Summary
Nucleosome-depleted-regions (NDRs) are ubiquitous on eukaryotic promoters. The formation of many NDRs can not be readily explained by previously proposed mechanisms. Here, we carry out a focused study on a physiologically important NDR in the yeast CLN2 promoter (CLN2pr). We show that this NDR does not result from intrinsically unfavorable histone-DNA interaction. Instead, we identified eight conserved factor binding sites, including that of Reb1, Mcm1 and Rsc3, that cause the local nucleosome depletion. These nucleosome depleting factors (NDFs) work redundantly, and simultaneously mutating all their binding sites eliminates CLN2pr NDR. The loss of the NDR induces unreliable “on / off” expression in individual cell cycles, but in the presence of the NDR, NDFs have little direct effect on transcription. We present bioinformatic evidence that the formation of many NDRs across the genome involve multiple NDFs. Our findings also provide significant insight into the composition and spatial organization of functional promoters.
Introduction
Genome-wide nucleosome mapping in budding yeast revealed a dominant promoter configuration: immediately upstream of the transcription start site (TSS), there is a stretch of DNA (on average ~150 bp) with very low nucleosome occupancy (nucleosome-depleted-regions, NDRs); while downstream of the TSS, there is nucleosome-rich-region containing highly-occupied and well-phased nucleosomes (Field et al., 2008; Kaplan et al., 2009; Lee et al., 2007; Mavrich et al., 2008a; Shivaswamy et al., 2008; Yuan et al., 2005). NDRs are detected in >90% of yeast promoters, and regulatory regions in higher eukaryotes (including drosophila and human) are also frequently nucleosome-deficient (Mavrich et al., 2008b; Schones et al., 2008).
Many transcription factor binding sites reside in NDRs, and such localization is likely to facilitate the binding of these factors (Morse, 2007). Change in the nucleosomal coverage of factor binding sites was shown to significantly affect the average level, cell-to-cell variability and dynamics of gene expression (Bai et al., 2010; Field et al., 2008; Floer et al., 2010; Lin et al., 2007; Tirosh and Barkai, 2008), as well as its sensitivity to activation signal and dynamic range (Lam et al., 2008). NDRs are also critical for the binding of the transcription machinery (Adkins et al., 2004; Zhang and Reese, 2007). In addition, it was proposed that NDRs affect the selection of start / termination site of transcription and define the boundary of polII’s promoter-proximal pausing (Jiang and Pugh, 2009). Consistent with their important function in transcription, NDRs are dynamically regulated during cell differentiation (Woo et al., 2010), and an abnormal NDR was found in the promoter of a tumor-supressor gene, MLH1, in cancer cells (Lin et al., 2007).
NDRs may have additional functions. NDRs were found to be highly enriched at replication origins, which may allow more efficient assembly of replication machinery (Gerbi and Bielinsky, 2002). Indeed, there is evidence that the efficiency of origin firing is correlated with the corresponding nucleosome density (Field et al., 2008). NDRs also contribute to the structural organization of chromatin. For instance, NDRs dictate the distribution of a histone variant, H2A.Z, which preferentially occupies the regions flanking the NDRs (Albert et al., 2007; Jin et al., 2009; Raisner et al., 2005). Nucleosomes in the vicinity of NDRs seem to be more precisely positioned than the distant ones, indicating that NDRs may serve as “barriers” to constrain the locations of neighboring nucleosomes. This so called “statistical positioning” was proposed to be one of the mechanisms that determine the nucleosome positioning genome-wide (Kornberg, 1981; Mavrich et al., 2008a).
Because of their functional and physiological significance, it is very important to understand how NDRs are generated and maintained. One theory is that NDRs are primarily determined by low intrinsic DNA-histone affinity (Segal et al., 2006). In support of this idea, many NDRs contain sequences that disfavor nucleosome formation (such as the relatively rigid polyA/T stretch), and NDRs observed in vivo can be partially reproduced by chromatin assembled in vitro (Kaplan et al., 2009; Zhang et al., 2009). However, there are substantial differences between the in vivo and in vitro nucleosome distribution, and in particular, the NDRs are less pronounced in the latter. Therefore, intrinsic DNA-histone affinity only contributes partially to NDR formation, and the importance and necessity of this contribution requires further elucidation.
There is evidence that certain DNA-binding factors and chromatin remodeling factors participate in generating NDRs. Factors such as Abf1 and Reb1 could antagonize nucleosome formation in the vicinity of their binding sites (De Winde et al., 1993; Fedor et al., 1988; Venditti et al., 1994). Consistently, the binding sites of Abf1 and Reb1 have the largest discrepancy between their in vivo and in vitro nucleosome occupancy (Kaplan et al., 2009). Several other essential DNA-binding factors, including Rap1 and Mcm1, may also have roles in nucleosome depletion, although their influence seems to be weaker (Badis et al., 2008; Yarragudi et al., 2004). A remodeling factor, RSC, was proposed to directly bind to DNA through its subunit Rsc3 and remove nucleosomes (Badis et al., 2008; Hartley and Madhani, 2009). Deleting one of the factors mentioned above increases nucleosome occupancy in a fraction of NDRs genome-wide (Badis et al., 2008; Hartley and Madhani, 2009). However, not all NDRs are affected by these deletions, and among the affected ones, the deletion tends to shrink the size of the NDR instead of completely eliminating it. It was proposed that the residual NDR could be caused by unfavorable intrinsic DNA / histone affinity, but it is also possible that multiple factors participate in the formation of a single NDR. A focused mutational analysis of the requirements for nucleosome depletion at specific NDRs will be required to resolve these issues.
In our previous work, we identified an NDR on a cell-cycle-regulated promoter, CLN2pr, in Saccharomyces cerevisiae (Figure 1A) (Bai et al., 2010). CLN2pr is activated by the transcription factor complex SBF (and possibly a close homolog, MBF), which is only active within a brief time window early in a cell cycle (Cross et al., 1994; Stuart and Wittenberg, 1994). Comparing with the more typical NDRs found on yeast promoters, which locate immediately upstream the TSS with an average length of ~150 bp, the CLN2pr NDR extends ~300 bp and its center is ~400 bp upstream the TSS, a configuration common to more dynamic promoters (Tirosh and Barkai, 2008). This NDR contains three SBF binding sites (SCBs), and such localization is critical to ensure reliable, once-per-cycle gene expression (Bai et al., 2010). Interestingly, this NDR does not contain a long polyA/T stretch and has relatively high G/C content, features that promote nucleosome formation (Iyer and Struhl, 1995; Tillo and Hughes, 2009). Besides SBF / MBF, the only factor known to bind the CLN2 NDR is Rme1 (Toone et al., 1995), and none of these factors have known function in nucleosome depletion. Badis et al. (2008) showed that CLN2 NDR is intact in the absence of Abf1, Reb1, Rap1, Tbf1 or Rsc3 (Figure S1). Therefore, the CLN2pr NDR is presently not explicable by any known NDR-promoting mechanisms. Here, we determine the contribution of intrinsic DNA-histone affinity and specific DNA-protein interactions in generating the CLN2pr NDR, investigate the functional consequences of NDR deficiency for CLN2 expression, and carry out bioinformatic analysis to establish the likely generality of our findings for many promoters across the yeast genome.
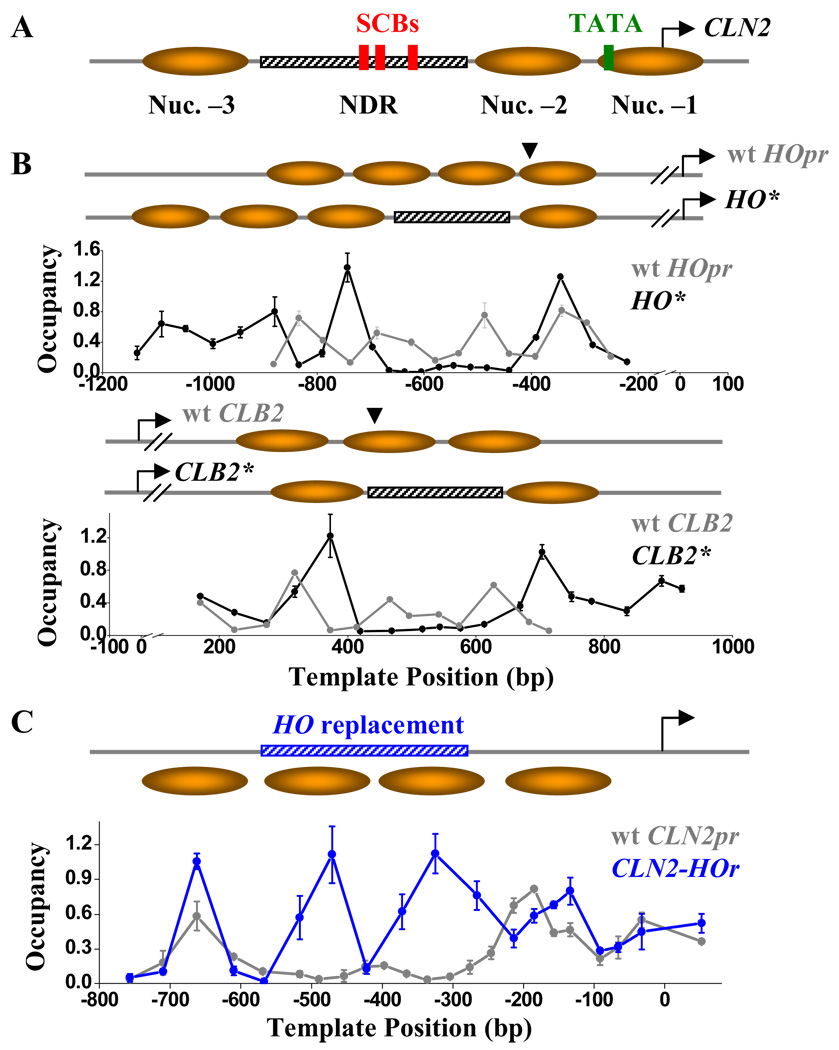
Figure 1. CLN2pr NDR is a property of local DNA sequence.
