Abstract
Approximately 50–80% of oligodendrogliomas demonstrate a combined loss of chromosome 1p and 19q. Chromosome 1p/19q deletion, appearing early in tumorigenesis, is associated with improved clinical outcomes, including response to chemotherapy and radiation. Although many hypotheses have been proposed, the molecular mechanisms underlying improved clinical outcomes with 1p/19q deletion in oligodendrogliomas have not been characterized fully. To investigate the molecular differences between oligodendrogliomas, we employed an unbiased proteomic approach using microcapillary liquid chromatography mass spectrometry, along with a quantitative technique called isotope-coded affinity tags, on patient samples of grade II oligodendrogliomas. Following conventional biochemical separation of pooled tumor tissue from five samples of undeleted and 1p/19q deleted grade II oligodendrogliomas into nuclei-, mitochondria-, and cytosol-enriched fractions, relative changes in protein abundance were quantified. Among the 442 total proteins identified, 163 nonredundant proteins displayed significant changes in relative abundance in at least one of the three fractions between oligodendroglioma with and without 1p/19q deletion. Bioinformatic analyses of differentially regulated proteins supported the potential importance of metabolism and invasion/migration to the codeleted phenotype. A subset of altered proteins, including the pro-invasive extracellular matrix protein BCAN, was further validated by Western blotting as candidate markers for the more aggressive undeleted phenotype. These studies demonstrate the utility of proteomic analysis to identify candidate biological motifs and molecular mechanisms that drive differential malignancy related to 1p19q phenotypes. Future analysis of larger patient samples are warranted to further refine biomarker panels to predict biological behavior and assist in the identification of deleted gene products that define the 1p/19q phenotype.
Keywords: 1p19q deletion, oligodendroglioma, microcapillary liquid chromatography mass spectrometry, BCAN, BEHAB
INTRODUCTION
The significance for correctly classifying gliomas has become increasingly apparent since the observation that subsets of recurrent anaplastic oligodendrogliomas responded dramatically to PCV (procarbazine, CCNU, vincristine) combination chemotherapy.1 This led to the realization that the combined deletion of 1p and 19q, present in 50–80% of oligodendroglial tumors,2–4 marked a subset with particularly favorable outcomes and enhanced treatment response. For WHO grade III patients, the robust response to alkylating chemotherapy noted in combined deletions translated to markedly increased survival of 70% at 10 years compared with virtually no survivors in patients with other molecular genetic profiles.3,5 For patients with WHO grade II oligodendrogliomas, the absence of the deletion is associated with a 60–70% 5-year survival compared with nearly 100% 5-year survival in deleted tumors.6,7 Although some conflicting data exists,8 the largest series of untreated grade II oligodendrogliomas demonstrates that 1p19q deletion status does not affect survival9 and underscores the importance of 1p19q deletion as a biomarker of tumor responsiveness to genotoxic therapies. Despite the important clinical implications, the gene products encoded by the 1p/19q loci or their downstream targets have not been identified and therefore the mechanism(s) responsible for the positive prognostic impact of this genotype are not known. In the current investigation, we profiled the differential expression of proteins from samples of grade II oligodendroglioma tumors of different 1p/19q deletion status. The technology utilized is a well-established unbiased proteomics approach, in conjunction with microcapillary liquid chromatography mass spectrometry (LC-MS) and isotope coded affinity tags (ICAT). Proteomics discovery was followed by conventional biochemical confirmation of candidate proteins.
MATERIALS AND METHODS
1. Case Selection
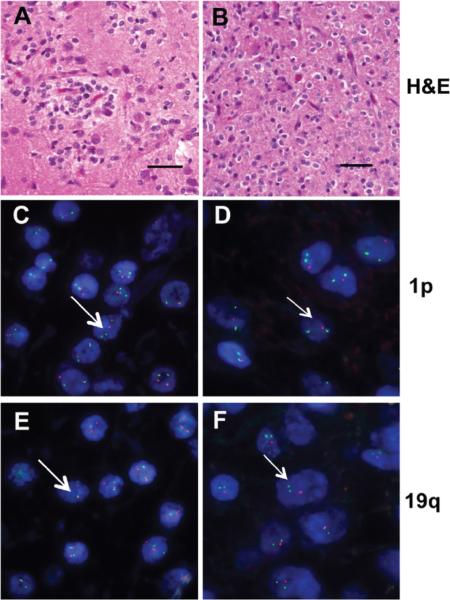
Our goal was to obtain 5 fresh frozen samples each for 1p and 19q codeleted and 1p and 19q nondeleted oligodendrogliomas. Candidate cases were identified from a database search of available fresh-frozen glioma tissue obtained at surgical resection with patient consent and according to an approved institutional human subjects protocol. After removal as en bloc tumor samples, fragments of solid tumor tissue free of gross hemorrhage or coagulation artifacts were placed immediately into liquid nitrogen and then stored at -80 °C. Adjacent sections of tumor tissue are fixed in formalin and used for routine neuropathology and FISH analysis. Candidate cases were then limited to those with diagnoses of oligodendroglioma, astrocytoma or mixed oligoastrocytoma (WHO grade II). At this point, slides were independently reviewed by 3 neuropathologists and only cases with a unanimous diagnosis of oligodendroglioma grade II were included in this study. Cases with existing informative analysis for 1p/19q status were immediately included along with additional cases where fluorescence in situ hybridization (FISH) studies were prepared from archived paraffin blocks. Figure 1 illustrates typical H&E histology from our patient group including diffuse infiltration by monomorphic cells with uniform round nuclei and perinuclear halos for both 1p/19q nondeleted (Figure 1A) and 1p/19q codeleted (Figure 1B) oligodendroglioma. Representative examples of FISH are also shown in Figure 1 for oligodendrogliomas with (Figures 1D, F) and without (Figure 1C, E) 1p/19 deletion.
Figure 1. H&E histology and FISH of 1p19q nondeleted or codeleted WHO grade II oligodendrogliomas.
Representative H&E photomicrographs and FISH analysis for 1p/19q nondeleted (A, C, E) and 1p/19q codeleted (B, D, F) are shown. The H&E sections (A, B) demonstrate typical morphologic features of WHO grade II oligodendrogliomas for both genotypes which are indistinguishable on histologic grounds. However, FISH analysis discriminates between these tumors based on the ratio of centromeric (green probes) and subtelomeric 1p (C, D) and 19q (E, F) specific probes (red signal), which is 2:2 in the nondeleted tumor (C, E) and 2:1 in the codeleted tumor (D, F). Representative cell nucleus is shown with long and short arrows, respectively. Scale bar 50 μM.
