Abstract
In fission yeast, as in many organisms, episomally replicating plasmid DNA molecules can be used for a wide variety of applications. However, replicating plasmids described previously are each propagated at a high copy number per cell. Plasmid fission yeast twenty (pFY20) contains the ura4+ gene for positive and negative selection, an origin of replication (ars1 ) and a stability element (stb). Although this plasmid does not have a centromere, it is propagated with a copy number of about two plasmids per haploid genome equivalent and it is transmitted with relatively high fidelity in mitosis and meiosis. This low-copy vector is useful for screens and mutational studies where overexpression (e.g. from high copy plasmids) is undesirable. We therefore constructed multiple partial-digest, size-fractionated, fission yeast genomic DNA libraries in pFY20 and in the cloning vector pBluescript KS+. These libraries have sufficient complexity (average of 2100 genome equivalents each) for saturation screening by complementation, plasmid shuffle or hybridization.
Keywords: Schizosaccharomyces pombe, fission yeast, plasmid vector, genomic library
Introduction
The fission yeast Schizosaccharomyces pombe is a genetically tractable organism that is easily cultured in the laboratory (Gutz et al., 1974). It shares many features with metazoans, such as centromere complexity and division by binary fission, and has long been a model organism for studies of eukaryotic cells (Leupold, 1950; Yanagida, 2002). As a few examples, fission yeast has yielded key information on the cell cycle, meiosis, and epigenetic regulation of centromere function (see e.g. reviews by Heit et al., 2006; Karlsson-Rosenthal and Millar, 2006; Wells et al., 2006). Moreover, because divergence between the fission yeast lineage and that of budding yeast Saccharomyces cerevisiae is ancient (Heckman et al., 2001), comparative analyses of these two yeast model organisms can reveal the relative conservation of key eukaryotic pathways.
The fission yeast genome sequence has been determined (Wood et al., 2002) and a variety of genetic and molecular tools are available (Forsburg and Rhind, 2006; Wixon and Wood, 2006). Most of the techniques that revolutionized the study of biological functions in budding yeast can also be applied to study fission yeast. However, there are no single-copy plasmids for fission yeast and of the existing multicopy plasmids some can be lost at a relatively high rate (Heyer et al., 1986; Siam et al., 2004). In certain cases, the analysis of genes carried by multicopy plasmids is useful (e.g. for cloning of genes by complementation or for the isolation of multicopy suppressors). In other cases, the presence of multiple copies of a gene is undesirable (e.g. where overexpression might be toxic or confound the phenotypes of mutations). We report here that the Escherichia coli–Sz. pombe shuttle vector pFY20 is propagated at a low copy number of about one to two plasmids per haploid genome equivalent. It is, nevertheless, transmitted with reasonably high fidelity. We also report on the construction and characterization of eight genomic DNA libraries, four of which are in pFY20 and four of which are in the standard cloning vector pBluescript KS+.
Materials and methods
Sz. pombe culture and genetic methods
Media, culture conditions, and genetic methods were as described by Gutz et al. (1974). Fission yeast strains were propagated in nitrogen base liquid or on nitrogen base agar minimal media (0.67% Difco yeast nitrogen base without amino acids, 1% glucose, 2% agar for solid media) supplemented where required with amino acids and bases (100 μg/ml). Plasmids were transformed by the lithium acetate procedure (Ito et al., 1983) and were maintained in a ura4-D18 (null) or ura4-294 (point) mutant background by selection for uracil prototrophy. Procedures used to induce mating, meiosis, preparation and plating of ascospores were as described (Gutz, 1971; Kon et al., 1997).