A) Genomic structure and nucleosome distribution on wt CLN2pr. The CLN2pr contains three well-positioned nucleosomes (shaded ovals) and NDR (hatched bar). The position of SCBs (red rectangles), TATA box (yellow rectangle) and TSS (arrow) are also shown. B) Nucleosome occupancy on the HO* (upper panel) and CLB2* (lower panel), as well as the inferred nucleosome positioning. HO* / CLB2* are constructed by inserting the CLN2pr NDR sequence into the HOpr / CLB2 ORF at the sites indicated by the arrows. The nucleosome occupancy on wt HOpr and CLB2 ORF are also shown in the corresponding panels. The position “0” on the plot represents the translation start site of HO and CLB2 gene. C) Nucleosome occupancy on the CLN2-HOr (blue curve) in comparison with that on the wt CLN2pr (gray). CLN2-HOr is a CLN2pr variant with the NDR replaced by a HOpr sequence (blue hatched bar). The position “0” on the plot represents the CLN2pr TSS (same for below).
Results
Sequences within the CLN2pr NDR are necessary and sufficient for nucleosome depletion
The NDR mechanisms proposed previously implicate that NDR is determined locally, either by its DNA mechanical property or by factors bound within the NDR. However, some chromatin structural features can be dictated through long-range interaction (e.g. silencing), rather than by the very local sequences. If this were the case, the sequence from the CLN2pr NDR might no longer be nucleosome-free when transplanted into another locus in the genome.
To test this possibility, we inserted the CLN2pr NDR sequence into two genomic loci that are originally nucleosomal (Figure 1B): −395 in the HOpr and +439 in the CLB2 ORF (HO* and CLB2*). The inserted CLN2pr sequence generates prominent NDRs in both of the new loci (Figure 1B). Conversely, replacing the CLN2pr NDR in the CLN2 locus with a HOpr sequence of the same length (CLN2-HOr, standing for CLN2-HO replacement) completely eliminates the NDR (Figure 1C). These results clearly show that nucleosome-depletion is determined by the sequence within the CLN2pr NDR, even when transplanted to different regulatory regions or ORFs that originally carried different histone variants and modifications (Albert et al., 2007; Pokholok et al., 2005).
Histone-DNA affinity is unlikely to be responsible for the CLN2pr NDR formation
To test whether the CLN2pr NDR sequence excludes nucleosome through unfavorable intrinsic DNA-histone interaction, we performed in vitro nucleosome assembly at a 1:1 histone:DNA ratio. We amplified a 2.1 kb genomic DNA fragment with the NDR sequence in the middle and assembled purified chicken histone onto it using salt dialysis, which allows nucleosomes to form at their most energetically favorable locations. We also performed in vitro assembly on the same template using purified Nap1 / ACF (Ito et al., 1997). These two methods for nucleosome assembly reflect thermodynamically or enzymatically favored sites for nucleosome assembly, and therefore should indicate whether the CLN2pr NDR is intrinsically antagonistic to nucleosome formation.
The two methods generated similar nucleosome distributions (Figure 2A,B), which are consistent with previously reported genome-wide in vitro assembly data in Kaplan et al. (2009) and Zhang et al. (2009), the former done at even lower histone:DNA ratios. Importantly, in vitro assembly in all of these studies completely fails to reproduce the in vivo CLN2pr NDR: this sequence is assembled into nucleosomes in vitro with density comparable to flanking sequences. We also used the mathematical model reported in Kaplan et al. (2009) to predict the nucleosome positioning on our sequence, and the prediction agrees well with the in vitro data but not with the in vivo data (Figure 2C). This suggests that the model does reflect intrinsic DNA-nucleosome affinity, but that this affinity cannot explain the CLN2pr NDR.
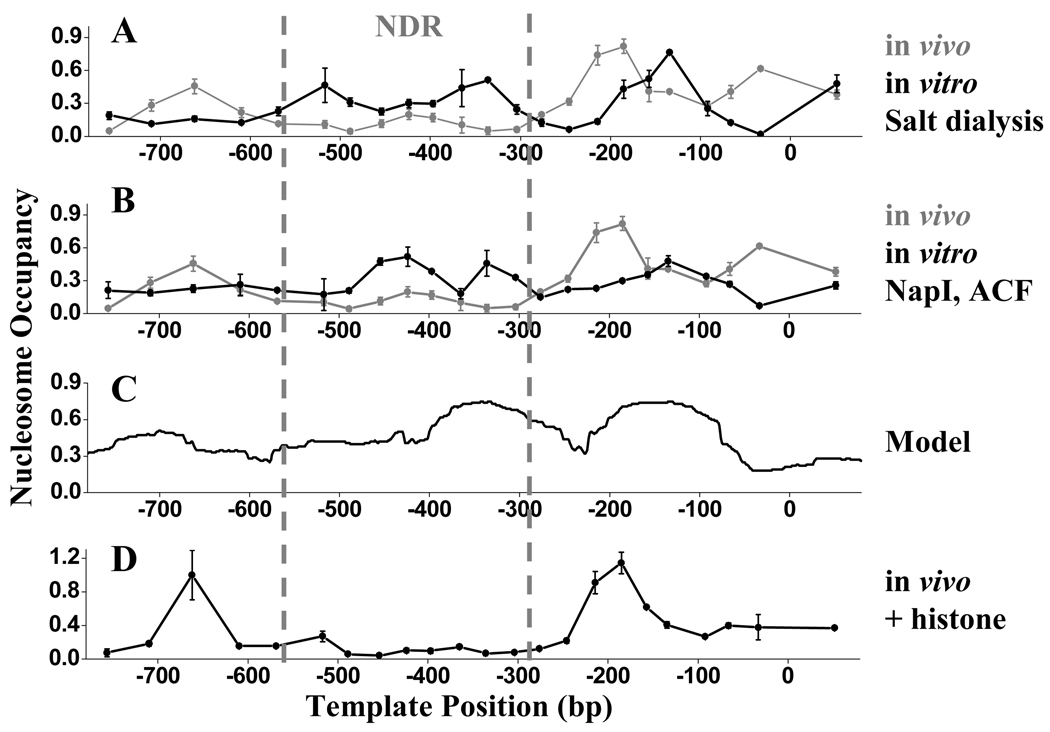
Figure 2. Histone-DNA affinity is unlikely to be responsible for the CLN2pr NDR formation.
A, B) In vivo (gray) and in vitro (black) nucleosome occupancy on the CLN2pr. The in vitro nucleosome assembly is carried out either by salt dialysis (A) or with purified NapI and ACF (B). C) Nucleosome occupancy on the CLN2pr predicted by a mathematical model from Kaplan et al. (2009). D) In vivo nucleosome occupancy on the CLN2pr in a strain with over-expressed histones.
If weak DNA-histone interaction leads to the formation of NDR, it may be compensated by higher histone concentration. Therefore as an independent test, we constructed a strain with over-expressed histones using high copy plasmid (pCC67) containing both HTA / HTB and HH3 / HH4 gene pair, which was shown to increase the average histone concentration by 3–5 fold (Clark-Adams et al., 1988). The nucleosome distribution on the CLN2pr did not change significantly in this strain. Particularly, the pronounced NDR was unaffected by excess histone (Figure 2D). All the evidence above argues against the notion that histone-DNA affinity is the key factor in determining the CLN2pr NDR.
Multiple sequence segments in CLN2pr NDR cause local nucleosome-depletion
Given the observations above, CLN2pr NDR is most likely generated by locally bound factors. As mentioned before, the factors that have been mapped to CLN2pr NDR, SBF / MBF and Rme1, have no known function in nucleosome depletion. In addition, both SBF / MBF and Rme1 binding are cell-cycle regulated (Frenz et al., 2001), whereas the CLN2pr NDR is constitutive (Bai et al., 2010). In a swi4−mbp1− double deletion strain in which SBF / MBF binding is completely abolished (Koch et al., 1993), the CLN2pr NDR remains intact (Figure S2A). Conversely, the ~10 copies of SBF binding sites in HOpr are unable to generate NDR (Takahata et al., 2009).
