Abstract
We recently identified an acidic-rich segment in the A1 domain of factor VIII (residues 110–126) that functions in the coordination of Ca2+, an ion necessary for cofactor activity (Wakabayashi et al., J. Biol. Chem. 279:12677–12684, 2004). Mutagenesis studies showed that replacement of residue Glu113 with Ala (E113A) yielded a factor VIII point mutant possessing increased specific activity as determined by a one-stage clotting assay. Mutagenesis at this site suggested that substitution with relatively small, nonpolar residues was well-tolerated, whereas replacement with a number of polar or charged residues appeared detrimental to activity. Ala-substitution resulted in the greatest enhancement yielding an ~2-fold increased specific activity. Time course experiments following reaction with thrombin revealed similar rates of activation and inactivation of E113A as observed for the wild type. Results from factor Xa generation assays showed minimal differences in kinetic parameters and factor IXa affinity for E113A and wild type factor VIIIa when run in the presence of synthetic phospholipid vesicles, whereas factor VIIIa E113A displayed an ~4-fold greater affinity for factor IXa compared with factor VIIIa wild type in reactions run on the platelet membrane surface. This latter effect may be attributed, in part to a 2-fold increased affinity of factor VIIIa E113A for the platelet membrane. Considering that low levels of factors VIIIa and IXa are generated during clotting in plasma, the increased cofactor specific activity observed for E113A factor VIII may result from its enhanced affinity for factor IXa on the physiological membrane.
Factor VIII functions as a cofactor for the serine protease factor IXa in the membrane-dependent conversion of zymogen factor X to the serine protease, factor Xa (1, 2). Deficiency or defects in factor VIII result in hemophilia A, which is characterized by significant reduction in factor Xa generation rates during the propagation phase of coagulation.
Metal ions play an important role in regulating factor VIII structure and activity. Factor VIII is inactivated by EDTA1, which facilitates dissociation of the factor VIII heavy (A1-A2-B domains) and light (A3-C1-C2 domains) chains (3, 4). Active factor VIII can be reconstituted by combining the isolated chains in the presence of Ca2+ or Mn2+ (5–7). While Ca2+ has little effect on inter-chain affinity (8), Ca2+ binding to both factor VIII chains is required for the generation of cofactor activity (9). That latter study also demonstrated a local conformational change in response to Ca2+ binding correlated with formation of the active cofactor. Factor VIII activity is also reconstituted following addition of Mn2+ (5). However, the Mn2+-binding site appeared non-identical to the Ca2+ binding site based upon differential competition by Tb3+ (10).
We recently employed alanine-scanning, site-directed mutagenesis to identify acidic residues within segment 110–126 of the A1 domain of factor VIII important in Ca2+ binding (11). Results from that study identified residues E110, D116, E122, D125 and D126, which are conserved in all known factor VIII sequences (12), as likely participating in Ca2+ coordination. Interestingly, residue E113, although not identified as contributing to Ca2+ coordination, and conserved in all known factor VIII sequences (12), was found to yield ~2-fold increases in specific activity and affinity for either Ca2+ or Mn2+ following replacement of the Glu residue with Ala.
In this report, we characterize this gain-of-function mutation at factor VIII E113 following saturation mutagenesis and activity and inter-molecular affinity assays employing synthetic and natural membrane surfaces. Results from these studies indicate that Ala represents the optimal residue at this position yielding maximal specific activity as determined using a plasma-based assay. A mechanism for this enhanced activity is suggested by the increased affinity of factor VIIIa E113A for factor IXa observed during factor Xa generation assays run on the platelet membrane surface, as well as an increased affinity of this factor VIII form for the physiological membrane surface.
Materials and Methods
Reagents
Phospholipid vesicles containing 20% phosphatidylserine (PS), 40% phosphatidylcholine (PC), and 40% phosphatidylethanolamine (PE) were prepared using octylglucoside as described previously (13). Washed gel-filtered human platelets were prepared according to a modification of a previously published method (14), and were maintained at 37°C and used within 3–4 hours after isolation. The reagents α-thrombin, factor IXaβ, factor X, and factor Xa (Enzyme Research Laboratories, South Bend, IN), hirudin and phospholipids (Sigma), the chromogenic Xa substrate S-2765 (N-α-benzyloxycarbonyl-D-arginyl-glycyl-L-arginyl-p-nitroanilide-dihydrochloride; DiaPharma, West Chester, OH), and D-Phe-Pro-Arg chloromethyl ketone (Calbiochem) were purchased from the indicated vendors. The B-domainless factor VIII (FVIIIHSQ) expression construct HSQ-MSAB-NotI-RENeo was a gift kindly provided by Dr. Pete Lollar and John Healey. The thrombin receptor hexapeptide, SFLLRN-amide, was synthesized using (9-fluorenyl)- methoxycarbonyl (FMOC) chemistry on an Applied Biosystems 430A Synthesizer and reverse phase HPLC purified to >99.9% homogeneity.