The shuttle vector pFY20
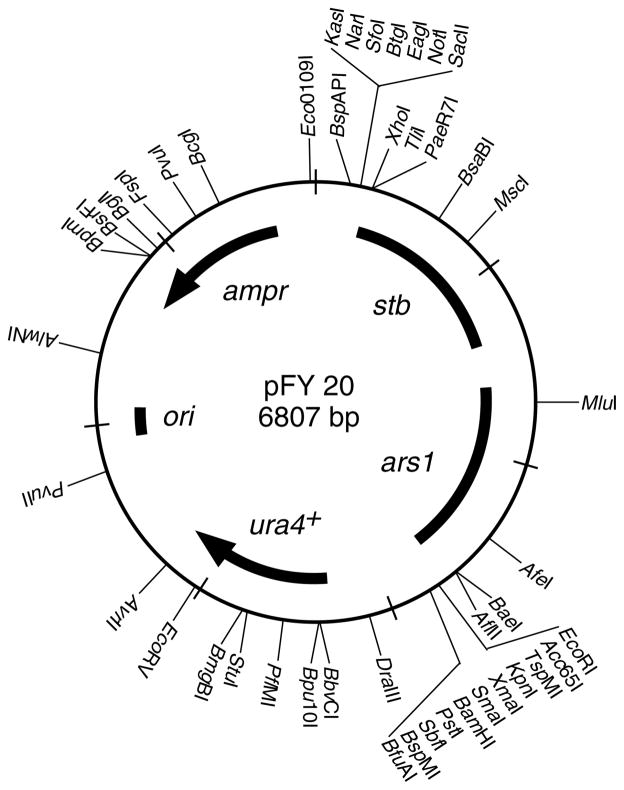
The pFY20 plasmid was constructed from portions of plasmids pura4 Sph and SPRp2 (Li et al., 1997). A map of pFY20 is provided in Figure 1 and the DNA sequence has been deposited in GenBank under Accession No. EU665 638.
Figure 1.
Map of pFY20. The positions of genes (ampr, ura4+), origins of replication (ori, ars1), a plasmid stability element (stb) and a subset of unique restriction sites are shown
Determination of plasmid copy number
Fission yeast cells harbouring or lacking plasmid pFY20 were grown to mid-log phase (5 × 106 cells/ml) under the appropriate selective conditions. Total DNA was prepared (Kon et al., 1998) and was analysed by two independent approaches. In the first approach, plasmids were propagated in a strain with a genomic ura4-294 (point mutant) allele and the relative abundance of the plasmid-borne ura4+ and chromosomal ura4-294 alleles were determined by Southern blotting after digestion with EcoR1. The autoradiograms were analysed using Kodak 1D image analysis software v. 3.5.4 (Eastman Kodak Co., New Haven, CT, USA) to determine the signal intensities of each band. Program parameters were set for automatic band detection and for the integration of the sum of each band intensity. The copy number of pFY20 in each sample was determined as the ratio of plasmid-borne ura4+ signal relative to that of the internal control, chromosomal ura4-294 signal. In the second approach, plasmids were propagated in a strain with a 1.8 kbp deletion (Grimm et al., 1988) spanning the chromosomal ura4 locus (ura4-D18 ). Total DNA was subject to real-time, quantitative PCR using TaqMan technology and an Applied Biosystems (Foster City, CA, USA) Prism 7700 fluorometric thermal cycler (Davidson et al., 2004a, 2004b). Primer Express software (v. 1.5; Applied Biosystems) was used to design TaqMan primers and probes. Oligonucleotides for ura4 were: 5′-TTGTAAACTCGGTAGCGATATCA-3′ (forward), 5′-GGCTTCGACAACAGGATTACG-3′ (reverse), and 5′-6FAM-TTGTTGGTCGTGGAGTCTATGGAGCTGG-TAMRA-3′ (TaqMan). Oligonucleotides for cam1 were: 5′-CGATGGTAATGGCACAATTGAT-3′ (forward), 5′-TTCGTTGTCGGTATCCTTCATTT-3′ (reverse), and 5′-6FAM-TACCGAATTTTTGACTATGATGGCCCGA-TAMRA-3′ (TaqMan). For each experiment the abundance of the plasmid-borne ura4+ DNA was determined relative to that of an internal, single-copy control gene in the genome, cam1+. The ratio values were normalized to those of cells expressing the single-copy ura4+ gene in the genome, but lacking pFY20, to yield the copy number of pFY20. For both the real-time PCR and Southern blotting approaches, the data are given mean ± SD from three or more independent experiments.