2. Patient Characteristics
Ten patients (n = 5/group), 6 male and 4 female, ranging in age from 28 to 65 with verified WHO grade II oligodendrogliomas by blinded neuropathologic review, were included in this study. In all but one case, the tumor samples were obtained at the time of initial diagnosis. The one recurrent sample (5D) was obtained 7 years after initial diagnosis. Two patients were lost to follow-up (1ND, 2ND). At the time of last follow-up for the remaining 8 patients (range 9–67 months), all patients are alive with only two patients (5D and 4ND) documented to have either clinical or radiographic progression. Patient 5D progressed 34 months from the time that tissue was acquired for this study (a second recurrence) that remained a grade II oligodendroglioma. Patient 4ND had disease progression 7 months after surgery and radiation based on the presence of MR contrast enhancement deep to his resection bed and a high choline peak on MR spectroscopy and Temozolomide chemotherapy was instituted. For all other patients, there has been no clinical or radiographic evidence of disease progression. Patient demographic data is listed in Table 1.
Table 1.
Clinical characteristicsa
| PT | Age | Sex | 1p19q deletion | Comment | Path | % Mib-1 | Loc | SX | Recc | C+ | Pminost-op XRT | TTP (mos) | Alive (Y/N) | Length F/U (mos) | Status at last F/U |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1D | 47 | F | + | 19q-relative deletion | O–II | 0.2 | R-Fr | SZ | N | NO | STR | none | Y | 45 | S |
| 2D | 48 | M | + | O–II | 1.2 | L-Fr | SZ | N | YESc | GTR | none | Y | 56 | S | |
| 3D | 37 | M | + | O–II | 0.9 | R-Fr | SZ | N | NO | GTR | none | Y | 45 | S | |
| 4D | 29 | F | + | O–II | 0 | L-Fr | SZ | N | YESc | GTR | none | Y | 67 | S | |
| 5D | 29 | M | + | O–II | 0.5 | R-Fr | P | Y | NO | GTR | none | Y | 34 | P | |
| 1UD | 65 | M | − | O–II | 0 | L-T | SZ | N | NO | GTR | LTF/U | NA | NA | NA | |
| 2UD | 34 | M | − | 19 polysomy | O–II | NA | R-Fr | HA | N | NO | GTR | LTF/U | NA | NA | NA |
| 3UD | 46 | F | − | 19qpolysomy | O–II | 1 | L-Fr | SZ | N | NO | STR | none | Y | 29 | S |
| 4UD | 34 | M | − | O–II | 0b | R-Fr | SZ | N | NO | STR | 7 | Y | 9 | P | |
| 5UD | 28 | F | − | 1, 19polysomy | O–II | 0.3 | L-Fr | SZ | N | NO | STR | none | |||
A: alive, C+: contrast enhancement, CTX: chemotherapy, D: deleted, EOR: extent of resection, Fr: frontal, FU: follow up, GTR: Gross total resection, HA: headache, LT F/U: lost to follow-up, NA: no data, OII: oligodendroglioma grade II, P: progression, path: pathology, recc: recurrence, S: stable, STR: subtotal resection, surv: survival, SX: symptoms, SZ: seizure, T: temporal, TTP: time to progression, UD: undeleted, XRT: irradiation.
Focal Mib-1 25%.
Minimal.
3. 1p/19q FISH
We prepared dual-color fluorescence FISH using commercially available probes and standard methods with minor variations. After sectioning, paraffin-embedded tissue was mounted on slides, dried at 50 °C, deparaffinized in xylenes and washed with ethanol followed by distilled water. Preparations for hybridization included immersing the slides sequentially in 90 °C citrate buffer (pH 6.0) for 20 min, distilled water (after cooling), 0.4% pepsin (P-7012, Sigma-Aldrich, St. Louis, MO) 15 min at 37 °C and 2× standard saline citrate (SSC) for 5 min. Slides were then air-dried. Ten to 20 μL of paired probe for 1p32/1q42 or 19q/19q was applied over the tissue section and coverslipped. Codenaturation was achieved at 37 °C in a humidified chamber for the overnight incubation period during which hybridization occurred. The next day, slides were removed, washed in 50% formamide/1× SSC solution, 2 washes of SSC for 2 min each. Slides were removed and allowed to air-dry. Ten to 20 μL of DAPI in Fluorgard (Insitus) was applied to each slide, which were then coverslipped using 10–20 μL of DAPI in Fluorgard (Insitus).
For each hybridization, a minimum of 100 nonoverlapping nuclei were assessed for numbers of green and red signals under 100× oil immersion using a Nikon E400 fluorescence microscope with appropriate filters (Olympus, Melville, NY). An interpretation of deletion was made when >50% of the nuclei harbored only 1 red signal. The deletion was considered a relative deletion, for example, two copies of 1p in association with increased copies (polysomy) of chromosome 1, and was defined by a ratio of target to reference signals of <0.8 when more that 20% of nuclei had 3 or more signals from the reference chromosome. FISH images were captured using appropriate filters and a SPOT camera (Diagnostic Instruments, Sterling Heights, MI). The resulting images were thresholded and combined using the manufacturer's software.
4. Sample Preparation before Proteomics Analysis
One of the major challenges of current proteomics technology relates to its limited dynamic range when a complex sample is analyzed. To circumvent this limitation, tissue fraction or isolation of organelles10–13 is usually required to gain an indepth proteome characterization. In this study, using conventional biochemical methods, frozen tumor tissue was homogenized gently in a buffer (20 mM HEPES (pH 7.5), 320 mM sucrose, protease inhibitor cocktail (Sigma), phosphatase inhibitors (0.2 mM Na3VO4 and 1 mM NaF), and 1 mM PMSF) with glass homogenizer. Then, the homogenates were centrifuged at 100× g for 30 s to remove large debris. Next, the suspension was centrifuged at 4 °C at 800× g for 10 min to produce a pellet (crude nuclear fraction) and a supernatant, which was further centrifuged at 10 000× g for 15 min to produce another pellet (mitochondria-enriched fraction) and a remainder cytosol-enriched fraction. The crude nuclear fraction was incubated with nuclear extraction buffer containing 0.3 M KCl, 20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 20% glycerol, and 0.1% Triton X-100 at 4 °C for 40 min first, and then centrifuged at 14 000× g for 15 min to obtain nucleienriched fraction. All three portions, that is, nuclei-, mitochondria- and cytosol-enriched fractions were frozen at -80 °C until proteomic analysis. Protein concentrations were determined by standard BCA method.