Construction, expression and purification of wild type and Glu113Ala factor VIII
Glu113Ala and wild-type factor VIII forms were constructed as a B-domainless factor VIII forms, stably expressed in BHK cells, and purified as described before (11). Resultant factor VIII forms were typically >90% pure as judged by SDS-PAGE. Specific activity of the factor VIII proteins were determined by one-stage clotting assays (see below) to measure activity and a combination of ELISA (see below) and direct protein determination by dye binding (15), densitometry scans of stained gels, and A280.
Saturation mutagenesis and the transient expression at Glu113 of factor VIII
Factor VIII possessing a mutation at Glu113 to every amino acid was constructed as described (11) and transiently expressed in COS-7 cells. The factor VIII expression vector constructs were transfected in exponentially growing COS-7 cells with ~80% confluency using FuGene6 (Roche, Indianapolis, IN) for 1 day on 6 well tissue culture plates. Medium was changed to AIM-V (Invitrogen) and the conditioned medium was collected after 2 days, spun at 13,000 rpm for 10 min and factor VIII activity was measured by one-stage clotting assay.
Enzyme-Linked Immunoadsorbant Assay
A sandwich ELISA was performed to measure the concentration of factor VIII proteins (16). The procedure employed ESH8 (anti-factor VIII light chain antibody; American Diagnostica) as a capture antibody and biotinylated R8B12 (anti-factor VIII A2 domain antibody; Green Mountain Antibodies) as the detection antibody. Thus, the epitopes for these antibodies are far-removed from the sites of mutagenesis. The amount of bound factor VIII was determined optically using a streptoavidin-linked horse radish peroxidase (Calbiochem) with the substrate, Ophenylenenediamine dihydrochloride (Calbiochem), as previously described (16). Purified commercial recombinant factor VIII (Kogenate, Bayer Corporation) was used as the standard to determine the concentration of the samples.
One-stage clotting assay
One-stage clotting assays were performed using substrate plasma chemically depleted of factor VIII (17) and assayed in a Diagnostica Stago clotting instrument. Plasma was incubated with APTT reagent (General Diagnostics) for 6 min at 37°C at which time a dilution of factor VIII was added to the cuvette. After 1 min the mixture was recalcified and time to clot formation determined and compared to a pooled normal plasma standard.
Thrombin activation assay
Wild type or E113A factor VIII (10 nM) was activated by thrombin (5 nM, 0.6 units/ml) in 20 mM HEPES, pH 7.2, 0.1 M KCl, 0.01% Tween 20, 0.01% BSA, 5 mM CaCl2. At indicated times aliquots were removed and treated with hirudin (2 units/ml). After appropriate dilution (1:100~1:5,000) activity was measured by clotting assay.
Factor Xa generation assays
The rate of conversion of factor X to factor Xa was monitored in a purified system (18) and performed at room temperature according to methods previously described (8, 9). Activity was determined as the amount of factor Xa generated (nM) per minute and converted to a value per nM factor VIII. The specific activities of factor VIII forms were determined using a modification of this assay. A factor VIII standard was made by diluting pooled normal plasma in 20 mM HEPES, pH 7.2, 0.1 M KCl, 0.01% Tween 20, 5 mM CaCl2, 0.01% BSA and 10 mM PSPCPE vesicles with factor VIII deficient hemophilic plasma (George King Biomedical Inc., Overland Park, KS) mixed to adjust the final concentration of total plasma in the reaction to 10%. Factor VIII wild type and E113A samples were prepared in the same manner. Factor VIII activation and subsequent Xa generation reactions were initiated by 0.5 nM thrombin, 10 nM factor IXa, and 100 nM factor X at 37°C. After 15 minutes the samples were reacted with the factor Xa chromogenic substrate (0.46 mM, S-2765), 50 mM EDTA, and 2 units/ml hirudin for 30 minutes at 37°C and optical density at 405 nm was measured by Vmax microtiter plate reader (Molecular Devices, Sunnyvale, CA).