Measurement of plasmid stability in mitosis and in meiosis
Established methods were used to determine what fraction of cells retained plasmid after passage through mitosis and meiosis (Heyer et al., 1986). For the analysis of mitotic stability, the cells were grown in liquid media under selective conditions, were plated at a suitable dilution on solid media under non-selective conditions, were incubated for 3 days at 32 °C to permit formation of colonies, were replica-plated to media lacking uracil, and the fraction of colonies capable of growth in the absence of uracil was determined. To analyse plasmid transmission in meiosis, plasmid-containing cells were first grown under selective conditions in liquid medium to stationary phase, two heterothallic haploid strains were mixed and plated on sporulation agar in the absence of selection for plasmid and, following mating and meiosis, the frequency of plasmid transmission was determined by comparing the titres of spore colonies on selective and non-selective media. In each set of experiments, the data are given as mean ± SD from three or more independent experiments.
Construction and amplification of genomic DNA libraries
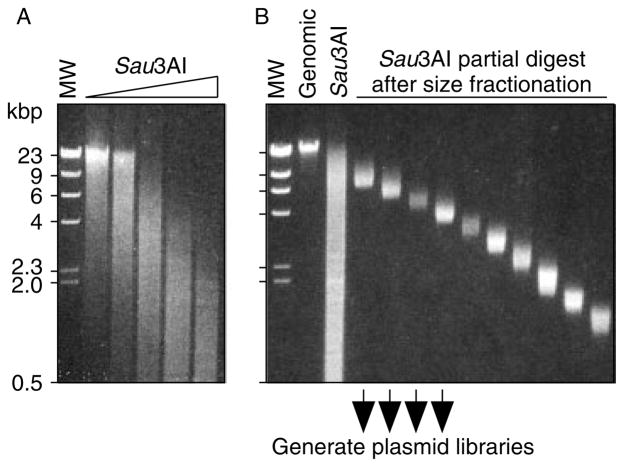
Construction of genomic DNA libraries in pFY20 and pBluescript KS+ were as described (Li et al., 1997). High molecular weight genomic DNA was prepared (Kon et al., 1998) from strain WSP 0206 (h− ade6-D1 ) and was digested in separate reactions with serial dilutions of Sau3AI for 30 min at 37 °C. DNA reaction products were pooled, fractionated by agarose gel electrophoresis, gel slices were excised, and the DNA was recovered by electroelution and precipitation with ethanol. Vectors pBluescript KS+ and pFY20 were linearized with BamHI. Genomic DNA molecules from each of four different size-fractionated, Sau3AI partial-digest preparations were ligated into the compatible, cohesive BamHI site of each vector.
Certain types of eukaryotic DNA elements, such as those that produce secondary and tertiary structures, can be ‘unclonable’ in conventional E. coli strains due to instability (Razin et al., 2001). Therefore, to ensure the broadest representation of fission yeast DNA sequences in the libraries, the ligation mixtures were introduced by electroporation into two different E. coli strains. These were DH1 (supE44 hsdR17 recA1 gyrA96 thi-1 relA1; Hanahan, 1983) and SURE {hsdR mcrA mcrB mrr endA1 recB recJ sbcC umuc::Tn5 uvrC supE44 thi-1 gyrA96 relA1 lac(F′ proAB lacIq Z[delta]M15 Tn10) λ−} (Stratagene, Agilent Technologies, Santa Clara, CA, USA). An aliquot of each transformation mixture was plated in serial dilutions on LB-ampicillin plates to determine the library titres, the remaining portion of each transformation mixture was grown in 1 l LB-ampicillin broth and the plasmids were purified by extraction and CsCl density-gradient centrifugation (Sambrook et al., 1989). Data for plasmids isolated from the DH1 and SURE strains were similar, and are therefore discussed in aggregate to save journal page space. Similarly, because the overall goal was to increase the representation of clones, the DNA samples amplified within E. coli strains DH1 and SURE were pooled prior to the analyses in fission yeast and for the distribution of the plasmid libraries to the research community.