5. Data Acquisition by Mass Spectrometry
A comparison of the relative abundance of protein profiles in the oligodendroglioma with and without combined 1p/19q deletion was achieved with the ICAT labeling technique that was initially described by Gygi et al14 and routinely utilized in our laboratory.15–18 To perform the ICAT experiment, 100 μg of mitochondria-, nuclei- or cytosol-enriched proteins from either deletion (+) or deletion (−) samples were reduced and the cysteine groups were biotinylated with a 5-fold molar excess of either heavy (13C) (deletion positive) or light (12C) (deletion negative) cleavable ICAT reagents. Next, the two-labeled samples were mixed and digested with trypsin (Promega, Madison, WI) overnight at 37 °C. Then, the digested peptide solution was passed consecutively over an ionic exchange column and a monomeric avidin column (Applied Biosystem, Foster City, CA). The biotinylated peptides were eluted with 0.3% trifluoroacetic acid (TFA) in 30% acetonitrile, and biotin was cleaved from the labeled peptides with concentrated TFA. Finally, the peptides were separated and analyzed on an LCQ DECA Plus Workstation (Thermo, Electron, San Jose, CA), using an automated, online two-dimensional LC separation (strong cation exchange (SCX) and then C18 column), followed by nanoelectrospray LC-MS.19 The eluted peptides from the SCX column (10–800 mM NH4Cl) were loaded onto one of the peptide traps, while peptides on the other trap and the PicoFrit column (5-μm BioBasic C18, 300-Å pore size, 75 μm × 10 cm, tip 15 μm, New Objective, Woburn, MA) were eluted with a 0–65% mobile phase B gradient for 60 min and then 65–85% B for 5 min. The solvents used for the reversed-phase column were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) solutions. The solvent for the sample pump was 0.1% formic acid in water (C). A flow rate of 75 μL/min before the split and 250 nL/min after the split was used for the MS pump, and a flow rate of 150 μL/min before splitting and 2 μL/min after splitting was used for the sample pump. The spray voltage was 1.8 kV, the capillary temperature was 150 °C, and 35 units of collision energy were used to obtain the fragment spectra. Two MS/MS spectra of the most intense peaks were obtained following each full-scan mass spectrum. The dynamic exclusion feature was enabled to obtain MS/MS spectra on coeluting peptides. Proteins from the mixture were later identified automatically using the computer program Sequest, which searched the MS/MS spectra against the human International Protein Index (IPI, v2.33) database.15–17,19 Search parameters for the cleavable ICAT-labeled samples used in this study were the following: +227.13 Da for static modification on cysteine residues labeled with cleavable ICAT, +9 Da for 13C isotopic ICAT-labeled cystein, +16 Da for oxidized methionine, +57 Da for carbamidomethyl; mass tolerance (1.5 Da; restriction on Cys-containing peptides. Potential peptides and proteins were further analyzed with two computer software programs, PeptideProphet and ProteinProphet based on statistical models.20 PeptideProphet uses various SEQUEST scores and a number of other parameters to calculate a probability score for eachidentified peptide. The peptides were then assigned a protein identification using the ProteinProphet software. Protein- Prophet allows filtering of large-scale data sets with assessment of predictable sensitivity and false-positive identification error rates. In our study, only proteins with a high probability of accuracy (<5% error rate) were selected. Quantification of the ratio of each protein (isotopically heavy [deletion positive] vslight [deletion negative]) was calculated using the ASAP Ratio program.21 The algorithm utilized for calculation of ASAP Ratios of signals recorded for the different isotopic forms of peptides of identical sequence are based on numerical and statistical methods, such as Savitzky-Golay smoothing filters, statistics for weighted samples, and Dixon's test for outliers, to evaluate protein abundance ratios and their associated errors. Information about these software tools and the software tools themselves can be found online at http://tools.proteomecenter.org/software.php and downloaded freely.
It should be emphasized that ICAT analysis was performed with pooled samples, with the following rationale: The sampling reproducibility is about 30% for LCQ instrument. This is largely because LCQ (or even a more advanced LTQ mass spectrometer) only captures a small fraction of the peptides detected by the first stage of MS analysis. Thus, it is expected that multiple runs with pooled samples can increase proteome coverage while decreasing interindividual variability. It cannot be stressed enough, however, despite the fact that the protein profile in each ICAT experiment may vary when complex protein mixtures are analyzed multiple times due to the reasons outlined above, that quantitative information is valid when the same peptide is detected simultaneously in paired samples.15,19 This is because MS looks for the corresponding peptide (e.g., light if heavy is captured) and does so with very high sensitivity.
6. Bioinformatics of Proteomics Data
Bioconductor package topGO22,23 was used to determine enhanced GO categories. This approach uses gene/protein lists and identifies GO categories by evidence of over-representation of significant genes/proteins.
7. Confirmation by Western Blot
The following antibodies were tested with the pooled samples first: Actin alpha (Sigma monoclonal, 1:1000), AP2M1 (GeneTex-GTX13992, 1:2000), Apolipoprotein (Chemicon-AB820, 1:2000), BEHAB (gift of Dr. Russell Matthews, SUNY Upstate; B6 1:25 000), Clathrin (Santa Cruz- sc-6579, 1:1000), GFAP (Sigma mouse monoclonal 1:2000 and DAKO polyclonal 1:50 000), NABC1 (BD Transduction- 611332, 1:500), SM22 alpha (Novocastra-NCL-L-SM22a, 1:1000), and transferrin (GeneTex-GTX72750, 1:5000). Of those, only BEHAB, Clathrin and transferrin demonstrated unique bands at the correct molecular sizes, which were followed by analysis in individual cases. For Western blot, typically, 10–20 μg of protein from mitochondria-, nuclei- or cytosol-enriched fractions from deletion-positive vs deletion-negative samples were subjected to 8–16% SDS-PAGE and transferred to PVDF, blocked, and probed overnight at 4 °C with primary antibodies described above, followed by peroxidase-conjugated secondary (1:20 000) and developed with enhanced chemiluminescence. Bands of interest were selected and analyzed by densitometry using Image J software. Grouped data was expressed as mean (SE. Changes between groups were analyzed by Student t test using GraphPad Prism 3.0 (San Diego, CA). All measurements were repeated at least 3 times in all experiments. P <0.05 was accepted as significant.