To start an unbiased search for factors that contribute to CLN2pr NDR formation, we divided the NDR sequence into four segments (Seg1–4), each ~80 bp in length, and deleted one segment at a time (Figure S2B). The remaining NDR sequence would be longer than 200 bp, more than enough to accommodate one nucleosome. If the deleted segment contains the key factor binding site(s) responsible for the NDR formation, we expect to see the rest of the NDR sequence become nucleosomal. Surprisingly, no matter which segment we deleted, the other three segments always remained nucleosome-free (Figure S2B).
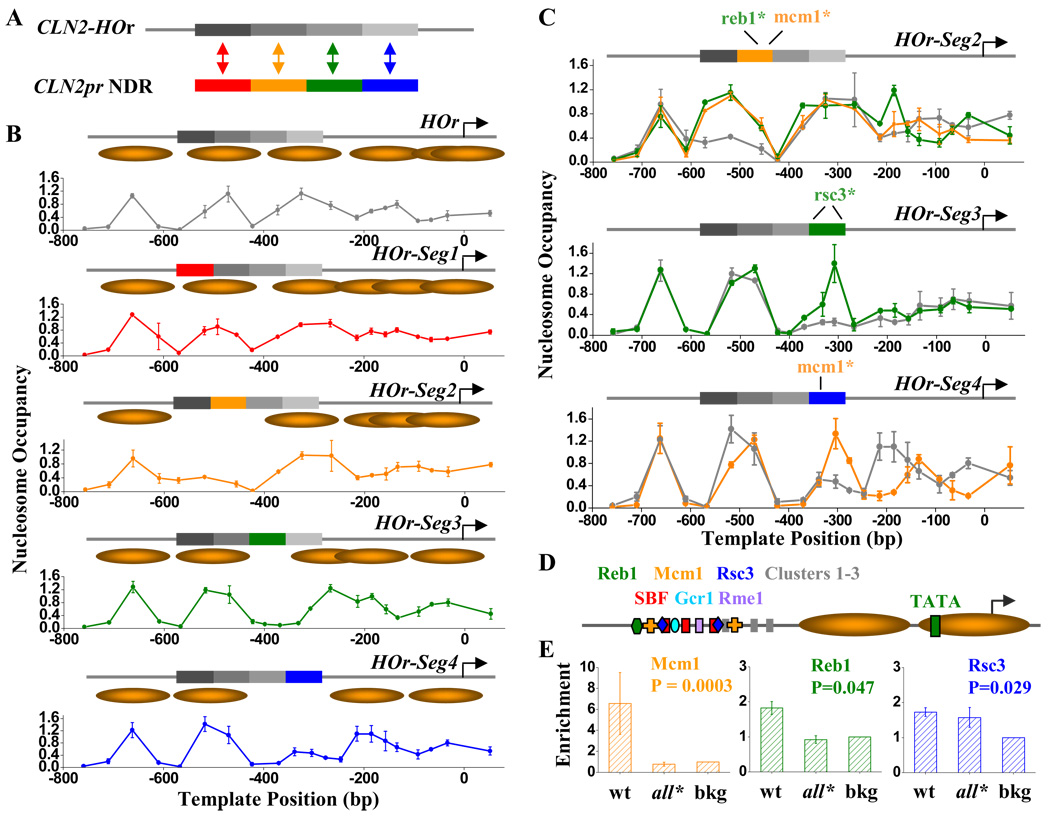
The results above suggested that there may be multiple factors bound on different segments, all of which lead to NDR formation. To test this idea, we constructed another series of promoter variants: we started from the CLN2-HOr sequence (in which the CLN2pr NDR sequence is replaced by normally nucleosome-covered DNA from the HO promoter; Figure 1C), and replaced the four CLN2pr NDR segments back, one at a time (HOr-Seg1–4; Figure 3A). Segments 2, 3 and 4, but not segment 1, formed short local NDRs (Figure 3B). Again, this result strongly suggests that CLN2pr NDR is not generated by a single factor. Instead, it is likely “coated” with many factors, and each of them contributes to the local nucleosome-depletion.
Figure 3. Multiple sequence-specific factors are responsible for nucleosome-depletion on CLN2pr NDR.
A) Construction of the HOr-Seg1–4 sequences. We divided the CLN2pr NDR sequence into 4 segments, each ~80 bp in length, and replaced them back into the CLN2-HOr (Figure 1C), one at a time. B) Nucleosome occupancy and inferred positioning on CLN2-HOr (repeated from Figure 1C) and HOr-Seg1–4 Seg2–4 each independently forms local NDRs. C) The measured nucleosome occupancy on the HOr-Seg2–4 sequences with mutated (colored curves) or wt Seg2–4 (gray curves). Mutations in the Reb1 or Mcm1 binding sites in the Seg2 (top panel), the two Rsc3 binding sites on Seg3 (middle panel), and the Mcm1 binding site on Seg4 (bottom panel), eliminates the local NDR. See Table S1 for detailed sequence / factor binding sites / mutation information. D) Summary for all the potential factor binding sites on CLN2pr NDR. The binding sites listed on the top row, but not the bottom row, contribute to the NDR formation. Cluster 1–3 represent three conserved sequence elements on Seg4 that lower the local nucleosome density, but the bound factors are yet to be identified (see text for details). E) ChIP measurement of the enrichment of Reb1, Mcm1 and Rsc3 on the wt CLN2pr and CLN2pr-all* (CLN2pr with mutations in the binding sites of Reb1, Mcm1, Rsc3 and Cluster 1–3). The enrichment is normalized to the background level (bkg). The binding of all three factors to the wt CLN2pr are significantly above the bkg (P value shown in panels), whereas Reb1 and Mcm1 binding to the CLN2pr-all* are eliminated.
Factor binding sites important for NDR formation on Seg2–4
To reduce the complexity generated by multiple factors, we looked for sequence elements critical for NDR formation on individual segments 2, 3 and 4. We scanned through the sequences for known factor binding sites and searched in literature for factors either CHIP to the CLN2 locus or functionally linked to Cln2 expression. We also examined sequence conservation among closely related yeast species with the rationale that the binding sites for factors functioning in NDR formation are likely to be more conserved than the neighboring neutral sequences. Consistent with this idea, Seg1, which has the least effect on nucleosome depletion, is the least conserved among the four segments (Table S1). After identifying the candidate binding sites, we performed mutational analysis to determine their function in NDR formation.
On Seg2 there are two well-conserved sequence clusters, which map to the binding motifs of Reb1 and Mcm1 respectively (Table S1). Mutations on either the Reb1 or Mcm1 binding site, each by 2–3 bases, completely eliminated the NDR on Seg2 (Figure 3C), strongly indicating that both Reb1 and Mcm1 bind to Seg2 and contribute to the local nucleosome depletion.
Seg3 is well conserved throughout. Besides the known SBF and Rme1 binding sites, sequence analysis also revealed conserved Gcr1 and Rsc3 consensus sites (Table S1). Gcr1 has been proposed to promote Cln2 expression (Willis et al., 2003) and RSC complex was found to associate with nucleosome –2 on CLN2pr (Floer et al., 2010). In Figure 3B, the NDR generated by Seg3 is rather short and can be confused with regular linker DNA. To avoid this problem, we moved Seg3 downstream so that it no longer borders the upstream nucleosome. At this new location, wt Seg3 causes strong, unambiguous nucleosome-depletion (gray curve in Figure 3C middle panel). In contrast, Seg3 with mutations of the two Rsc3 sites is nucleosomal. Mutations in the Gcr1 / Rme1 or SBF binding sites, however, have little effect on nucleosome density (Figure S2C). Therefore, RSC is likely to be primarily responsible for the NDR formation on Seg3. This result also confirms that not all DNA binding factors have significant nucleosome depletion activity.
Seg4 contains a conserved Mcm1 binding site (Table S1), and similar to our observation on Seg2, a 2 bp mutation in this Mcm1 site completely eliminates the NDR (Figure 3C). There are other highly conserved sequence clusters on Seg4, which are potential binding sites for unidentified factors (Clusters 1–3; Table S1). Mutations in one of these three clusters increase the local nucleosome density to an intermediate level, while mutations in two clusters (such as cluster 2 and 3) bring a nucleosome onto the Seg4 with full occupancy (Figure S2D). These observations are consistent with the scenario that Clusters 1–3 are bound by factors weakly antagonizing nucleosome formation, and the simultaneous presence of all these factors and Mcm1 are required to achieve significant nucleosome depletion on Seg4.
Reb1, Mcm1 and Rsc3 bind physically to CLN2pr NDR
The experiments above identified potential Reb1, Mcm1 and Rsc3 binding sites in the CLN2pr NDR (Figure 3D). Previous genome-wide ChIP studies on Reb1 and RSC (Ng et al., 2002) complex did not find significant binding of these factors in the CLN2pr region, and there are conflicting results with respect to Mcm1 binding (Harbison et al., 2004; Workman et al., 2006). Genome-wide ChIP experiments tend to be unreliable and different datasets are often inconsistent with each other (Bean et al., 2005), so we performed a small scale ChIP experiment to specifically look for these factors on the CLN2pr.