Characterization of libraries
Characteristics of the plasmid libraries were defined both prior to and after the identification of specific clones by hybridization screening (Kon et al., 1997). The fraction of recombinants and physical characteristics of clones were determined using a combination of standard molecular biology tools, including restriction mapping, PCR and DNA sequence analysis.
Results and discussion
Features of the fission yeast vector pFY20
A schematic diagram of ‘plasmid fission yeast 20’ (pFY20) is provided in Figure 1. The bacterial ori and ampr elements support propagation in E. coli, whereas the ars and ura4+ elements support propagation in fission yeast. For the ura4+ marker one can apply both positive selection (growth of ura4 mutant cells in the absence of uracil) and negative selection (growth on medium that contains 5-fluoroorotic acid, 5-FOA) (Grimm et al., 1988). The latter provides a way to select for cells that have lost the pFY20 plasmid, which is useful for certain types of genetic screens and for plasmid shuffle analyses of essential genes (e.g. Kiely et al., 2000; Paluh and Clayton, 1996).
There are no single copy fission yeast plasmids, due in part to the fact that the centromeres are too large to be placed within plasmid vectors, and where defined the existing plasmids are usually propagated at a copy number of 10s to 100s per cell (Siam et al., 2004). Despite their high copy number, fission yeast plasmids are typically lost at a fairly high rate in the absence of selection. However, symmetric segregation can be improved by the inclusion of a 1.3 kbp stability (stb) element (Heyer et al., 1986), so the stb element was included in pFY20. This element was isolated as a non-centromeric, orphan fragment of fission yeast DNA, and we found that it overlaps with the 3′ end of the snf 22+ gene (Yamada et al., 2004). The snf 22+ promoter and 5′ coding capacity are missing, so it is unlikely that the Snf22 protein itself has a role in plasmid stability. While beyond the scope of this study, mutational analyses of the stb element might reveal discrete, cis-acting DNA sequences that confer enhanced plasmid stability.
Copy number of pFY20
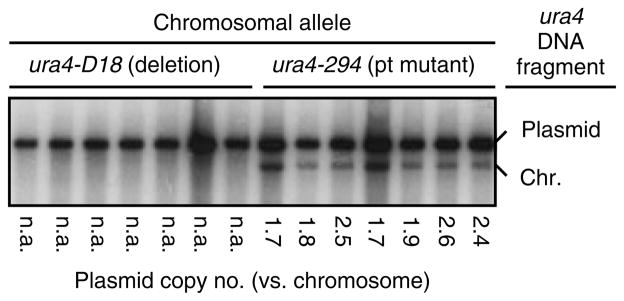
The fission yeast ura4+ gene was included as a selectable marker within pFY20. We introduced the pFY20 plasmid into cells with a point mutation in the chromosomal copy of the ura4 gene (ura4-294), purified total DNA, and conducted Southern blot analysis of the ura4 DNA fragments (Figure 2). The abundance of the plasmid-borne ura4+ DNA in each sample was then compared to that of single-copy chromosomal ura4-294, which revealed a pFY20 copy number of 2.1 ± 0.4 per haploid genome equivalent. We conclude that pFY20 is maintained at a low copy number per cell. This result was unanticipated, so we sought to confirm plasmid copy number by another approach.
Figure 2.