RESULTS
1. Quantitative Analysis of Protein Profiles in Three Cellular Fractions
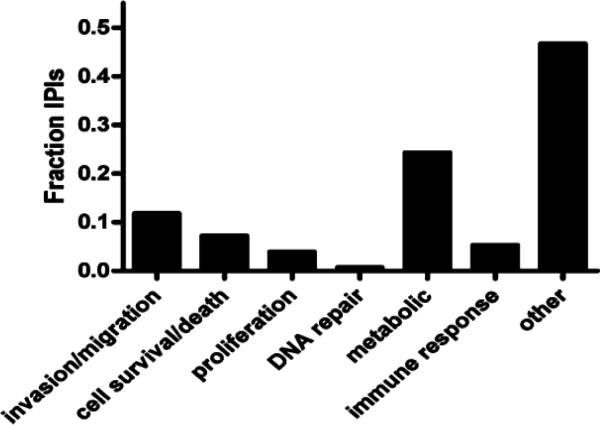
One of the essential steps in proteomics is fractionation of a complex sample to achieve a better identification of proteins of low abundance.15,24,25 Though not without significant caveats when fractionating human frozen tissue, this approach also increases the likelihood of identifying proteins with changes in specific subcellular compartments while their total cellular levels remain the same during a disease process. In this study, cytosol-, mitochondria- and nucleienriched fractions were prepared from the pooled homogenate of five cases of oligodendroglioma with vs without 1p/19q deletion, using a technique well established in our lab.15,25,26 For every comparison, each of the two groups was labeled with a specific ICAT reagent and the samples were then mixed for relative quantifications. A total of 333 and 109 nonredundant proteins (a total of 442) were identified by more than two peptides and a single peptide only, respectively, from the combined fractions. Among them (some of which were overlapped among different fractions), 210, 191, and 329 proteins were identified in the cytosol-, mitochondria-, nuclei-enriched fraction, respectively. Notably, proteins identified by a single peptide should be considered provisional according to the guidelines of most proteomics experts.27 A full list of proteins identified in human oligodendroglioma is shown in Appendix I (Supporting Information). Among the total proteins identified, 163 nonredundant proteins displayed significant changes in relative abundance in at least one of the three fractions between oligodendroglioma with and without 1p/19q deletion; 102 of them were identified by two or more peptides. A complete list of these proteins identified by more than two peptides can be found in Appendix II (Supporting Information). This investigation represents the first extensive characterization of the “proteome” of oligodendroglioma. With so many proteins or possible proteins involved, interpretation of the results at this early stage is difficult. One of the approaches in the proteomic field to reduce the data set is Gene Ontology (GO) analysis,25,28–30 which typically demonstrates molecular functions, cellular locations and involved biological processes of the identified proteins. Since cancer malignancy and treatment responsiveness are related to tumor cell invasion/migration, cell survival/death, proliferation, DNA repair, metabolism, and (evasion of) immune response, we hypothesized that proteins involved in these processes would be relevant to the distinct phenotypes of 1p/19q genotypes in oligodendroglioma. We therefore quantified the representation of differentially expressed proteins in each of these selected categories. As seen in Figure 2, approximately half of all differentially expressed proteins belonged to one of these categories. Furthermore, of the proteins belonging to one of these categories, approximately 70% were related to metabolism or invasion/ migration. We then mapped the genes corresponding to each protein to its chromosome location (Appendix II, Supporting Information) to determine whether we could identify potential candidate deleted genes. Of all proteins, two mapped to the 1p arm and 3 mapped to the 19q arm (Appendix III, Supporting Information). Of these, all had greater expression in nondeleted tumors indicating that they are unlikely to represent candidate deleted genes. For the proteins with changes, further GO analysis was preformed to identify over-represented categories using the cumulative hypergeometric distribution method.23 In this analysis, protein folding, fatty acid catabolic process/betaoxidation, glucose catabolic process/glycolysis, generation of precursor metabolites and energy, vesicle-mediated transport, synaptic vesicle transport/endocytosis, protein polymerization, ferric iron transport and positive regulation of myelination were identified as the top-ranked GO biological process categories or most statistically over-represented biological functions. Furthermore, hyaluronic acid binding/sugar binding, actin binding/(unfolded) protein binding, (peptidyl-prolyl cis-trans) isomerase activity, enoyl-CoA hydratase activity/3-hydroxyacyl- CoA dehydrogenase activity, GTPase activity, purine ribonucleotide binding, lipid transporter activity and ferric iron transmembrane transporter activity were identified as the top-ranked GO molecular function categories (summarized in Table 2). A more detailed list of top-ranked proteins is shown in Table 3, which contains proteins from the top-ranked GO categories that were identified with high confidence, that is, identified by more than two peptides.
Figure 2. Differentially expressed proteins are members of Gene Ontology groups related to malignancy.
Differentially expressed proteins identified in proteomic analysis were annotated as being involved in tumor cell invasion/migration, cell survival/death, proliferation, DNA repair, metabolism and immune response according to their Gene Ontology classifications. On the basis of these annotations, we quantified the representation of differentially expressed proteins in each of these selected categories. Approximately half of all differentially expressed proteins belonged to one of these categories, and of these, approximately 70% were related to metabolism or invasion/migration.
Table 2.
Gene ontology categories over-represented in proteomic analysis
| Biological process | Molecular function |
|---|---|
| protein folding | hyaluronic acid binding/sugar binding |
| fatty acid catabolic process/beta-oxidation | actin binding/(unfolded) protein binding |
| generation of precursor metabolites and energy | (peptidyl-prolyl cis-trans) isomerase activity |
| vesicle-mediated transport | enoyl-CoA hydratase activity/3-hydroxyacyl-CoA dehydrogenase activity |
| synaptic vesicle transport/endocytosis | GTPase activity |
| protein polymerization | purine ribonucleotide binding |
| ferric iron transport | lipid transporter activity |
| positive regulation of myelination | ferric iron transmembrane transporter activity |
Table 3.
Representative proteins with changes from top-ranked gene ontology categoriesa
| Accession | Protein name | Protein symbol | Unique peptides | Adjusted ratio mean (undel:del) | Adjusted ratio (SD) |
|---|---|---|---|---|---|
| IPI00455383 | Clathrin heavy chain 1, splice isoform 2 | CLTC | 19 | 0.6 | 0.09 |
| IPI00022463 | Serotransferrin precursor | TF | 18 | 2.56 | 0.88 |
| IPI00022977 | Creatine kinase, B chain | CKB | 8 | 0.62 | 0.17 |
| IPI00456623 | Brevican core protein precursor, splice isoform 1 (brain-enriched hyaluronan-binding protein) | BCAN | 7 | 2.81 | 0.84 |
| IPI00383573 | Septin 5 | SEPT5 | 5 | 0.47 | 0.06 |
| IPI00472718 | Peptidylprolyl isomerase A isoform 2 | PPIA | 4 | 2.2 | 3.51 |
| IPI00031522 | Trifunctional enzyme alpha subunit, mitochondrial precursor | HADHA | 4 | 0.61 | 0.22 |
| IPI00220663 | Hexokinase type I, splice isoform 2 | HK1 | 4 | 0.57 | 0.14 |
| IPI00163187 | FSCN1 protein | FSCN1 | 4 | 2.41 | 0.89 |
| IPI00472102 | 60 kDa heat shock protein, mitochondrial precursor | HSPD1 | 3 | 0.6 | 0.44 |
| IPI00024993 | Enoyl-CoA hydratase, mitochondrial precursor | ECHS1 | 3 | 0.41 | 0.26 |
| IPI00027770 | Synaptophysin | SYP | 3 | 0.33 | 0.15 |
| IPI00021854 | Apolipoprotein A-II precursor | APOA2 | 2 | 3.13 | 1.06 |
| IPI00032406 | DnaJ homologue subfamily A member 2 | DNAJA2 | 2 | 0.5 | 0.22 |
| IPI00019912 | Peroxisomal multifunctional enzyme type 2 | HSD17B4 | 2 | 0.28 | 0.44 |
| IPI00219018 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 2 | 1.84 | 0.62 |
| IPI00377087 | Gelsolin isoform b | GSN | 2 | 1.55 | 0.27 |
| IPI00018146 | 14-3-3 protein tau | YWHAQ | 2 | 0.42 | 0.6 |
| IPI00015029 | Telomerase-binding protein p23 | PTGES3 | 2 | 0.59 | 0.07 |
| IPI00027434 | Rho-related GTP-binding protein RhoC | RHOC | 2 | 0.59 | 0.34 |
| IPI00186299 | Synapsin-2, splice isoform 2 | SYN2 | 2 | 0.62 | 0.06 |
| IPI00025311 | Breast carcinoma amplified sequence 1 (novel amplified in breast cancer 1) | BCAS1 | 2 | 1.68 | 0.35 |
Undel:del: ratio of undeleted to deleted, SD: standard deviation.