Using strains expressing myc-tagged Reb1, Mcm1 and Rsc3, we measured the binding of these factors on the wt CLN2pr, as well as several randomly selected genomic loci as our background. All of the three factors were shown to be significantly more enriched on the CLN2pr than the background (P value <0.05; Figure 3E). The fold enrichment observed for Reb1 and Rsc3 was small, but typical of the fold enrichment observed in published genome-wide ChIP experiments. This likely reflects relatively low but still authentic specificity of binding of these factors. Therefore, our result is consistent with recent finding that low affinity sites for NDF can also contribute to nucleosome depletion (Goh et al., 2010; Ganapathi et al., 2010).
To additionally show that the binding of the factors is specific to the predicted binding sites in the CLN2pr NDR, we constructed a mutated version of CLN2pr by simultaneously disrupting all the sequence elements contributing to NDR formation, including the Reb1, Mcm1, Rsc3 sites and Cluster 1–3 (CLN2pr-all*), and carried out the same ChIP measurement. Binding of Reb1 and Mcm1 to CLN2pr-all* was reduced to background (Figure 3E). However, binding of Rsc3 to CLN2pr-all* was not reduced significantly. This could be due to other mechanisms or sites for Rsc3 binding, such as the previously reported binding of Rsc3 to the nucleosome −2 on the CLN2pr (Floer et al., 2010). Overall, these results strongly support that Reb1, Mcm1 and probably Rsc3 bind to the identified binding sites in the CLN2pr NDR.
The formation of NDR in the context of wt CLN2pr
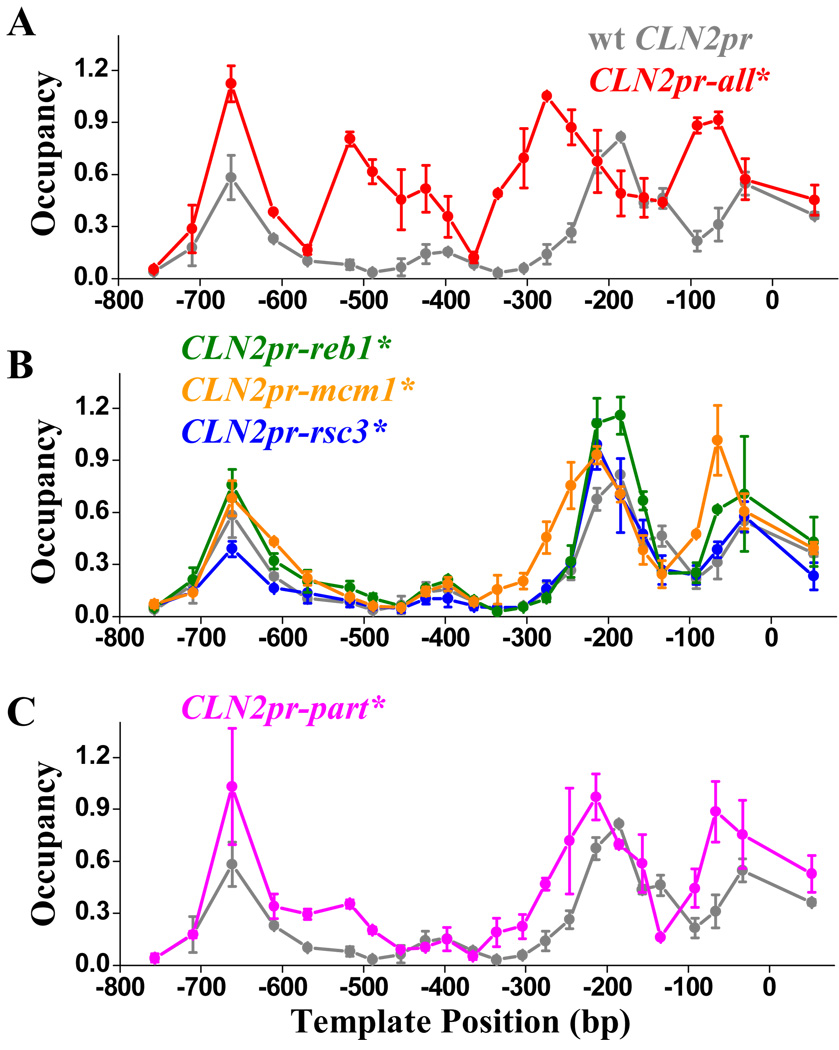
In the experiments above, we studied local NDR formation on individual CLN2pr NDR segments adjacent to HOpr sequence, in order to isolate the effects of individual binding sites. We next investigated NDR formation in the context of the full-length CLN2pr. Figure 4A shows that the simultaneous disruption of Reb1, Mcm1 and Rsc3 binding sites together with Cluster 1–3 (CLN2pr-all*) completely eliminates the ~300 bp NDR, confirming that these factors are jointly responsible for the CLN2pr NDR formation. To address the possible redundancy of these factor binding sites, we generated mutant CLN2 promoters with subsets of factor binding sites inactivated. Figure 4B shows that NDR remains intact when the binding site(s) of Reb1 or Rsc3 are mutated (CLN2pr-reb1* and -rsc3*), and mutations in the two Mcm1 binding sites only slightly shrank the NDR (CLN2pr-mcm1*). Disruption of a larger subset (Reb1 + Mcm1 + Clusters 1–3; CLN2pr-part*) results in nucleosome invasion from both the upstream and downstream end, but the remaining Rsc3 sites keep the middle part of NDR nucleosome-free (Figure 4C). This reflects significant redundancy of these factors in NDR formation. As a result, the wt CLN2pr NDR is much more robust against sequence perturbation than its individual sub-segments.
Figure 4. The NDR formation in the context of wt CLN2pr.
A–C) The measured nucleosome occupancy on the CLN2pr-all* (reb1* + mcm1* + rsc3* + cluster 1–3*) (A), CLN2pr-reb1*, -mcm1*, -rsc3* (B) and CLN2pr-part* (reb1* + mcm1* + cluster 1–3*) (C) vs. that on wt CLN2pr (gray curves in A–C). Simultaneous mutations in the Reb1, Mcm1, Rsc3 binding sites and Clusters 1–3 eliminates NDR (CLN2pr-all*); mutations in a subset of the binding sites either have no apparent effect on CLN2pr NDR (CLN2pr-reb1* and -rsc3*), or mildly shrink the NDR (CLN2pr-mcm1* and -part*).
If we compare the nucleosome distribution on CLN2pr in close yeast species (Tsankov et al., 2010), the highly conserved region close to the SCBs are always nucleosome-depleted. In contrast, the upstream part of the NDR with low sequence conservation becomes more and more nucleosomal in more distant species (Figure S3). This could be due to a change in the intrinsic DNA property (e.g. the GC content of this region gradually increases from 37% in S.cer to 53% in S.bay), and / or that this region in S.cer CLN2pr contains some weak and unconserved nucleosome-repelling elements.
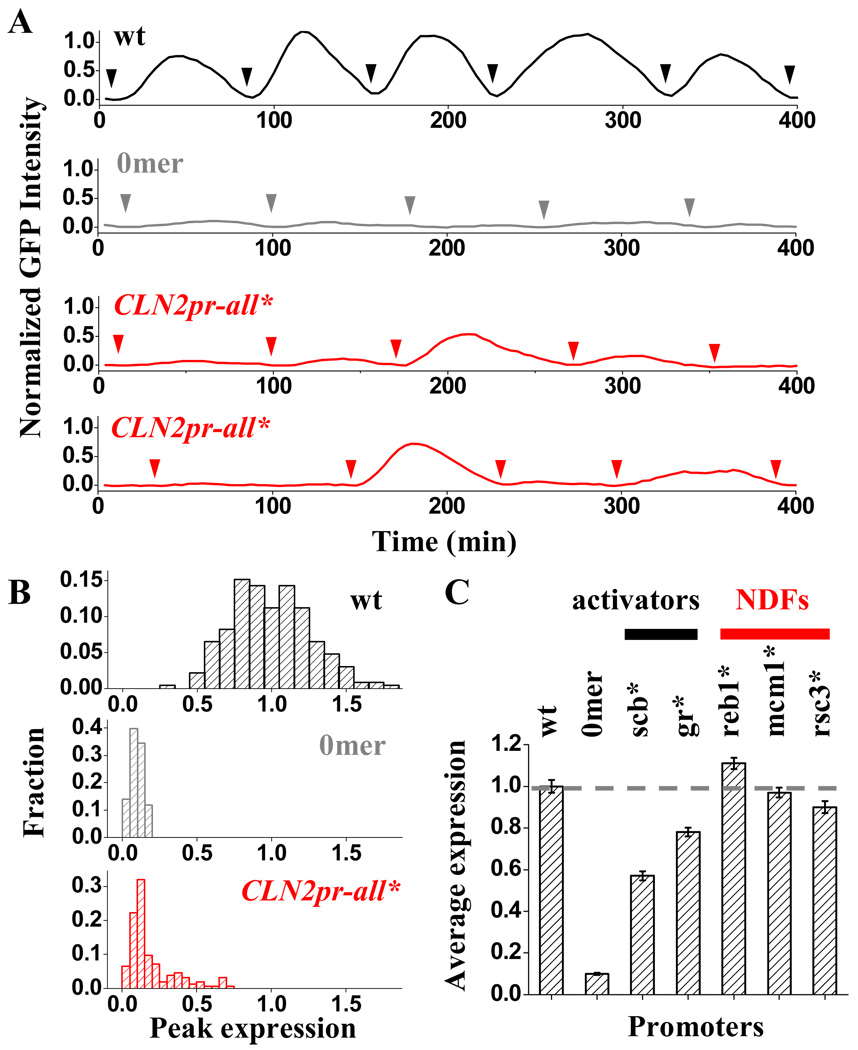
CLN2pr-all* induces “on / off” gene expression in individual cell cycles
To understand how NDR elimination affects the promoter’s transcriptional activity, we constructed a strain with CLN2pr-all* driving the expression of unstable GFP (Mateus and Avery, 2000) and monitored the expression level in single cells using time-lapse fluorescence microscopy (Bean et al., 2006; Charvin et al., 2008). Previously, we used this method to show that GFP driven by wt CLN2pr exhibits periodic change in intensity, which occurs reliably once per cell cycle (Figure 5A,B) (Bai et al., 2010; Bean et al., 2006; Skotheim et al., 2008). With all the SBF binding sites in the CLN2pr deleted (CLN2pr-del), the GFP signal becomes essentially flat (Figure 5A,B). Thus this assay reliably detects SBF-dependent, cell-cycle-regulated CLN2 expression.