Factor VIII cofactor parameters in the presence of synthetic phospholipid vesicles
Indicated concentrations (0–15 nM) wild type or E113A factor VIII were activated by thrombin (20 nM, 2.4 units/ml) for 1 minute followed by the incubation with 1 nM factor IXa for 2 minutes in 20 mM HEPES, pH 7.2, 0.1 M KCl, 0.01% Tween 20, 0.01% BSA, 5 mM CaCl2, and 10 μM PSPCPE vesicles. The activity of formed factor Xase complex was measured by factor Xa generation assay as described above and was plotted as a function of factor VIII concentration. The data were fitted to the quadratic equation shown below by non-linear least square regression and affinity to factor IXa was estimated.
| eq. 1 |
where A is activity (nM/min/nM factor IXa), IXa is the applied factor IXa concentration (1 nM) in the reaction mixture, VIII is the factor VIII concentration, Kd is the dissociation constant, and k is a constant. Michaelis-Menten constants for factor Xase were estimated as follows. Wild type or E113A factor VIII (20 nM) was activated by thrombin and incubated with 1 nM factor IXa for 1 min. Reactions were initiated by addition of factor X (0–300 nM). Data were fitted to the Michaelis-Menten equation shown below by non-linear least square regression and factor Xase enzyme parameters were estimated.
| eq. 2 |
where A is activity (nM/min/nM factor IXa concentration), [FX] is the factor X concentration, Vmax is the maximum reaction velocity, and Km is the Michaelis-Menten constant.
Factor Xa generation in the presence of platelets
Factor VIII (0–40 nM for factor VIIIa titration and 12 nM for factor X titration) in the presence of factor IXa (1 nM) was activated by thrombin (10 nM, 1 unit/ml) at 37°C for 2 minutes. Thrombin was inhibited by subsequent addition of hirudin (6 units/ml), immediately before reactions were added to platelets (5 × 107 platelets/ml). Where indicated, platelets were activated by 50 M SFLLRN-amide at 37°C for 5 minutes in HEPES-Tyrodes-BSA buffer and 5 mM CaCl2. Reactions were initiated by addition of factor X to the final concentrations indicated in the figure legends and stopped after 2 minutes at 37°C by addition of 10 mM EDTA. Progress curves were fit to the Michaelis-Menten equation (eq. 2).
Measurement of factor VIII/VIIIa binding to activated platelets
Wild type and E113A factor VIII were radiolabeled with Bolton-Hunter reagent to a specific activity of ~600–1,000 cpm/ng and were characterized structurally and functionally as described previously (14). Platelets (3–4 × 108 platelets/ml) were incubated in HEPES-Tyrodes buffer supplemented with BSA (2 mg/ml), 5 mM CaCl2, and mixtures of labeled and unlabeled factor VIII proteins as previously described (14). All binding experiments were performed in 1.5 ml Eppendorf plastic centrifuge tubes at 37°C for 20 minutes. After incubation, aliquots (100 μl) were removed and centrifuged at 12,000 × g in a Beckman Microfuge E, through a mixture of silicone oils (Dow-Corning 500 and Dow Corning 200 mixed 4:1, vol:vol) to separate platelets from unbound proteins (19). The 125I constant in both the platelet pellets and the supernatants was determined by counting gamma-emission in a Wallac Gamma Counter (Gaithersburg, MD) using the 125I energy window. The number of binding sites and Kd values were estimated by non-linear least square regression using the equation.
| eq. 3 |
where B is a concentration of bound factor VIII or VIIIa converted from radioactivity, [FVIII] is the factor VIII or VIIIa concentration, Bmax is the maximum factor VIII concentration able to bind to platelet surface, and Kd is the dissociation constant. Binding capacity was estimated by multiplying the Bmax value by Avogadro s number and dividing by the number of platelets.
Statistical Analysis
Nonlinear least-squares regression analysis was performed by Kaleidagraph (Synergy, Reading, PA) to obtain parameter values and standard deviations.
