of General Virology. 2003;84(11):3021–3027. doi: 10.1099/vir.0.19291-0. [DOI] [PubMed] [Google Scholar]
- 127.Deng F, Wang R, Fang M, et al. Proteomics analysis of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus identified two new occlusion-derived virus-associated proteins, HA44 and HA100. Journal of Virology. 2007;81(17):9377–9385. doi: 10.1128/JVI.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peng KE, Wu M, Deng F, et al. Identification of protein-protein interactions of the occlusion-derived virus-associated proteins of Helicoverpa armigera nucleopolyhedrovirus. Journal of General Virology. 2010;91(3):659–670. doi: 10.1099/vir.0.017103-0. [DOI] [PubMed] [Google Scholar]
- 129.Vanarsdall AL, Pearson MN, Rohrmann GF. Characterization of baculovirus constructs lacking either the Ac 101, Ac 142, or the Ac 144 open reading frame. Virology. 2007;367(1):187–195. doi: 10.1016/j.virol.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li G, Wang J, Deng R, Wang X. Characterization of AcMNPV with a deletion of ac68 gene. Virus Genes. 2008;37(1):119–127. doi: 10.1007/s11262-008-0238-9. [DOI] [PubMed] [Google Scholar]
- 131.McCarthy CB, Theilmann DA. AcMNPV ac143 (odv-e18) is essential for mediating budded virus production and is the 30th baculovirus core gene. Virology. 2008;375(1):277–291. doi: 10.1016/j.virol.2008.01.039. [DOI] [PubMed] [Google Scholar]
- 132.Ke J, Wang J, Deng R, Wang X. Autographa californica multiple nucleopolyhedrovirus ac66 is required for the efficient egress of nucleocapsids from the nucleus, general synthesis of preoccluded virions and occlusion body formation. Virology. 2008;374(2):421–431. doi: 10.1016/j.virol.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 133.Belyavskyi M, Braunagel SC, Summers MD. The structural protein ODV-EC27 of Autographa californica nucleopolyhedrovirus is a multifunctional viral cyclin. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(19):11205–11210. doi: 10.1073/pnas.95.19.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Braunagel SC, He H, Ramamurthy P, Summers MD. Transcription, translation, and cellular localization of three Autographa californica nuclear polyhedrosis virus structural proteins: ODV-E18, ODV-E35, and ODV-EC27. Virology. 1996;222(1):100–114. doi: 10.1006/viro.1996.0401. [DOI] [PubMed] [Google Scholar]
- 135.Chen HQ, Chen KEP, Yao Q, Guo ZJ, Wang LL. Characterization of a late gene, ORF67 from Bombyx mori nucleopolyhedrovirus. FEBS Letters. 2007;581(30):5836–5842. doi: 10.1016/j.febslet.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 136.Simón O, Gutiérrez S, Williams T, Caballero P, López-Ferber M. Nucleotide sequence and transcriptional analysis of the pif gene of Spodoptera frugiperda nucleopolyhedrovirus (SfMNPV) Virus Research. 2005;108(1-2):213–220. doi: 10.1016/j.virusres.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 137.Pijlman GP, Pruijssers AJP, Vlak JM. Identification of pif-2, a third conserved baculovirus gene required for per os infection of insects. Journal of General Virology. 2003;84(8):2041–2049. doi: 10.1099/vir.0.19133-0. [DOI] [PubMed] [Google Scholar]
- 138.Faulkner P, Kuzio J, Williams GV, Wilson JA. Analysis of p74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. Journal of General Virology. 1997;78(12):3091–3100. doi: 10.1099/0022-1317-78-12-3091. [DOI] [PubMed] [Google Scholar]
- 139.Zhou W, Yao L, Xu H, Yan F, Qi Y. The function of envelope protein p74 from Autographa californica multiple nucleopolyhedrovirus in primary infection to host. Virus Genes. 2005;30(2):139–150. doi: 10.1007/s11262-004-5623-4. [DOI] [PubMed] [Google Scholar]
- 140.Haas-Stapleton EJ, Washburn JO, Volkman LE. P74 mediates specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to primary cellular targets in the midgut epithelia of Heliothis virescens larvae. Journal of Virology. 2004;78(13):6786–6791. doi: 10.1128/JVI.78.13.6786-6791.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yao L, Zhou W, Xu H, Zheng Y, Qi Y. The Heliothis armigera single nucleocapsid nucleopolyhedrovirus envelope protein P74 is required for infection of the host midgut. Virus Research. 2004;104(2):111–121. doi: 10.1016/j.virusres.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 142.Braunagel SC, Russell WK, Rosas-Acosta G, Russell DH, Summers MD. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(17):9797–9802. doi: 10.1073/pnas.1733972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kikhno I, Gutiérrez S, Croizier L, Crozier G, López Ferber M. Characterization of pif, a gene required for the per os infectivity of Spodoptera littoralis nucleopolyhedrovirus. Journal of General Virology. 2002;83(12):3013–3022. doi: 10.1099/0022-1317-83-12-3013. [DOI] [PubMed] [Google Scholar]
- 144.Ohkawa T, Washburn JO, Sitapara R, Sid E, Volkman LE. Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. Journal of Virology. 2005;79(24):15258–15264. doi: 10.1128/JVI.79.24.15258-15264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fang M, Nie Y, Harris S, Erlandson MA, Theilmann DA. Autographa californica multiple nucleopolyhedrovirus core gene ac96 encodes a per os infectivity factor (pif-4) Journal of Virology. 2009;83(23):12569–12578. doi: 10.1128/JVI.01141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Braunagel SC, Elton DM, Ma H, Summers MD. Identification and analysis of an Autographa californica nuclear polyhedrosis virus structural protein of the occlusion-derived virus envelope: ODV-E56. Virology. 1996;217(1):97–110. doi: 10.1006/viro.1996.0097. [DOI] [PubMed] [Google Scholar]
- 147.Sparks WO, Harrison RL, Bonning BC. Autographa californica multiple nucleopolyhedrovirus ODV-E56 is a per os infectivity factor, but is not essential for binding and fusion of occlusion-derived virus to the host midgut. Virology. 2011;409(1):69–76. doi: 10.1016/j.virol.2010.09.027. [DOI] [PubMed] [Google Scholar]
- 148.Cohen DPA, Marek M, Davies BG, Vlak JM, van Oers MM. Encyclopedia of Autographa californica nucleopolyhedrovirus genes. Virologica Sinica. 2009;24(5):359–414. [Google Scholar]
- 149.Jiang Y, Deng F, Rayner S, Wang H, Hu Z. Evidence of a major role of GP64 in group I alphabaculovirus evolution. Virus Research. 2009;142(1-2):85–91. doi: 10.1016/j.virusres.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 150.Herniou EA, Jehle JA. Baculovirus phylogeny and evolution. Current Drug Targets. 2007;8(10):1043–1050. doi: 10.2174/138945007782151306. [DOI] [PubMed] [Google Scholar]
- 151.de Andrade Zanotto PM, Krakauer DC. Complete genome viral phylogenies suggests the concerted evolution of regulatory cores and accessory satellites. PLoS ONE. 2008;3(10) doi: 10.1371/journal.pone.0003500. Article ID e3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Goodman D, Ollikainen N, Sholley C. Baculovirus phylogeny based on genome rearrangements. In: Proceedings of the International Conference on Comparative Genomics, vol. 4751; 2007; pp. 69–82. Lecture Notes in Computer Science. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplementary text: explains in detail alternative bioinformatic approaches used to validate the recognition of core genes. It also contains a detailed table showing the numbers of ORF homologous within the family Baculoviridae.