Figure 5. Transcriptional activity of CLN2pr-all* and other CLN2pr mutants.
A) Example traces of GFP expression driven by wt CLN2pr (black), CLN2pr-del (gray) and CLN2pr-all* (red). Each trace represents the GFP intensity as a function of time in a single yeast cell over multiple cell generations. The arrows indicate the time points of cell division. In the case of CLN2pr-all*, two traces of the GFP measurements are shown. B) Histogram of the peak expression per cell cycle for the wt, CLN2pr-del and CLN2pr-all*. The wt CLN2pr is “on”, and the CLN2pr-del is “off” every cell cycle. In contrast, CLN2pr-all* switches between “on” and “off” cycles with a ~20% “on” probability. C) Normalized average expression levels of CLN2pr variants with only a subset of binding site(s) mutated. gr* represents the mutation in Gcr1 and Rme1 binding sites. Based on the functions of the factors in transcriptional activation vs. NDR formation, we can separate them into two categories: activator vs. nucleosome depleting factors (NDFs).
In contrast, CLN2pr-all* induces unreliable “on / off” expression: GFP activation was observed in some cell cycles, but undetectable in the others (Figure 5A). The corresponding histogram of CLN2pr-all* expression contains a Gaussian-like distribution centered at the basal level of CLN2pr-del (“off” cycles), with a tail (~20%) on the side of higher expression (“on” cycles) (Figure 5B). Since this tail is not present in the histogram for CLN2pr-del, it should reflect sporadic activator-dependent transcriptional activation of the CLN2pr-all* promoter. This finding is consistent with our previous results in which SBF binding sites were placed at ectopic locations with or without associated NDR (Bai et al., 2010), and extend the findings to a largely native CLN2pr context.
Two classes of regulatory factors bind the CLN2 promoter
The factor binding sites mutated on the CLN2pr-all* could affect transcription directly, or through the change in nucleosome occupancy. To decouple these two potential effects, we took advantage of the promoter variants in Figure 4B where the binding site(s) of individual factor are disrupted without significantly perturbing the NDR. CLN2pr-reb1*, -mcm1* and -rsc3* all activate reliably, once-per-cell-cycle (data not shown), and their average expression levels only differ slightly from that of the wt CLN2pr (within 10%; Figure 5C). This shows that, in the presence of NDR, the direct influence of Reb1, Mcm1 and Rsc3 on transcription is very minor, and the dramatic “on / off” transcription is more likely due to the loss of NDR.
In contrast, mutations in the SBF and Gcr1 / Rme1 binding sites significantly reduced the CLN2pr expression (Figure 5C). Interestingly, unlike Reb1, Mcm1 and Rsc3, these factors do not have significant effect on the NDR formation (Figure S2C). Therefore, we can divide the factors bound on CLN2pr NDR into two functionally distinct groups (Figure 5C): “activators” that mainly function to recruit transcriptional machinery and activate gene expression, and “nucleosome-depleting factors” (NDFs) that mainly function to generate the NDR and indirectly activate transcription by allowing access of the first class of activators.
The formation of many NDRs in yeast likely involves multiple factors
Does the formation of other yeast NDRs involve the NDFs? Our first hint comes from the evolution of promoter sequence. Functional promoter elements are subject to selective pressure and thus evolve more slowly than the neutral sequences. In the CLN2pr, both the binding sites for activators and NDFs are well-conserved among close yeast species, while the neighboring sequence under nucleosome −3 is much less conserved (Figure S4A). Consistently, the nucleosomal sequence in the HOpr URS2 is not very conserved except for the functional SCB elements (Figure S4B). Such contrast in sequence conservation between the NDR and nucleosomal region is not unique to these two examples. Yeast promoters often contain multiple conserved elements beyond the known functional sites, and many of these elements locate in the NDR (several examples shown in Figure S4C). Previous study has shown that NDR sequence in general is more conserved than the neighboring sequences (Yuan et al., 2005). It is intriguing to speculate that many of these conserved elements could be binding sites of NDFs.
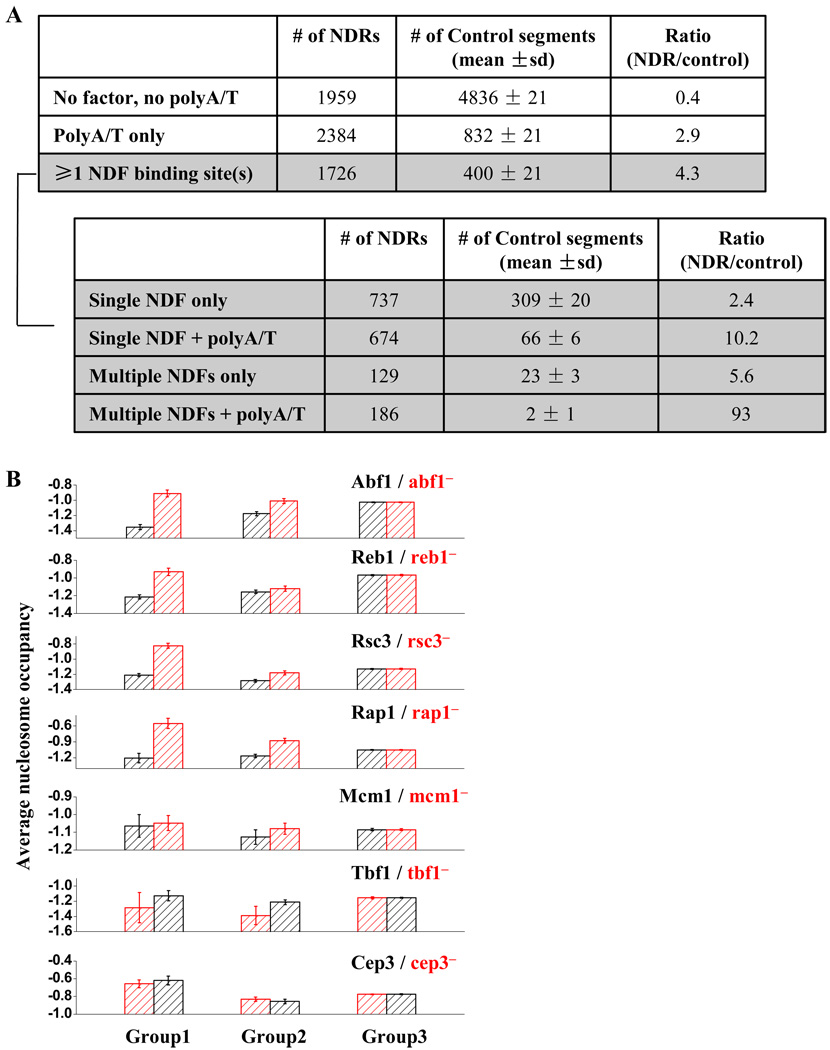
To look for direct evidence that NDFs play a general role in yeast NDR formation, we scanned through the genome-wide NDR sequences for consensus binding sites of the known nucleosome-antagonizing factors, including Abf1, Mcm1, Rap1, Reb1 and Rsc3. We also looked for polyA/T (n≥7) in the NDR sequences, which potentially disfavors nucleosome formation through intrinsic histone-DNA affinity (the cutoff of 7 is chosen because polyA/T (n≥7) in yeast genome occurs at a frequency higher than random expectation based on the overall frequency of A/T) (Dechering et al., 1998). The search results are summarized in Figure 6A. Out of the ~6000 NDRs identified in yeast, we found ~1700 NDRs (~30%) contain at least one NDF binding site(s), a much higher proportion than the non-NDR control sequences. With more NDFs identified, this number is likely to increase further. Therefore, the statistics shown in Figure 6A is consistent with a wide-spread role of NDFs in genome-wide NDR formation. Among these ~1700 NDRs, about half of them also contain polyA/T, suggesting a frequent synergy between NDFs and intrinsic low histone-DNA affinity in promoting NDR formation.
Figure 6. Genome-wide analyses for NDR formation.