Results
Saturation mutagenesis at position E113
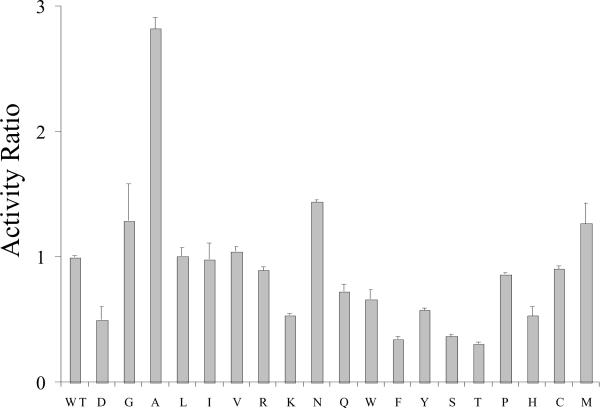
In a recent report we showed that a point mutation at A1 domain residue E113 to Ala (E113A) yielded a factor VIII form with an ~2-fold increased specific activity as determined by one-stage clotting assays (11). In order to determine whether Ala represented the optimal residue at this position for activity, we performed saturation mutagenesis at this site. Factor VIII constructs were prepared in which every amino acid was encoded at position 113. Factor VIII point mutants were transiently expressed in COS cells and factor VIII activity in the conditioned media was assayed using a one-stage clotting assay. A wild type control was expressed with each set of mutations and its activity value was set at unity. Results presented in Figure 1 showed that factor VIII:E113A possessed the apparent greatest increase in activity relative to wild-type (~3-fold). Levels of the expressed proteins were insufficient to accurately quantitate factor VIII concentration, hence specific activity values. Thus the following observations are limited by this constraint. Substitution with Gly, Asn or Met at this position yielded a modest activity increases (<50%), whereas several substitutions including Leu, Ile, Val, Pro, Cys and Arg showed little if any effect. On the other hand, a number of residues including Lys, Gln, Trp, Tyr, Pro, His, Asp, Phe, Ser, and Thr appeared to be somewhat detrimental to activity. Given the limitation in this analysis, these results tend to suggest that while substitutions at position 113 are not typically well-tolerated, selected mutations appear to result in modest increases in cofactor activity. Thus residue 113 appears to contribute, directly or indirectly to factor VIII function.
Figure 1.
Clotting activity following saturation mutagenesis at E113. Factor VIII expression vector constructs (HSQ-MSAB-NotI-RENeo) for the indicated point mutations were transfected into exponentially growing Cos-7 cells and the expressed factor VIII was collected and activity was measured using a one-stage clotting assay. Activity is presented relative to a transfected wild type control normalized to a value = 1.
Functional characterization of factor VIII:E113A
Further analyses of factor VIII:E113A and comparisons to wild type factor VIII used purified proteins obtained from stable expressing BHK cell lines as described in Methods. Factor VIII:E113A concentration and subunit composition were equivalent to wild type as confirmed by SDS-PAGE analysis and Coomassie staining (results not shown). Specific activity, as determined by one-stage clotting assay and ELISA, was rigorously tested over a range of factor VIII concentrations (0.1 to 2 nM) and for several separate preparations of the proteins. Results indicated that enhanced specific activity values, 9.8 ± 0.9 units/μg compared with 4.8 ± 0.5 units/μg for E113A and wild type factor VIII, respectively (this study and (11)), were observed for all preparations of E113A and at all concentrations tested (data not shown). Interestingly, kinetic data obtained in our earlier study (11) examining Ca2+ affinity by factor Xa generation assays performed in the presence of synthetic phospholipid vesicles suggested the E113A mutant possessed a modest (~10%) activity increase compared with wild type. Indeed, determination of specific activity values substituting the chromogenic, factor Xa generation assay for the one-stage clotting assay yielded values of 11.3 ± 1.8 and 10.9 ± 1.7 units/μg for E113A and wild type, respectively, confirming that the activities assessed by this method showed little if any difference.