Copy number of pFY20 replicating in fission yeast. Total DNA samples from plasmid-bearing cells, of the indicated chromosomal genotypes, were digested with EcoRI and subjected to Southern blot analysis to detect ura4 DNA fragments. The plasmid copy number in each sample was calculated as the ratio of signal intensities for plasmid-borne and chromosomal (single-copy) ura4 genes. The sample with the ura4D18 (null) allele in the chromosome serves as a control for specificity and size. n.a., not applicable (no genomic copy of ura4 was present, so no ratio could be calculated)
We used real-time, quantitative (TaqMan) PCR analysis (Davidson et al., 2004a) to determine the abundance of the plasmid-borne ura4+ DNA, relative to single-copy controls in the chromosome (see Materials and methods). This revealed a pFY20 copy number of 1.0 ± 0.3 per haploid genome equivalent. Intriguingly, the copy numbers as determined by real-time PCR were reproducibly about 50% lower than the copy numbers as determined by Southern blotting. Two not mutually exclusive possibilities could explain this discrepancy. First, Southern blotting might inflate the plasmid: chromosome signal intensity ratios, because the plasmid DNA fragment of ura4 available for transfer to membrane and for hybridization to probe is larger than that of the chromosomal ura4 DNA fragment. Second, real-time PCR might deflate the plasmid: chromosome signal intensity ratios, because the chromosomal DNA (sheared, linear DNA) might be a slightly better template for the initial rounds of PCR amplification than the plasmid-borne DNA (predominantly intact, circular DNA). Notwithstanding these possibilities, the data from the two independent experimental approaches are largely concordant — pFY20 is propagated with a low copy number of about one to two copies per haploid genome equivalent.
Since pFY20 was present at only one to two copies per genome equivalent, it was possible that the plasmid had integrated into the genome. However, three independent results indicate that pFY20 replicates as a free episome. First, Southern blotting of multiple isolates revealed no evidence for integration into genomic DNA (e.g. Figure 2). Second, parallel analysis of those strains revealed high-frequency loss of plasmid under non-selective conditions (see below). This is as expected for a low copy episome that lacks a centromere and would be unexpected for integrated copies, which should be propagated stably under non-selective conditions. Third, undigested total DNA obtained from yeast cells can readily transform E. coli and, following recovery from bacteria, the plasmid DNA molecules are indistinguishable from those used initially to transform yeast (data not shown). We conclude that pFY20 replicates as a free, low-copy episome in the majority of transformants, although it remains possible that it could integrate into the genome at a frequency that is below our limits of detection.
Transmission efficiency and stability of pFY20
Standard methods were used to determine the efficiency with which pFY20 is transmitted. In mitotic cells, 49 ± 7.9% of colonies retained pFY20 after plating on non-selective media and, following meiosis in the absence of selection, 35 ± 3.0% of spore colonies retained pFY20. The rates of mitotic plasmid loss are consistent with the finding that pFY20 has an average copy number of about one or two per haploid genome equivalent. If there are two plasmids present at the time of mitosis and directed, bipolar segregation occurs (as for replicated chromosomes or cen plasmids in budding yeast), then each daughter cell would receive one plasmid. If random partitioning occurs, then statistically 25% of daughter cells would fail to receive at least one copy of the plasmid. The observed loss frequency is higher than this value, so pFY20 does not seem to harbour any centromere-like elements which confer directed, bipolar segregation. Rather, partitioning of pFY20 is likely achieved predominantly by a random, or nearly-random, process.
Genomic DNA libraries pRSPL (replicating) and pSPL (non-replicating)
For construction of libraries, genomic DNA was digested with a restriction endonuclease, Sau3AI, which cuts frequently in the fission yeast genome. We pooled DNA from a series of partial digests to optimize the randomness of cleavage, the heterogeneity of fragments, and the coverage of the genome (Figure 3A). Once a uniform distribution of fragment sizes was obtained, the DNA molecules were fractionated into individual pools with discrete fragment size ranges (Figure 3B). Four of these fragment pools were cloned independently into the BamHI restriction sites of the E. coli–Sz. pombe shuttle vector pFY20 and the E. coli cloning vector pBluescript KS+. Our rationale for using both vectors was that the former would permit cloning by complementation and plasmid shuffle in fission yeast, whereas the latter (a smaller, very high-copy number bacterial vector) would be better suited for cloning by colony hybridization in E. coli. This approach produced eight genomic DNA libraries which are designated by shorthand notation. Those constructed using pFY20 were named ‘plasmid replicating Sz. fission yeast library’ (pRSPL), whereas those constructed in pBluescript KS+ were named ‘plasmid Sz. fission yeast library’ (pSPL). The numeral indicates the average size of the DNA fragments inserted (e.g. pSPL4.3 has an average insert size of 4.3 kbp).