2. Validation of Candidate Proteins with Western Blot Analysis
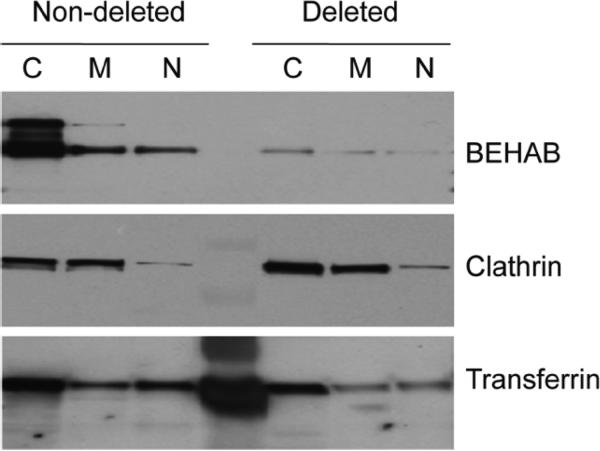
It cannot be emphasized enough that candidate proteins identified by proteomics need to be confirmed/ validated before their biological functions are investigated extensively. This is because the identification of proteins by all mass spectrometric analysis is inferred from a theoretical database (human proteome in this case) that is currently incomplete, meaning that the identification of any given protein could be incorrect.24,31 As the first step in evaluating the significance of candidate proteins for their roles in 1p/19q deletion, we focused on a subset of the proteins listed in Table 3 that have already been implicated in the pathogenesis of various cancers, including gliomas. Another critical determining factor in selecting protein candidates is whether antibodies are available. The proteins selected as our first panel of candidates for confirmation included brevican core protein precursor/ brain-enriched hyaluronan-binding protein (BCAN), clathrin heavy chain 1 (CLTC), serotransferrin (TF), AP-2 complex subunit mu-1 (AP2M1), apolipoprotein A-II precursor (APOA2), glial fibrillary acidic protein (GFAP) and breast carcinoma amplified sequence 1/Novel amplified in breast cancer 1 (BCAS1). Of these, only BCAN, CLTC and TF demonstrated unique bands at the correct molecular size. Representative Western blot data for each of the pooled samples of cellularfractionated proteins for BCAN, CLTC and TF are shown in Figure 3. This was followed by analysis of protein expression by Western blot using whole cell lysates from each individual case. The comparison of mean expression ratios (1p/19q undeleted versus 1p/19q deleted) as calculated by ASAP analysis or densitometry of bands from Western blot analysis of whole cell lysates extracted from individual cases is summarized in Table 4. Representative Western blot and densitometry data for BCAN from individual samples is shown in Figure 4. Full-length BCAN (~150–160 kDa) is cleaved into two smaller fragments (~50–60 kDa and ~90–100 kDa) of which the full-length and cleaved 90–100 kDa form are recognized by the B6 antibody used here.32,33 Semiquantitative analysis of BCAN band intensity in Western blots indicated that the expression level of both uncleaved BCAN (data not shown) and cleaved BCAN was significantly decreased by about 3-fold in oligodendrogliomas without 1p/19q deletion versus those with deletions, that is, largely consistent with our proteomic findings (ASAP ratios). Ratios of TF expression in undeleted versus deleted samples detected by ASAP and Western blot were also concordant (2.56 versus 2.2, respectively). The small sample size and incomplete or short follow-up periods prevented correlations being drawn between the relative expression levels for BCAN or TF in individual samples and clinical course. Detection of CLTC was decreased in nondeleted samples based on ASAP analysis (ratio 0.6 undeleted to deleted; Table 4) and Western blot analysis of cellular fractions pooled from all samples (Figure 3). However, the undeleted-to-deleted ratio (0.9) in detection of CLTC in individual samples by Western blot was not significant.
Figure 3. Expression of selected proteins in different fractions of pooled undeleted and deleted WHO grade II oligodendrogliomas.
Proteins isolated from cytosolic (C), mitochondrial (M) and nuclear (N) fractions were pooled from individual tumors according to their 1p/19q status and equivalent amounts for each sample (30 μg) were run on Western blots. Blots were probed with antibodies to proteins identified in proteomic analysis as being significantly upregulated (BCAN and transferrin) or downregulated (clathrin) in nondeleted samples. The level of protein detected by Western blot was consistent with differences in protein expression based on proteomic analysis.
Table 4.
Validation of selected proteinsa
| Protein name | Protein description | WB | ASAP |
|---|---|---|---|
| BEHAB (BCAN) Clathrin | Splice isoform 1 of Brevican core protein precursor | 1.9454 | 2.81 |
| Clathrin | Clathrin Heavy Chain 1 | 0.9072 | 0.6 |
| Transferrin | Serotransferrin | 2.203 | 2.56 |
WB: ratio of mean densitometry values of undeleted:deleted samples from Western blot, ASAP: automated statistical analysis of protein abundance ratios.
Figure 4. BCAN is upregulated in 1p/19q nondeleted versus 1p/ 19q codeleted WHO grade II oligodendrogliomas.
Equivalent amounts of proteins derived from individual whole cell lysates of each of the 1p/19q undeleted and 1p/19q codeleted WHO grade II oligodendrogliomas used for proteomic analysis were run on Western blots. The blots were probed with antibodies to BCAN (B6) and beta actin as a loading control. B6 antibody identifies both the full length isoforms (upper bands; ~150–160 kDa) and cleaved products (lower bands; ~90–100 kDa) of BCAN.32 Representative Western blots are shown with beta actin controls (lower panel). Quantification of signal intensities of cleaved BCAN normalized to beta actin using densitometry demonstrated significantly increased mean signal intensities in nondeleted versus deleted grade II oligodendrogliomas (p < 0.05).
DISCUSSION
The present study is the most extensive characterization of the proteome of oligodendroglioma to date and demonstrated that analysis of unbiased quantification of differential protein expression can identify candidate molecular motifs and potential biomarkers for clinically distinct subgroups. Analysis of differentially expressed proteins using biased and unbiased bioinformatic techniques identified motifs such as metabolism and invasion/migration that suggested fundamental differences between the two tumor types. Among differentially expressed individual proteins, several, including BCAN, an extracellular matrix protein implicated in glioma invasiveness and tumor progression, were validated as upregulated in the biologically more aggressive undeleted grade II oligodendrogliomas. While the identities of specific gene products responsible for the phenotype of 1p/19q-deleted oligodendrogliomas remain elusive, proteomic techniques employed here markedly increase the repertoire of differentially expressed proteins that can be identified. This supports the potential of this approach to complement gene mapping and genomics approaches to identify molecular mechanisms underlying the unique clinical phenotype of 1p/19q codeleted oligodendrogliomas.