Thrombin activation and activity decay of factor VIII:E113A
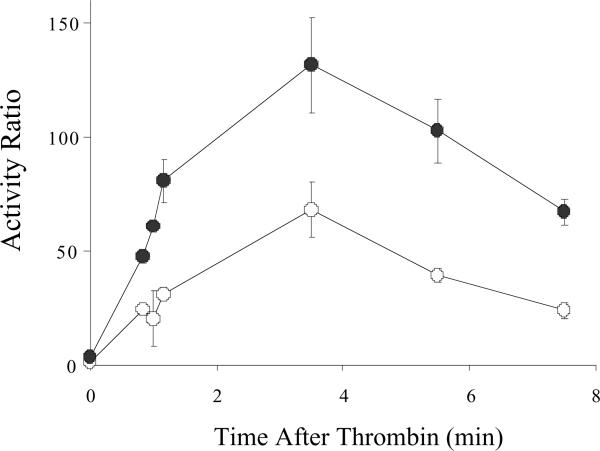
E113A and wild type factor VIII forms were activated by thrombin and changes in activity were monitored over time using a one-stage clotting assay (Figure 2). Both forms were activated ~40-fold which occurred over a similar time course. Furthermore, at all time points factor VIIIa derived from the E113A factor VIII possessed about twice the activity as wild type factor VIIIa. In addition, both activated forms decayed at similar rates suggesting that this mutation did not alter in the affinity of the A2 subunit within the factor VIIIa molecule.
Figure 2.
Factor VIII activity following activation by thrombin. Factor VIII E113A (closed circles) and wild type (open circles) (10 nM each) were activated by thrombin (5 nM, 0.6 units/ml) and activity was monitored by one-stage clotting assay over the indicated time course. Activity is expressed as a ratio to the non-activated factor VIII activity at time 0. Data points represent the average of two separate determinations.
Kinetic parameters for factor Xa generation in the presence of synthetic phospholipid vesicles
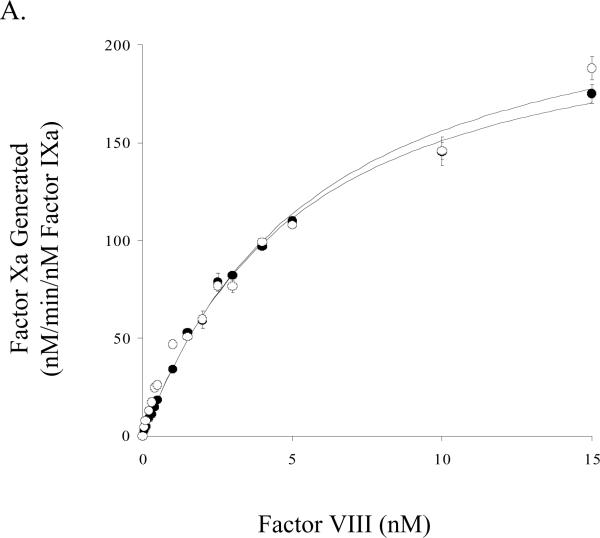
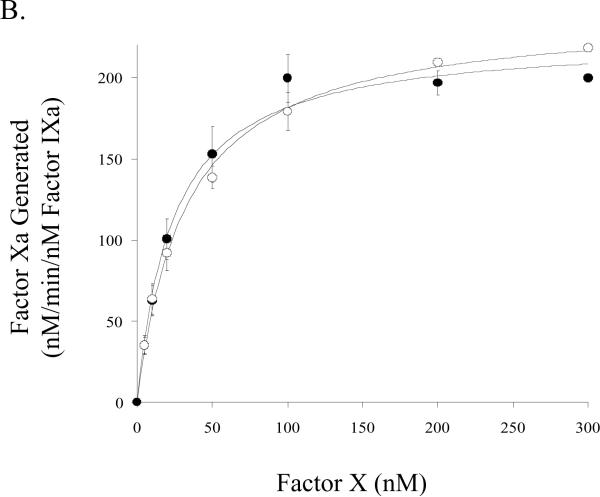
Factor Xa generation assays performed using purified reagents were used to assess kinetic and binding parameters for the E113A and wild-type factor VIII forms. Titration of factor IXa (1 nM) and saturating (see below) factor X (300 nM) with variable concentrations of the factor VIIIa forms were performed to yield functional Kd values to assess affinity of factor VIIIa for factor IXa in forming the factor Xase complex. Results from these experiments showed maximal rates of factor Xa generation (Vmax) were not significantly greater using factor Xase comprised of the E113A factor VIIIa form (Figure 3A, Table 1). The derived affinity values for the factor VIIIa-factor IXa interaction suggested no difference for the E113A factor VIIIa and wild type factor VIIIa forms (Table 1). Using fixed concentrations of factors VIIIa and IXa in the presence of phospholipid vesicles and titrating factor X, we determined a modestly increased Km value for E113A compared to wild type (Table 1).