A) The number of NDRs that contain NDF consensus site(s) and / or PolyA/T, in comparison with the non-NDR control. The ~1700 NDRs containing at least one NDF binding sites are further dissected based on the number of NDF binding sites as well as the presence of PolyA/T. B) For each NDF (factor A; including Abf1, Reb1, Rap1, Rsc3 and Mcm1), the genome-wide NDRs are divided into three groups based on their sequences. Group1: factor A only, Group2: factor A + other nucleosome-repelling elements, Group3: no factor A. The average nucleosome occupancy of NDRs in these three groups are plotted before (black) and after (red) the deletion of factor A. NDRs in Group2 tends to be more “resistant” upon factor A deletion. For comparison, similar test is applied to factors with little or no nucleosome-depleting activity, including Tbf1 and Cep3. Data size (# of NDRs in Group1–3 for each factor): Abf1: 157, 187, 5725; Reb1:238, 287, 5544; Rsc3: 346, 511, 5212; Rap1: 15, 85, 5969; Mcm1: 29, 76, 5964; Tbf1: 41, 133, 5895; Cep3: 83, 207, 5779).
Compared to the non-NDR control, NDRs that contain multiple nucleosome-repelling elements (including multiple NDF binding sites with or without polyA/T, or single NDF binding site with polyA/T) are particularly enriched (Figure 6A). To examine the functional consequence of these multiple elements, we carried out the following test: for each NDF (factor A, including Abf1, Reb1, Rsc3, Rap1 and Mcm1), we divided genome-wide NDRs into three groups: 1) NDR containing only the binding site(s) of factor A, 2) NDR containing the factor A binding site(s) and other nucleosome-antagonizing elements, including the binding sites for other NDFs and / or polyA/T, 3) NDR with no factor A binding site. We then used the data in Badis et al. (2008) to examine how these three groups respond to the deletion of factor A. If multiple elements contribute to the NDR formation and they are somewhat redundant, we would expect that Group 2 should be more resistant to factor A deletion than Group 1. Figure 6B shows that this is indeed the case: upon a factor deletion, the increase of nucleosome density in the Group 1 NDRs tends to be much more significant than that of Group 2. These data support the idea that the formation of many NDRs in yeast involves multiple factors. The same trend is not observed for Tbf1 and Cep3 (Figure 6B), two control factors that have little or no nucleosome-repelling function (Badis et al., 2008).
In Figure 6B, we also noticed that upon the NDF inactivation, even the Group 1 NDRs still have relatively low nucleosome density (scored as a significantly negative number, compared to the genome-wide average, which is close to 0). This could be due to incomplete inactivation of the NDFs and /or additional nucleosome-repelling elements that are yet to be identified.
Discussion
Formation of CLN2pr NDR requires concerted action from multiple factors
NDR is a dominant and conserved feature in eukaryotic promoters, but its formation mechanism is not entirely understood. Here we studied in detail the formation of the NDR on CLN2pr. We found that this NDR is not due to unfavorable intrinsic histone-DNA interaction; instead, it is generated by multiple factors locally bound to the NDR. In total, we discovered 8 potential factor binding sites contributing to the local nucleosome depletion. This list may not even be complete: 1) in our mutation series, we did not test every conserved sequence clusters in the NDR, 2) binding sites related to NDR formation may not always be well-conserved, 3) some factors may bind and further stabilize NDR, but their contribution could be too small to be detected in our assay. Nevertheless, disrupting these binding sites simultaneously is sufficient to eliminate the CLN2pr NDR.
The NDRs generated by these factors have several features. First, they tend to be local (NDRs rarely extend beyond 100–200 bp from the binding sites, e.g. see Figure 3). Such locality is not always the case for nucleosome-depletion. On HOpr, the binding of the activator Swi5 can lead to a “wave” of downstream nucleosome depletion extending >1kb (Takahata et al., 2009); upon the binding of heat shock factor, nucleosomes on the entire Drosophila Hsp70 gene (>2kb) could be rapidly disrupted (Petesch and Lis, 2008). This difference presumably reflects different nucleosome-depletion mechanisms. Second, the extent of the NDR generated by certain factors depends on the neighboring sequence. For example, the NDRs formed by Seg3 in two sequence contexts are of very different lengths (compare Figure 3B to 3C). Third, different factors can have accumulative effects (more than one factor is required to achieve nucleosome depletion on HOr-Seg2 and HOr-Seg4), and multiple factors with closely-spaced binding sites are utilized to generate a long and robust NDR. Finally, although not the case for CLN2pr, our bioinformatic analysis indicates that formation of many NDRs may benefit from synergy between NDF and low intrinsic histone-DNA interaction. One such case is the RPS28A promoter in yeast, where both Abf1 and PolyA/T stretch contribute to the formation of NDR (Lascaris et al., 2000).
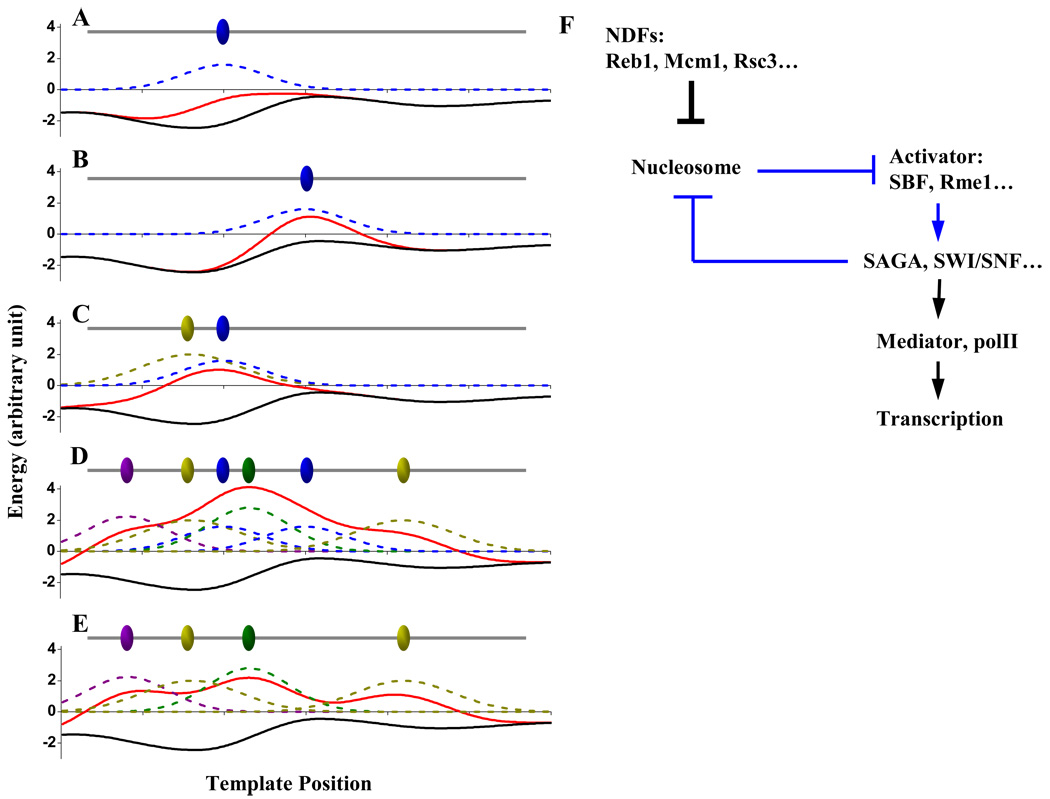
Simple energy model for NDR formation
The observations above are consistent with a simple energy model for NDR formation (cartoon illustrated in Figure 7). Because intrinsic histone-DNA affinity is sequence-dependent, nucleosome formation on naked DNA has a non-flat energy landscape (black curves in Figure 7A–E). This “basal” energy curve shown in Figure 7 is negative on the entire stretch of DNA, representing a net energy gain during nucleosome assembly no matter where the nucleosome forms. Bound factors can create a local energy penalty on the landscape so that the nucleosomes in this vicinity are less energetically favorable (e.g. blue curve in Figure 7A). NDR will form at locations where the overall energy landscape (red curve) exceeds 0. Whether this factor can create a NDR depends on the amplitude of the penalty, which is determined by the factor occupancy as well as its nucleosome-repelling activity. It also depends on the location of the binding site: a factor is more likely to generate a NDR when bound in a region with weak histone-DNA interaction (comparing Figure 7AB). Figure 7A provides an explanation for the observation that though NDFs repel nucleosomes, not all of their consensus binding sites are located in NDRs (Figure S5). The scenario in Figure 7B is consistent with the finding that Reb1-PolyT can generate NDR on randomly selected, originally nucleosomal loci (Raisner et al., 2005).
Figure 7. Models for NDR formation and transcription activation of CLN2pr.
A–E) Simple energetic model for the formation of NDR. Ovals: bound factors on the DNA. Black curves: basal energy landscape for the nucleosome assembly on naked DNA. Blue, yellow, green and purple dashed curves: local energy penalties against nucleosome assembly generated by the factors. Red curves: the overall energy landscape. When the red curve goes above 0, nucleosome assembly becomes energetically unfavorable and NDR will form. See text for details. F) Reaction pathway for transcription activation on CLN2pr. See text for details.