Figure 3.
Preparation of genomic DNA inserts for construction of libraries. (A) EtBr-stained, agarose gel analysis of genomic DNA digested with different concentrations of Sau3AI. (B) Analysis of pooled, partial-digest genomic DNA prior to and after size fractionation. Size-fractionated samples were cloned into the BamHI restriction sites of pBluescript KS+ and pFY20, respectively
Because some eukaryotic DNA elements can be unstable during propagation in E. coli, the eight libraries were transformed and amplified in two different E. coli strains, one of which has a genotype that ostensibly supports the recovery of molecules that are ‘unclonable’ in conventional strains (see Materials and methods). Each of the libraries had a titre of >1 × 106 independent transformants and within each library 50% or more of the plasmid clones surveyed harboured a genomic DNA insert (Table 1). From the library titres, the recombinant fractions, the average sizes of the inserts and the size of the fission yeast genome (Wood et al., 2002), we calculate that the libraries each contain 900–4200 genome equivalents of fission yeast DNA (Table 1).
Table 1.
Characteristics of genomic DNA libraries
| Library name1 | Insert size range (kbp) | Plasmids in library2 | Recombinant fraction (%)3 | Sz. pombe genome equivalents4 |
|---|---|---|---|---|
| Non-replicating libraries in pBluescript KS+ | ||||
| pSPL4.3 | 3.9–4.7 | 14.8 × 106 | 70 | 3600 |
| pSPL5.3 | 4.7–5.8 | 9.0 × 106 | 60 | 2300 |
| pSPL7.0 | 5.8–8.2 | 8.9 × 106 | 50 | 2500 |
| pSPL10.1 | 8.2–12.0 | 10.4 × 106 | 50 | 4200 |
| Replicating libraries in pFY20 | ||||
| pRSPL4.3 | 3.9–4.7 | 4.3 × 106 | 80 | 1200 |
| pRSPL5.3 | 4.7–5.8 | 3.6 × 106 | 60 | 920 |
| pRSPL7.0 | 5.8–8.2 | 3.2 × 106 | 50 | 900 |
| pRSPL10.1 | 8.2–12.0 | 3.8 × 106 | 50 | 1500 |
Nomenclature: pSPL, plasmid Sz. fission yeast library cloned in pBluescript KS+ (non-replicating in Sz. pombe); pRSPL, plasmid replicating Sz. fission yeast library cloned in pFY20 (replicates in Sz. pombe). Numbers indicate average insert size (kbp).
Each library was amplified in two different E. coli strains (see Materials and methods). The values displayed are the combined total from those two strains.
Based upon the analysis of restriction-digested DNA from 10 independent clones.
Calculated based upon a genome size of 12.5 Mbp, the average insert size, and the recombinant fraction of each library.
A formula based upon the Poisson distribution can be used to calculate the probability that a random library of a given titre and insert size will contain any particular gene of interest (Moore, 1998). Based upon this formula, there is a >99% probability that each of the pSPL and pRSPL libraries contains any given gene of interest. Moreover, the cloning methods and the library titres indicate that if an individual gene is isolated multiple times, then there is a high probability that each plasmid isolate will contain a distinct, independently-derived fragment of fission yeast DNA.
Coverage, complexity and chimeras
To further validate the libraries, we asked whether they meet two expectations: first, can target genes be recovered at the expected frequency?; second, are multiple isolates of the same gene distinct? To answer these questions we used colony hybridization to identify clones in the pFYL5.3 library that harbour the atf 1+ (mts1+) gene (Kon et al., 1997), then we characterized the inserts.
In two screens with a total of ~82 000 colonies we identified 15 clones that harboured the atf 1+ gene. Therefore the observed frequency of atf 1+containing clones (~1/5450) was close to the frequency expected (~1/3950), based upon the average size of the inserts, the recombinant fraction and the library titre. It seems likely that the modest difference between the observed and expected values is due to a fraction of atf 1+-containing colonies (28%) that give a false-negative result during primary and secondary colony hybridization. We and our colleagues have obtained similar results for other screens of these libraries (data not shown). We conclude that individual genes are represented in the libraries at or near to the frequency expected, which confirms the degree of coverage predicted from the library titres (Table 1).