Figure 3.
Factor VIII activity determined by a factor Xa generation assay in the presence of phospholipid A) Titration of factor IXa with factor VIIIa. Indicated concentrations of factor VIII:E113A (closed circles) or wild type (open circles) were activated with thrombin (20 nM, 1 unit/ml) and reacted with factor IXa (1 nM) and phospholipid vesicles (10 μM). Factor X (300 nM) was added and rates of factor Xa generation were measured. B) Titration of factor Xase complex with factor X. Reactions were as described above using factor IXa (1 nM), factor VIIIa form (20 nM), and phospholipid vesicles (10 μM). Reactions were initiated with the indicated concentration of factor X.
Table 1.
Comparison of factor VIII cofactor parameters measured in the presence of membrane surfaces
| wild type | E113A | |||||
|---|---|---|---|---|---|---|
| Surface | Phospholipid Vesicles | Activated Platelets | Non-activated Platelets | Phospholipid Vesicles | Activated Platelets | Non-activated Platelets |
| Factor IXa binding affinity (Kd in nM) | 4.6 ± 0.3 | 20.3 ± 5.1 | 25.6 ± 2.5 | 5.1 ± 0.7 (p>0.05)* | 6.0 ± 1.4 (p<0.02)* | 5.7 ± 0.6 (p<0.01) |
| Vmax (nM·min−1) | 225 ± 7.7 | 23.8 ± 2.9 | 3.1 ± 0.2 | 240 ± 4.7 (p>0.05) | 18.9 ± 1.8 (p>0.05) | 2.5 ± 0.1 (p<0.02) |
| Km (nM) | 23.8 ± 3.1 | 14.3 ± 0.8 | 16.7 ± 7.2 | 32.3 ± 2.2 (p<0.05) | 18.0 ± 1.1 (p<0.02) | 41.9 ± 16.8 (p>0.05) |
Kinetic parameters for factor Xa generation in the presence of platelets
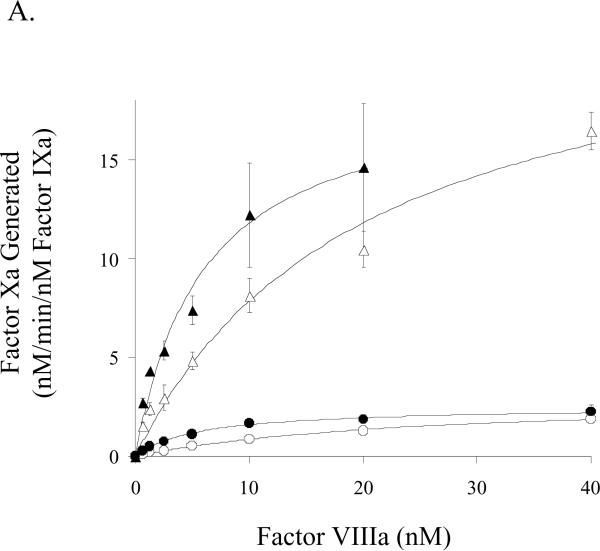
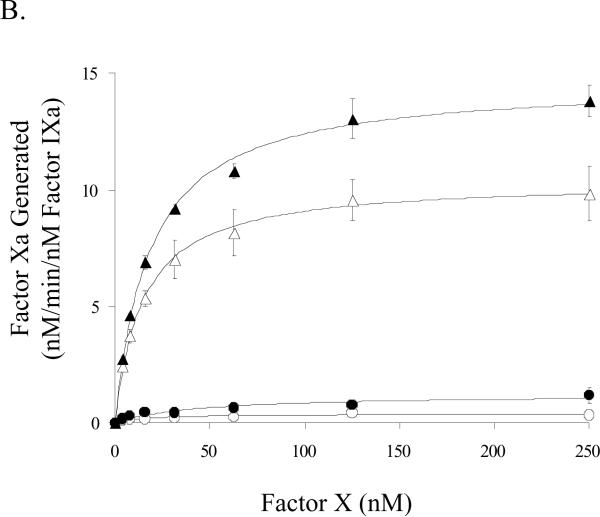
We also performed factor Xa generation assays to assess kinetic and binding parameters for the E113A and wild-type factor VIII forms in the presence of activated and non-activated platelets. Surprisingly, E113A factor VIIIa affinity for factor IXa, while similar to that observed for wild-type factor VIIIa on the synthetic membrane vesicles, was 3–4-fold higher than that observed for wild type factor VIIIa when measured on either activated or non-activated platelets (Fig. 4A, Table 1). E113A factor VIIIa showed a slightly higher Km value (~30 %) compared with wild type when factor Xa generation assays were performed in the presence of activated platelets (Table 1). A greater