Of the 333 identified proteins (by more than two peptides), 102 displayed significant differences in expression between the undeleted and deleted tumors. By comparison, the only other proteomic analysis of oligodendroglioma has identified 19 proteins.34 In this study, proteins were selected based on levels of detection in two-dimensional gel electrophoresis and identification by liquid chromatography/mass spectrometry.34 In our study, the use of multiple dimensional chromatographies, that is, biochemical fractionation that produced cytosol-, mitochondria- and nuclei-enriched fractions, followed by strong cation exchange as well as reverse phase separation of proteins before MS analysis are major differences that allowed for more robust and specific identification of proteins. Of interest, 7 of the 19 proteins reported by Okamoto et al (2007)34 were also identified in our study, but of these 7, only 4 demonstrated significant changes and only one, glyoxalase I, had concordant changes in expression relative to 1p19q status as reported by Okamoto et al, 2007.34 The disparities between the two studies can be attributed to many factors, including differences in patient populations, technology and databases utilized in proteomics profiling. One important issue in the 2-D gel-based approach is comigration of multiple proteins at the same spot, particularly when the samples are unfractionated, making it exceedingly difficult to determine which one is changing in relative abundance.
Gene expression array studies used to profile oligodendrogliomas consistently identify a neuronal or proneural gene expression signature in 1p/19q codeleted oligodendroglio-mas.35–37 Similarly, we found over-representation of proteins in the GO biological process category of “synaptic vesicle transport/endocytosis” in the present study. Specific proteins with increased expression in codeleted tumors included synaptophysin and synapsin 2. In addition, a robust gene marker of codeletion, the neuronal intermediate filament protein alpha-internexin (INA),38 was identified here as a differentially expressed protein. Therefore it appears that the codeletion of 1p/19q is associated with neuronal gene and protein expression signatures in oligodendroglioma. These findings support the relevance of our data and solidify the concept that global expression analysis can be employed to identify biomarker panels relevant to differential malignancy. The mechanistic importance of a neuronal signature for increased treatment responsiveness warrants further study.
The current analysis identified 163 nonredundant differentially expressed proteins. This large number of identified proteins presents a significant challenge to synthesize the data into meaningful output. We used two separate but complementary approaches for data analysis. First, we analyzed the distribution of proteins within arbitrarily selected GO categories known to reflect malignancy and treatment responsiveness (invasion/migration, cell survival/death, proliferation, DNA repair, metabolism, and evasion of immune response). We found that approximately 50% of differentially expressed proteins belonged to one (or more) of these oncologically relevant GO categories with approximately 25% of the differentially expressed proteins linked to metabolic processes. We also performed unbiased analysis to identify over-represented GO biological process (BP) and molecular function (MF) categories and identified GO categories of “glucose catabolic process/glycolysis” and “generation of precursor metabolites and energy” as significantly over-represented. The robust representation of metabolic and glycolytic proteins is in keeping with the known importance of “metabolic reprogramming” to malignant phenotype through functions that include promoting evasion of apoptosis and resistance to chemotherapy.39
Following metabolism, our analysis demonstrated that proteins linked to invasion/migration comprised the second most frequently represented biological process among those arbitrarily picked to represent hallmark malignant properties. We pursued this finding by selecting the brain-specific lectican BCAN for further analysis. Full-length BCAN is cleaved by ADAMTS-5 to form a pro-invasive isoform that enhances glioma cell motility and invasion.32,33,40–42 We detected increased full-length and cleaved BCAN in undeleted tumor samples, the latter indicating that the differential expression of processed BCAN could contribute to 1p19q differential oligodendroglial malignancy based on enhanced tumor invasiveness. Of additional interest, fascin 1 (FSCN1), an actinbinding protein implicated in human glioma invasion, higher tumor grade and poor prognosis,43–45 was also upregulated in undeleted oligodendrogliomas. Glioma invasiveness is associated with activation of cell survival mechanisms that abrogate therapeutic responses in glioma cells46 and therefore the increased expression of BCAN cleavage products and FCSN1 in nondeleted oligodendrogliomas indicates that differential invasiveness may contribute to the 1p/19q-dependent clinical phenotypes of oligodendrogliomas.
Additional differentially expressed proteins including serotransferrin (TF) and clathrin heavy chain proteins (CLTC) were selected from the highest ranked GO categories for validation in Western blot analysis of individual tumor samples. The quantitative proteomics result for TF (ASAP ratio for undeleted:deleted of 2.56) was validated in Western blot where the ratio of mean expression values for undeleted:deleted individual tumors samples was 2.20. TF delivers iron to cells taken up through TF receptor-mediated endocytosis. Consistent with its role as a potential contributor to the more malignant phenotype of undeleted oligodendrogliomas, increased iron uptake increases growth of experimental gliomas47 and TF receptor expression levels correlate with increased glioma grade.48 The validation of differential expression of selected proteins that function to promote glioma malignancy in undeleted tumors supports the potential of the current proteomic approach to define biomarkers and mechanisms relevant to the 1p19q phenotype. However, as was the case for CLTC, differential expression of proteins identified in ASAP proteomic analysis are not always be validated by other techniques such as Western blot. This disparity may reflect the semiquantitative nature of Western blot analysis. Alternatively, proteins quantified by mass spectrometry are based on peptides, which are subject to post-translational modifications, leading to changes in one or more peptides quantitatively when determined by mass spectrometer, whereas proteins assessed by Western blot are based on antibodies that typically do not differentiate proteins with or without post-translational modifications. Finally, because the human database is currently incomplete, one could argue that the protein identified by mass spectrometry might not be the same protein assessed by Western blot, pointing out, again, the importance of validation of individual proteins identified by proteomic analysis.
Conclusions
This study demonstrated the feasibility of applying quantitative proteomic approaches to indentify biomarkers and address the underlying mechanisms that define the distinct clinical phenotypes of human oligodendrogliomas stratified by 1p/19q status. Our data suggested that in addition to the neuronal motifs noted in gene expression array studies, metabolic reprogramming and differential invasiveness are potential consequences of the 1p/19q deletion that contribute to their differential clinical phenotypes. The validation of BCAN and TF as differentially expressed potential biomarkers for the 1p/ 19q phenotype provides important rationale to extend this analysis to a larger sample size. These future studies can generate more robust biomarker panels and interrogate the functional importance of specific mechanisms preliminarily identified here that may contribute to 1p/19q-related prognosis and treatment responsiveness. Finally, these studies are expected to provide complementary information that will assist in the identification of the deleted gene products that drive the clinical phenotype.
Supplementary Material
Acknowledgment
We thank Rosemary Kimmel for editorial assistance, Alexander M. Spence and Daniel L. Silbergeld for clinical support, and Dr. John Silber for tumor bank support. This work is supported by NIH T32 NS0007144 Clinical Neuroscience Training Grant and the Department of Neurological Surgery Bridge Fund, University of Washington.
Footnotes
Supporting Information Available: Appendix I, Proteins identified by 2 or more peptides. Appendix II, Proteins displayed significant changes and identified by 2 or more peptides. Appendix III, Summary of genes mapped to chromosome 1 and chromosome 19.