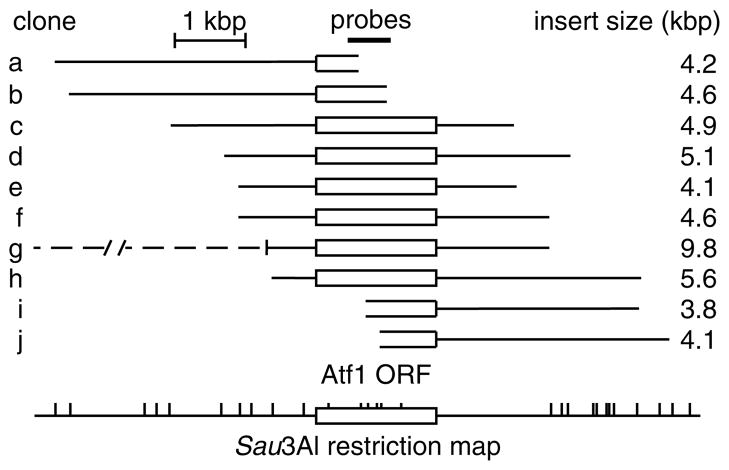
Physical analysis of independently-isolated, atf 1+-containing plasmid clones revealed the expected complexity (Figure 4). Nine of 10 clones characterized in detail contained only DNA that was contiguous to that of the atf 1 + ORF. One insert was found to be chimeric, as it contained a DNA insert that was about twice the expected size and that was derived from two non-contiguous portions of the genome. Such chimeras were expected from the cloning strategy employed. After eliminating chimeras, the average size of each insert post-screening (4.6 ± 0.6 kbp) was indistinguishable statistically from the average size of DNA fragments used for generation of libraries (5.3 kbp). Moreover, each insert was distinct (Figure 4), so independently-isolated clones have the degree of complexity predicted from the library titres (Table 1).
Figure 4.
Representation and complexity of clones. The indicated atf1 probes were used for colony hybridization screening of library pSPL5.3. Clones confirmed after secondary screening were analysed by a combination of restriction mapping, PCR and DNA sequencing. Diagram indicates relative positions of the atf 1+ ORF (box), DNA contiguous to the atf 1+ ORF in the genome (lines), non-contiguous DNA in chimeric clones (dashed line) and DNA recognition sites for Sau3AI restriction enzyme
Concluding remarks
The pFY20 plasmid is a relatively small shuttle vector, with multiple cloning sites, that is maintained at a low copy number per fission yeast cell. Despite the low copy number and the lack of a centromere, pFY20 partitions with reasonably high efficiency in mitosis and meiosis and one can select directly for plasmid retention or plasmid loss. A series of size-fractionated, genomic DNA libraries in pFY20 (replicating in fission yeast) and pBlue-script (non-replicating in fission yeast) have sufficient representation and coverage to permit saturation screening by complementation, plasmid shuffle or hybridization.
Acknowledgments
Our research is supported by funds from the National Institute of General Medical Sciences (GM081766) and the National Institute of Environmental Health Sciences (ES013787) at the National Institutes of Health. We thank Gerry Smith for his support and our colleagues for their encouragement, insight and critical reading of the manuscript.