For two closely-bound factors, their combined energy contribution could make nucleosome formation energetically unfavorable even though either individual factor could not (Figure 7C). By this mechanism, long NDR can be generated by multiple factors (Figure 7D). The overall energy from these factors can be over-compensated so that NDR can remain intact in the absence of certain factors (comparing Figure 7DE), like what we observed on CLN2pr.
The model described above is likely to be over-simplified. Nucleosome positioning is not only affected by the local formation energy, but also by the steric exclusion from the neighboring nucleosomes. The binding of the factors could be cooperative so that the energy contribution from each factor depends on the presence of other factors. The energy model implies that histone turnover and factor binding / dissociation reach equilibrium, which may not be true, and the nucleosome positioning may depend on the rates of these events. Nonetheless, the simple model seems to provide an intuitive explanation for diverse experimental data.
Nucleosome-depleting factors can be critical for promoter activity
In addition to NDR formation mechanism, our data also provide significant insight for how to construct a functional promoter. Previous studies on random promoter libraries in eukaryotic cells have shown that, at least in some cases, this question is not well understood (Ligr et al., 2006). In general, TATA box (or equivalent polII initiation site) and activator / repressor binding sites are considered to be the key elements for promoter function. However, although CLN2pr-all* contain intact TATA box and activator binding sites, it induces unreliable, “on or off” transcriptional activation.
The bimodal behavior may be explained by the model shown in Figure 7F. When activator binding sites are nucleosome-embedded, the activator and nucleosome mutually inhibit each other: nucleosomes prevent efficient binding of the activators, and the bound activators can lead to the eviction of the nucleosomes (blue arrows in Figure 7F). Such a double negative feedback loop has the potential to generate bimodal promoter activity (Ferrell, 2002; Gardner et al., 2000). Why does the transition from “off” to “on” state only occur in a fraction of cell cycles? There are two possibilities: 1) The double inhibition between activators and nucleosomes may not be homogeneous in all cell cycles. For instance, nucleosomes in different cell cycles may have different occupancy / positioning / modifications, and some of these configurations may be more effective in preventing activators from binding than others. 2) The transition between “off” to “on” state may be slow, and in some cell cycles, it may miss the transient SBF activation pulse. Further experiments are required to differentiate between these possibilities.
For most genes in the genome, unreliable activation is likely to be undesirable and entails a fitness cost. On CLN2pr, this problem is solved by NDFs. Unlike the common transcriptional coactivators such as SAGA, SWI/SNF and mediator, the NDFs bind directly to DNA, and their binding precedes that of the activators (Figure 7F). We show that NDFs have minor direct effect on transcription; instead, their main function is likely to create a constitutive NDR so that the activators can bind to their cognate sites without competing with nucleosomes.
Another important aspect of the promoter architecture is the spatial organization of the factors. On CLN2pr, the activator binding sites are located in the middle of the NDR, surrounded by the densely spaced NDFs (Figure 3D). What would happen if the positioning is rearranged? The 3merNuc promoter we used in our previous study is a good example (Bai et al., 2010). This CLN2pr variant contains the binding sites of a subset of the NDFs and three artificially-generated SCBs, and the two sets of binding sites are spatially separated so that the SCBs are no longer protected against nucleosome. Like CLN2pr-all*, 3merNuc promoter exhibits bimodal activation. Therefore, reliable activation not only depends on the presence of NDFs, but also their proximity to the functional sites. These findings may lead to an understanding of the mechanisms by which promoter architecture can independently promote gene expression reliability and amplitude.
Experimental Procedures
Strains and Plasmids
Standard methods were used to construct the strains and plasmids. The strains are listed in Table S2, and they are all W303-congenic. To construct the HO* / CLB2* (Figure 1B), we cloned the sequence of HOpr / CLB2 ORF into a plasmid, inserted the CLN2pr NDR segment (−503 to −295 relative to TSS) at −395 in HOpr and 439 in CLB2 ORF, and integrated into the HO / CLB2 locus selected by the URA marker. Because our nucleosome mapping is based on a qPCR method (see below), it is critical to keep the sequence under analysis as a single copy in the yeast genome. To eliminate the wt HOpr / CLB2 ORF, we generated the corresponding ura− “popout” strain through FOA selection. We also eliminated the CLN2pr NDR sequence in the CLN2 locus.
For the rest of the strains, we started from plasmid pLB02 containing wt CLN2pr-GFP-CLN2pest and mutated the wt CLN2pr as desired. The CLN2pr-del1–4 deletes the CLN2pr NDR from −571 to −508 (del1), −507 to −433 (del2), −432 to −353 (del3) and − 352 to −271 (del4). The CLN2-HOr is constructed by replacing the entire CLN2pr NDR (−591 to −271) with a HOpr segment (−610 to −290 relative to the +1 of HO ORF). The HOr-Seg1–4 is generated by replacing one of the four segments in CLN2pr NDR (−591 to −512, −511 to −433, −432 to −353, −352 to −271,respectively) back into the CLN2-HOr (Figure 3A). The detailed sequence mutations on individual binding sites are shown in Table S1. In the Seg3, the upstream putative Rsc3 binding site partially overlaps with one SCB. To disrupt this Rsc3 site without eliminating the SCB, it was mutated as “CaCGAAA”, which is another SCB consensus. Reversely, the sequence was mutated as “CGCGgAA” to disrupt the SCB but not the Rsc3 site. We integrated these plasmids into the wt strain at the CLN2 locus, and this strain can be subjected to transcription analysis using time-lapse fluorescence microscopy. For the nucleosome analysis, we again selected the popout strains that contain the mutated CLN2pr but not the wt one.
Strains for the ChIP experiment bearing c-myc-tagged DNA-binding factors were a gift from Young and Struhl labs (Harbison et al., 2004; Ng et al., 2002). Construction of CLN2pr-all* strains in these backgrounds was performed using the same FOA popout selection method as described above.
In vivo nucleosome mapping
We used the same method described in Bai et al. (2010). Nucleosomes were mapped by qPCR followed by MNase assay (Kent and Mellor, 1995; Sekinger et al., 2005). The PCR product was designed to be ~100 bp, and the neighboring PCR primers are ~30–50 bp apart. In the nucleosome map (such as Figure 1B), the x axis represents the mid point of the PCR product. We used the nucleosome −1 on the PHO5pr as the standard to scale the occupancy from 0 to 1(Sekinger et al., 2005). The error bars on the nucleosome occupancy in Figure 1C, 3 and 4 represent the s.e. from two independent MNase measurements (biological replicates). All the MNase experiments are highly reproducible. The error bars in Figure 1B and 2 represent the s.e. from two independent qPCR measurements (technical replicates).
In vitro nucleosome assembly and mapping
We PCR amplified the ~2.1kb genomic DNA near the CLN2pr using the following two primers: GGGCGATCTTCTAACCTCAT, and GACCATAAAATCTTCCGCCAGTA. We followed the well-established protocols for salt dialysis (Lee and Narlikar, 2001) and NapI / ACF mediated in vitro assembly (Ito et al., 1997). We used the same procedure (MNase followed by qPCR) to map the in vitro assembled nucleosomes. To compare with the in vivo results, the in vitro data were normalized so that the two datasets have the same average nucleosome density over the CLN2pr.
Sequence analysis
We searched for the consensus factor binding sites on the CLN2pr NDR sequence using TRANSFAC (Matys et al., 2003) and YEASTRACT (Monteiro et al., 2008) databases. For the sequence alignment in Table S1, we compared the S.cerevisiae sequence with those from other yeast species (S.paradoxus, S.mikatae, S.bayanus and S. kudriavzevii). These species are distant enough to have many silent mutations, but close enough so that their CLN2pr sequences can be aligned. In the alignment, the conserved bases are assigned as “1”, and the others as “0”, and this digitized data was low-pass-filtered by a 10 bp window to generate the plots in Figure S4.
Timelapse fluorescence microscopy and data analysis
We used the same methods as described in Bai et al. (2010). Briefly, we grew yeast micro-colonies in between a glass surface and a small piece of agar gel at 30°C. Phase and fluorescent images were acquired every 4 minutes for ~8 hours with auto-focusing. During this period, the majority of the cells grow in the horizontal plane constrained by the glass and gel (we stop analyzing data once the cells grow out of focus). The timelapse image data were then analyzed using Matlab programs (Charvin et al., 2008). Occasionally we observed cells arrested in mitosis due to photo damage (<5% of the whole population), and these cells were discarded in the analysis. The GFP intensity vs time curves shown in Figure 5A were smoothed, then corrected by subtracting a baseline connecting flanking troughs, and finally normalized by the average peak GFP intensity of wt CLN2pr. The cell division time was determined by the shrinkage of My01 ring at the budneck (labeled with mcherry).