References
- (1).Cairncross JG, Macdonald DR. Successful chemotherapy for recurrent malignant oligodendroglioma. Ann. Neurol. 1988;23(4):360–4. doi: 10.1002/ana.410230408. [DOI] [PubMed] [Google Scholar]
- (2).Cairncross G, Berkey B, Shaw E, Jenkins R, Scheithauer B, Brachman D, Buckner J, Fink K, Souhami L, Laperierre N, Mehta M, Curran W. Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J. Clin. Oncol. 2006;24(18):2707–14. doi: 10.1200/JCO.2005.04.3414. [DOI] [PubMed] [Google Scholar]
- (3).Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, Ramsay DA, Louis DN. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J. Natl. Cancer Inst. 1998;90(19):1473–9. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- (4).Reifenberger G, Louis DN. Oligodendroglioma: toward molecular definitions in diagnostic neuro-oncology. J. Neuropathol. Exp. Neurol. 2003;62(2):111–26. doi: 10.1093/jnen/62.2.111. [DOI] [PubMed] [Google Scholar]
- (5).Ino Y, Betensky RA, Zlatescu MC, Sasaki H, Macdonald DR, Stemmer-Rachamimov AO, Ramsay DA, Cairncross JG, Louis DN. Molecular subtypes of anaplastic oligodendroglioma: implications for patient management at diagnosis. Clin. Cancer Res. 2001;7(4):839–45. [PubMed] [Google Scholar]
- (6).Smith JS, Perry A, Borell TJ, Lee HK, O'Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, Jenkins RB. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J. Clin. Oncol. 2000;18(3):636–45. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- (7).Walker C, du Plessis DG, Joyce KA, Fildes D, Gee A, Haylock B, Husband D, Smith T, Broome J, Warnke PC. Molecular pathology and clinical characteristics of oligodendroglial neoplasms. Ann. Neurol. 2005;57(6):855–65. doi: 10.1002/ana.20496. [DOI] [PubMed] [Google Scholar]
- (8).Kanner AA, Staugaitis SM, Castilla EA, Chernova O, Prayson RA, Vogelbaum MA, Stevens G, Peereboom D, Suh J, Lee SY, Tubbs RR, Barnett GH. The impact of genotype on outcome in oligodendroglioma: validation of the loss of chromosome arm 1p as an important factor in clinical decision making. J. Neurosurg. 2006;104(4):542–50. doi: 10.3171/jns.2006.104.4.542. [DOI] [PubMed] [Google Scholar]
- (9).Weller M, Berger H, Hartmann C, Schramm J, Westphal M, Simon M, Goldbrunner R, Krex D, Steinbach JP, Ostertag CB, Loeffler M, Pietsch T, von Deimling A. Combined 1p/ 19q loss in oligodendroglial tumors: predictive or prognostic biomarker. Clin. Cancer Res. 2007;13(23):6933–7. doi: 10.1158/1078-0432.CCR-07-0573. [DOI] [PubMed] [Google Scholar]
- (10).Bergquist J, Gobom J, Blomberg A, Roepstorff P, Ekman R. Identification of nuclei associated proteins by 2D-gel electrophoresis and mass spectrometry. J. Neurosci. Methods. 2001;109(1):3–11. doi: 10.1016/s0165-0270(01)00395-8. [DOI] [PubMed] [Google Scholar]
- (11).Fountoulakis M, Berndt P, Langen H, Suter L. The rat liver mitochondrial proteins. Electrophoresis. 2002;23(2):311–28. doi: 10.1002/1522-2683(200202)23:2<311::AID-ELPS311>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- (12).Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999;17(7):676–82. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- (13).Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12(8):1231–45. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999;17(10):994–9. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- (15).Jin J, Meredith GE, Chen L, Zhou Y, Xu J, Shie FS, Lockhart P, Zhang J. Quantitative proteomic analysis of mitochondrial proteins: relevance to Lewy body formation and Parkinson's disease. Brain Res. Mol. Brain Res. 2005;134(1):119–38. doi: 10.1016/j.molbrainres.2004.10.003. [DOI] [PubMed] [Google Scholar]
- (16).Zhang J, Goodlett DR, Peskind ER, Quinn JF, Zhou Y, Wang Q, Pan C, Yi E, Eng J, Aebersold RH, Montine TJ. Quantitative proteomic analysis of age-related changes in human cerebrospinal fluid. Neurobiol. Aging. 2005;26(2):207–27. doi: 10.1016/j.neurobiolaging.2004.03.012. [DOI] [PubMed] [Google Scholar]
- (17).Zhang J, Goodlett DR, Quinn JF, Peskind E, Kaye JA, Zhou Y, Pan C, Yi E, Eng J, Wang Q, Aebersold RH, Montine TJ. Quantitative proteomics of cerebrospinal fluid from patients with Alzheimer disease. J. Alzheimers Dis. 2005;7(2):125–33. doi: 10.3233/jad-2005-7205. and discussion 173–80. [DOI] [PubMed] [Google Scholar]
- (18).Zhang R, Tremblay TL, McDermid A, Thibault P, Stanimirovic D. Identification of differentially expressed proteins in human glioblastoma cell lines and tumors. Glia. 2003;42(2):194–208. doi: 10.1002/glia.10222. [DOI] [PubMed] [Google Scholar]
- (19).Zhou Y, Gu G, Goodlett DR, Zhang T, Pan C, Montine TJ, Montine KS, Aebersold RH, Zhang J. Analysis of alphasynuclein- associated proteins by quantitative proteomics. J. Biol. Chem. 2004;279(37):39155–64. doi: 10.1074/jbc.M405456200. [DOI] [PubMed] [Google Scholar]
- (20).Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- (21).Li XJ, Zhang H, Ranish JA, Aebersold R. Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal. Chem. 2003;75(23):6648–57. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- (22).Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22(13):1600–7. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- (24).Abdi F, Quinn JF, Jankovic J, McIntosh M, Leverenz JB, Peskind E, Nixon R, Nutt J, Chung K, Zabetian C, Samii A, Lin M, Hattan S, Pan C, Wang Y, Jin J, Zhu D, Li GJ, Liu Y, Waichunas D, Montine TJ, Zhang J. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J. Alzheimers Dis. 2006;9(3):293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- (25).Shi M, Jin J, Wang Y, Beyer RP, Kitsou E, Albin RL, Gearing M, Pan C, Zhang J. Mortalin: a protein associated with progression of Parkinson disease. J. Neuropathol. Exp. Neurol. 2008;67(2):117–24. doi: 10.1097/nen.0b013e318163354a. [DOI] [PubMed] [Google Scholar]
- (26).Jin J, Hulette C, Wang Y, Zhang T, Pan C, Wadhwa R, Zhang J. Proteomic identification of a stress protein, mortalin/mthsp70/ GRP75: relevance to Parkinson disease. Mol. Cell. Proteomics. 2006;5(7):1193–204. doi: 10.1074/mcp.M500382-MCP200. Oligodendrogliomas With and Without 1p/19q Deletion research articles Journal of Proteome Research • Vol. 9, No. 5, 2010 2617. [DOI] [PubMed] [Google Scholar]