References
- Davidson MK, Shandilya HK, Hirota K, et al. Atf1–Pcr1–M26 complex links stress-activated MAPK and cAMP-dependent protein kinase pathways via chromatin remodeling of cgs2+ J Biol Chem. 2004a;279:50857–50863. doi: 10.1074/jbc.M409079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson MK, Young NP, Glick GG, Wahls WP. Meiotic chromosome segregation mutants identified by insertional mutagenesis of fission yeast Schizosaccharomyces pombe; tandem-repeat, single-site integrations. Nucleic Acids Res. 2004b;32:4400–4410. doi: 10.1093/nar/gkh767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Kohli J, Murray J, Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Gutz H. Site-specific induction of gene conversion in Schizosaccharomyces pombe. Genetics. 1971;69:331–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King RC, editor. Handbook of Genetics. Plenum; New York: 1974. pp. 395–446. [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR, et al. Molecular evidence for the early colonization of land by fungi and plants. Science. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- Heit R, Underhill DA, Chan G, Hendzel MJ. Epigenetic regulation of centromere formation and kinetochore function. Biochem Cell Biol. 2006;84:605–618. doi: 10.1139/o06-080. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Sipiczki M, Kohli J. Replicating plasmids in Schizosaccharomyces pombe: improvement of symmetric segregation by a new genetic element. Mol Cell Biol. 1986;6:80–89. doi: 10.1128/mcb.6.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson-Rosenthal C, Millar JB. Cdc25: mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 2006;16:285–292. doi: 10.1016/j.tcb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Kiely J, Haase SB, Russell P, Leatherwood J. Functions of fission yeast orp2 in DNA replication and checkpoint control. Genetics. 2000;154:599–607. doi: 10.1093/genetics/154.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N, Krawchuk MD, Warren BG, et al. Transcription factor Mts1/Mts2 Atf1/Pcr1, Gad7/Pcr1 activates the M26 meiotic recombination hotspot in Sz. pombe. Proc Natl Acad Sci USA. 1997;94:13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N, Schroeder SC, Krawchuk MD, Wahls WP. Regulation of the Mts1–Mts2-dependent ade6-M26 meiotic recombination hotspot and developmental decisions by the Spc1 mitogen-activated protein kinase of fission yeast. Mol Cell Biol. 1998;18:7575–7583. doi: 10.1128/mcb.18.12.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leupold U. Die vererbung von homothallie und heterothallie bie Schizosaccharomyces pombe. C R Trav Lab Carlsberg Ser Physiol. 1950;24:381–480. [Google Scholar]
- Li YF, Numata M, Wahls WP, Smith GR. Region-specific meiotic recombination in Schizosaccharomyces pombe: the rec11 gene. Mol Microbiol. 1997;23:869–878. doi: 10.1046/j.1365-2958.1997.2691632.x. [DOI] [PubMed] [Google Scholar]
- Moore DD. Construction of recombinant DNA libraries. In: Asubel FM, Brent R, Kingston RE, et al., editors. Current Protocols in Molecular Biology. Wiley; Chichester: 1998. pp. 5.0.1–5.11.2. [Google Scholar]
- Paluh JL, Clayton DA. Mutational analysis of the gene for Schizosaccharomyces pombe RNase MRP RNA, mrp1, using plasmid shuffle by counterselection on canavanine. Yeast. 1996;12:1393–1405. doi: 10.1002/(SICI)1097-0061(199611)12:14%3C1393::AID-YEA29%3E3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Razin SV, Ioudinkova ES, Trifonov EN, Scherrer K. Non-clonability correlates with genomic instability: a case study of a unique DNA region. J Mol Biol. 2001;307:481–486. doi: 10.1006/jmbi.2000.4372. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring harbor, NY: 1989. [Google Scholar]
- Siam R, Dolan WP, Forsburg SL. Choosing and using Schizosaccharomyces pombe plasmids. Methods. 2004;33:189–198. doi: 10.1016/j.ymeth.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Wells JL, Pryce DW, McFarlane RJ. Homologous chromosome pairing in Schizosaccharomyces pombe. Yeast. 2006;23:977–989. doi: 10.1002/yea.1403. [DOI] [PubMed] [Google Scholar]
- Wixon J, Wood V. Tools and resources for Sz. pombe: a report from the 2006 European Fission Yeast Meeting. Yeast. 2006;23:901–903. doi: 10.1002/yea.1421. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Yamada T, Mizuno KI, Hirota K, et al. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M. The model unicellular eukaryote, Schizosaccharomyces pombe. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-comment2003. [comment] [DOI] [PMC free article] [PubMed] [Google Scholar]