ChIP measurement
We followed a well-established ChIP protocol (Dedon et al., 1991) with minor variations. Briefly, we grew cells in YPD to OD~0.4. Chromatin was crosslinked in vivo in 1% formaldehyde for 15 minutes, followed by cell lysis. Crosslinked chromatin was sheared using the Covaris S2 ultrasonicator to generate DNA fragments of average size of 300bp. Immunoprecipitation with 9E10 anti-myc antibody was performed using Millipore Magna ChIP G kit as per manufacturer protocol. We analyzed isolated DNA by qPCR using primers for a ~100bp PCR product in the immediate vicinity of the factor binding sites; for each sample, three qPCR reactions were performed with two-fold serial dilutions of the template. Enrichment was calculated by dividing qPCR yields of the immunoprecipitated sample to that of total DNA (input); background was calculated as average enrichment of DNA from three to four randomly selected genomic loci (CLB2 ORF, KIP1pr, GAL1 ORF and ASE1 ORF) that were predicted not to be bound by the immunoprecipitated factors. Finally, enrichment of CLN2pr DNA was normalized to background. The error bars represent the standard error from three independently performed immunoprecipitations (biological replicates).
Genome-wide NDR analysis
For the histogram presented in Figure S5, we searched the whole yeast genome for consensus factor binding sites: Reb1: 'TTACCCG'; Abf1: 'RTCAYTNNNNACG'; Rsc3: 'CGCGC'; Mcm1: 'TTCCNNWWNNGGAA'; Rap1: ‘CAYCCRTRCA’ and Swi4: 'CRCGAAA'. We recorded the genomic locations of these consensus sites, and looked for the corresponding nucleosome density in the genome-wide nucleosome mapping data (Lee et al., 2007).
For Figure 6A, we selected the genome-wide NDRs using the data in Lee et al. (2007). First, we picked out region where nucleosome density was lower than −1 (log2 scale); second, to account for the fluctuations in the density measurement, the neighboring low density regions were lumped together if the distance between them was smaller than 100 bp; finally, the low density region was considered as a NDR if its length was longer than 50 bp. With these thresholds, we detected in total 6069 NDRs across the genome. We obtained the sequences in these NDRs from SGD, and search for consensus factor binding sites and polyA/T. Based on the search results, the NDRs are divided into different categories, and Figure 6A listed the number of NDRs in each category. For the control, we randomly selected 6069 nucleosomal genomic segments with identical lengths as the NDRs, and carried out the same sequence search. We repeated the control 5 times, and listed the mean and standard deviation in Figure 6A.
For Figure 6B, we divided the genome-wide NDRs into three groups (see text) and used the data in Badis et al. (2008) to calculate the average nucleosome density for each group before and after the deletion of certain factors. Because each data set in Badis et al. were internally normalized, we added an offset to the average nucleosome density so that the density of group3 was not changed upon the factor deletion.
Highlights.
The nucleosome-depleted-region on CLN2pr is generated by multiple factors (NDFs).
These NDFs redundantly contribute to nucleosome depletion.
Both NDFs and activators are required for full CLN2pr activity.
The formation of many NDRs in the yeast genome may involve multiple NDFs.
Supplementary Material
Acknowledgement
The authors acknowledge Robert Roeder, Zhanyun Tang, Jon Widom, Irene Moore, Fred Winston, Richard Young, Nancy Hannett, Kevin Struhl, Carol Baisden, Trey Ideker and Dwight Kuo for kindly providing reagents, plasmids and strains for this work; Lance Langston, Irene Moore, Simon Elsaesser, Jung-Ae Kim for technical advice; Eric Siggia for in-depth discussion; and all the members of the F. Cross laboratory for insightful comments. This work was supported by the National Institute of Health (F.R.C.) and the Damon Runyon cancer research fellowship (L.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Charvin G, Siggia ED, Cross FR. Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev Cell. 18:544–555. doi: 10.1016/j.devcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, Cross FR. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics. 2005;171:49–61. doi: 10.1534/genetics.105.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, Cross FR. Coherence and timing of cell cycle start examined at single-cell resolution. Mol Cell. 2006;21:3–14. doi: 10.1016/j.molcel.2005.10.035. [DOI] [PubMed] [Google Scholar]
- Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS ONE. 2008;3:e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams CD, Norris D, Osley MA, Fassler JS, Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988;2:150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- Cross FR, Hoek M, McKinney JD, Tinkelenberg AH. Role of Swi4 in cell cycle regulation of CLN2 expression. Mol Cell Biol. 1994;14:4779–4787. doi: 10.1128/mcb.14.7.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winde JH, Van Leeuwen HC, Grivell LA. The multifunctional regulatory proteins ABF1 and CPF1 are involved in the formation of a nuclease-hypersensitive region in the promoter of the QCR8 gene. Yeast. 1993;9:847–857. doi: 10.1002/yea.320090805. [DOI] [PubMed] [Google Scholar]
- Dechering KJ, Cuelenaere K, Konings RN, Leunissen JA. Distinct frequency-distributions of homopolymeric DNA tracts in different genomes. Nucleic Acids Res. 1998;26:4056–4062. doi: 10.1093/nar/26.17.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedon PC, Soults JA, Allis CD, Gorovsky MA. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal Biochem. 1991;197:83–90. doi: 10.1016/0003-2697(91)90359-2. [DOI] [PubMed] [Google Scholar]
- Fedor MJ, Lue NF, Kornberg RD. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988;204:109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Ferrell JE., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenz LM, Johnson AL, Johnston LH. Rme1, which controls CLN2 expression in Saccharomyces cerevisiae, is a nuclear protein that is cell cycle regulated. Mol Genet Genomics. 2001;266:374–384. doi: 10.1007/s004380100515. [DOI] [PubMed] [Google Scholar]
- Ganapathi M, Palumbo MJ, Ansari SA, He Q, Tsui K, Nislow C, Morse RH. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1161. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Gerbi SA, Bielinsky AK. DNA replication and chromatin. Curr Opin Genet Dev. 2002;12:243–248. doi: 10.1016/s0959-437x(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Goh WS, Orlov Y, Li J, Clarke ND. Blurring of high-resolution data shows that the effect of intrinsic nucleosome occupancy on transcription factor binding is mostly regional, not local. PLoS Comput Biol. 2010;6:e1000649. doi: 10.1371/journal.pcbi.1000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent NA, Mellor J. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 1995;23:3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- Kornberg R. The location of nucleosomes in chromatin: specific or statistical. Nature. 1981;292:579–580. doi: 10.1038/292579a0. [DOI] [PubMed] [Google Scholar]
- Lam FH, Steger DJ, O'Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascaris RF, Groot E, Hoen PB, Mager WH, Planta RJ. Different roles for Abf1p and a T-rich promoter element in nucleosome organization of the yeast RPS28A gene. Nucleic Acids Res. 2000;28:1390–1396. doi: 10.1093/nar/28.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Narlikar G. Assembly of nucleosomal templates by salt dialysis. Chapter 21, Unit 21 26. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb2106s54. [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Ligr M, Siddharthan R, Cross FR, Siggia ED. Gene expression from random libraries of yeast promoters. Genetics. 2006;172:2113–2122. doi: 10.1534/genetics.105.052688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, Egger G, Gal-Yam EN, Jones PA. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus C, Avery SV. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast. 2000;16:1313–1323. doi: 10.1002/1097-0061(200010)16:14<1313::AID-YEA626>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008a;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008b;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro PT, Mendes ND, Teixeira MC, d'Orey S, Tenreiro S, Mira NP, Pais H, Francisco AP, Carvalho AM, Lourenco AB, et al. YEASTRACT-DISCOVERER: new tools to improve the analysis of transcriptional regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:D132–D136. doi: 10.1093/nar/gkm976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse RH. Transcription factor access to promoter elements. J Cell Biochem. 2007;102:560–570. doi: 10.1002/jcb.21493. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C. Cell cycle-dependent transcription of CLN2 is conferred by multiple distinct cis-acting regulatory elements. Mol Cell Biol. 1994;14:4788–4801. doi: 10.1128/mcb.14.7.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S, Yu Y, Stillman DJ. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell. 2009;34:405–415. doi: 10.1016/j.molcel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Hughes TR. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone WM, Johnson AL, Banks GR, Toyn JH, Stuart D, Wittenberg C, Johnston LH. Rme1, a negative regulator of meiosis, is also a positive activator of G1 cyclin gene expression. EMBO J. 1995;14:5824–5832. doi: 10.1002/j.1460-2075.1995.tb00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010;8:e1000414. doi: 10.1371/journal.pbio.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditti P, Costanzo G, Negri R, Camilloni G. ABFI contributes to the chromatin organization of Saccharomyces cerevisiae ARS1 B-domain. Biochim Biophys Acta. 1994;1219:677–689. doi: 10.1016/0167-4781(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Willis KA, Barbara KE, Menon BB, Moffat J, Andrews B, Santangelo GM. The global transcriptional activator of Saccharomyces cerevisiae, Gcr1p, mediates the response to glucose by stimulating protein synthesis and CLN-dependent cell cycle progression. Genetics. 2003;165:1017–1029. doi: 10.1093/genetics/165.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarragudi A, Miyake T, Li R, Morse RH. Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:9152–9164. doi: 10.1128/MCB.24.20.9152-9164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhang H, Reese JC. Exposing the core promoter is sufficient to activate transcription and alter coactivator requirement at RNR3. Proc Natl Acad Sci U S A. 2007;104:8833–8838. doi: 10.1073/pnas.0701666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.