- (27).Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312(5771):212–7. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- (28).Pan S, Shi M, Jin J, Albin RL, Lieberman A, Gearing M, Lin B, Pan C, Yan X, Kashima DT, Zhang J. Proteomics identification of proteins in human cortex using multidimensional separations and MALDI tandem mass spectrometer. Mol. Cell. Proteomics. 2007;6(10):1818–23. doi: 10.1074/mcp.M700158-MCP200. [DOI] [PubMed] [Google Scholar]
- (29).Pan S, Zhu D, Quinn JF, Peskind ER, Montine TJ, Lin B, Goodlett DR, Taylor G, Eng J, Zhang J. A combined dataset of human cerebrospinal fluid proteins identified by multidimensional chromatography and tandem mass spectrometry. Proteomics. 2007;7(3):469–73. doi: 10.1002/pmic.200600756. [DOI] [PubMed] [Google Scholar]
- (30).Shi M, Bradner J, Bammler TK, Eaton DL, Zhang J, Ye Z, Wilson AM, Montine TJ, Pan C, Zhang J. Identification of glutathione S-transferase pi as a protein involved in Parkinson disease progression. Am. J. Pathol. 2009;175(1):54–65. doi: 10.2353/ajpath.2009.081019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhang J. Proteomics of human cerebrospinal fluid - the good, the bad, and the ugly. Proteomics Clin. Appl. 2007;1(8):805–819. doi: 10.1002/prca.200700081. [DOI] [PubMed] [Google Scholar]
- (32).Viapiano MS, Bi WL, Piepmeier J, Hockfield S, Matthews RT. Novel tumor-specific isoforms of BEHAB/brevican identified in human malignant gliomas. Cancer Res. 2005;65(15):6726–33. doi: 10.1158/0008-5472.CAN-05-0585. [DOI] [PubMed] [Google Scholar]
- (33).Viapiano MS, Hockfield S, Matthews RT. BEHAB/brevican requires ADAMTS-mediated proteolytic cleavage to promote glioma invasion. J. Neurooncol. 2008;88(3):261–72. doi: 10.1007/s11060-008-9575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Okamoto H, Li J, Glasker S, Vortmeyer AO, Jaffe H, Robison RA, Bogler O, Mikkelsen T, Lubensky IA, Oldfield EH, Zhuang Z. Proteomic comparison of oligodendrogliomas with and without 1pLOH. Cancer Biol. Ther. 2007;6(3):391–6. doi: 10.4161/cbt.6.3.3731. [DOI] [PubMed] [Google Scholar]
- (35).Ferrer-Luna R, Mata M, Nunez L, Calvar J, Dasi F, Arias E, Piquer J, Cerda-Nicolas M, Taratuto AL, Sevlever G, Celda B, Martinetto H. Loss of heterozygosity at 1p-19q induces a global change in oligodendroglial tumor gene expression. J. Neurooncol. 2009;95(3):343–54. doi: 10.1007/s11060-009-9944-y. [DOI] [PubMed] [Google Scholar]
- (36).Mukasa A, Ueki K, Ge X, Ishikawa S, Ide T, Fujimaki T, Nishikawa R, Asai A, Kirino T, Aburatani H. Selective expression of a subset of neuronal genes in oligodendroglioma with chromosome 1p loss. Brain Pathol. 2004;14(1):34–42. doi: 10.1111/j.1750-3639.2004.tb00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Ducray F, Idbaih A, de Reynies A, Bieche I, Thillet J, Mokhtari K, Lair S, Marie Y, Paris S, Vidaud M, Hoang-Xuan K, Delattre O, Delattre JY, Sanson M. Anaplastic oligodendrogliomas with 1p19q codeletion have a proneural gene expression profile. Mol. Cancer. 2008;7:41. doi: 10.1186/1476-4598-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Ducray F, Criniere E, Idbaih A, Mokhtari K, Marie Y, Paris S, Navarro S, Laigle-Donadey F, Dehais C, Thillet J, Hoang- Xuan K, Delattre JY, Sanson M. alpha-Internexin expression identifies 1p19q codeleted gliomas. Neurology. 2009;72(2):156–61. doi: 10.1212/01.wnl.0000339055.64476.cb. [DOI] [PubMed] [Google Scholar]
- (39).Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13(6):472–82. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- (40).Hu B, Kong LL, Matthews RT, Viapiano MS. The proteoglycan brevican binds to fibronectin after proteolytic cleavage and promotes glioma cell motility. J. Biol. Chem. 2008;283(36):24848–59. doi: 10.1074/jbc.M801433200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Matthews RT, Gary SC, Zerillo C, Pratta M, Solomon K, Arner EC, Hockfield S. Brain-enriched hyaluronan binding (BEHAB)/brevican cleavage in a glioma cell line is mediated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family member. J. Biol. Chem. 2000;275(30):22695–703. doi: 10.1074/jbc.M909764199. [DOI] [PubMed] [Google Scholar]
- (42).Nakada M, Miyamori H, Kita D, Takahashi T, Yamashita J, Sato H, Miura R, Yamaguchi Y, Okada Y. Human glioblastomas overexpress ADAMTS-5 that degrades brevican. Acta Neuropathol. (Berlin) 2005;110(3):239–46. doi: 10.1007/s00401-005-1032-6. [DOI] [PubMed] [Google Scholar]
- (43).Gunal A, Onguru O, Safali M, Beyzadeoglu M. Fascin expression [corrected] in glial tumors and its prognostic significance in glioblastomas. Neuropathology. 2008;28(4):382–6. doi: 10.1111/j.1440-1789.2008.00889.x. [DOI] [PubMed] [Google Scholar]
- (44).Hwang JH, Smith CA, Salhia B, Rutka JT. The role of fascin in the migration and invasiveness of malignant glioma cells. Neoplasia. 2008;10(2):149–59. doi: 10.1593/neo.07909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Peraud A, Mondal S, Hawkins C, Mastronardi M, Bailey K, Rutka JT. Expression of fascin, an actin-bundling protein, in astrocytomas of varying grades. Brain Tumor Pathol. 2003;20(2):53–8. doi: 10.1007/BF02483447. [DOI] [PubMed] [Google Scholar]
- (46).Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J. Clin. Oncol. 2003;21(8):1624–36. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- (47).Chirasani SR, Markovic DS, Synowitz M, Eichler SA, Wisniewski P, Kaminska B, Otto A, Wanker E, Schafer M, Chiarugi P, Meier JC, Kettenmann H, Glass R. Transferrinreceptor- mediated iron accumulation controls proliferation and glutamate release in glioma cells. J. Mol. Med. 2009;87(2):153–67. doi: 10.1007/s00109-008-0414-3. [DOI] [PubMed] [Google Scholar]
- (48).Recht LD, Griffin TW, Raso V, Salimi AR. Potent cytotoxicity of an antihuman transferrin receptor-ricin A-chain immunotoxin on human glioma cells in vitro. Cancer Res. 1990;50(20):6696–700. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.