apparent increase in this parameter (~3 fold) was observed for E113A factor VIIIa compared with wild type on the unactivated platelet. However, the relatively large standard deviation of the experiment, likely resulting from low signal to background values, failed to show a statistical difference. The Vmax value obtained by titrating E113A factor VIIIa into 1 nM factor IXa was not significantly different compared with that for wild type factor VIIIa. The Vmax estimations in Figure 4B obtained by titrating substrate factor X were performed at cofactor concentrations (12 nM) below the Kd for factor IXa interaction with wild type factor VIIIa. Thus the greater apparent Vmax values observed with the E113A factor VIIIa (14.7 and 1.2 nM/min on activated and non-activated platelets, respectively) compared with wild type factor VIIIa (10.4 and 0.4 nM/min on activated and non-activated platelets, respectively) are likely a reflection of a higher concentration of factor Xase due to the higher affinity of E113A factor VIIIa for factor IXa.
Figure 4.
Factor VIII activity determined by a factor Xa generation assay in the presence of platelets. A) Titration of factor IXa with factor VIIIa. Indicated concentrations of factor VIII:E113A (closed symbols) or wild type (open symbols) were activated with thrombin (10 nM, 0.6 units/ml) and reacted with factor IXa (1 nM) and activated (triangles) or non-activated platelets (circles). Factor X (250 nM) was added and rates of factor Xa generation were measured. B) Titration of factor Xase complex with factor X. Reactions were as described above using factor IXa (1 nM), factor VIIIa form (12 nM), and activated (triangles) or non-activated platelets (circles). Reactions were initiated with the indicated concentration of factor X.
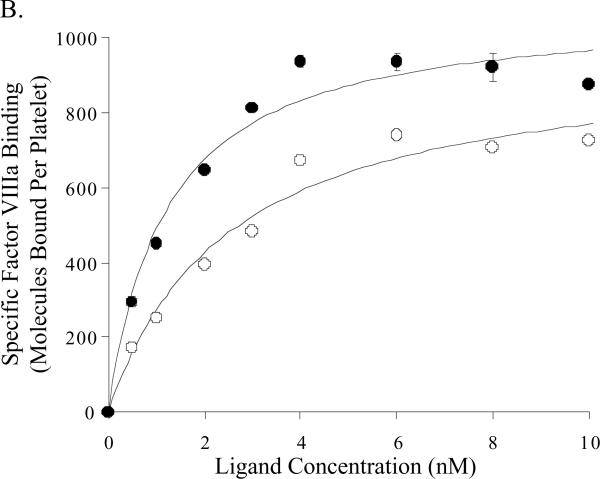
Factor VIII:E113A binding on activated platelets
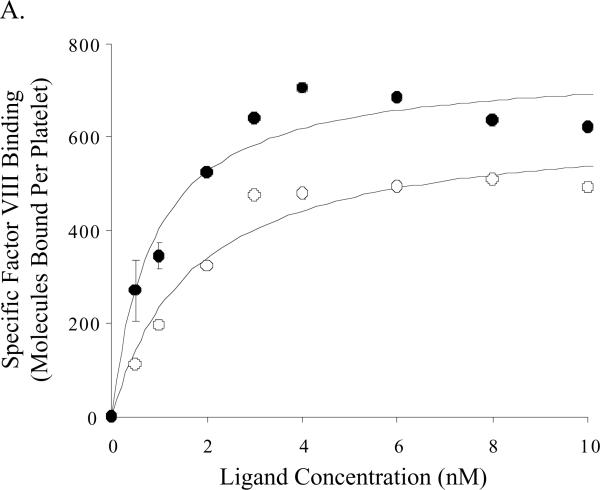
Binding parameters for wild type and E113A factor VIII/VIIIa on activated platelets were assessed using 125I-labeled proteins. Binding affinities of the E113A proteins for platelets were increased ~2-fold compared with the wild type proteins (Figure 5, Table 2) and this difference was significant for the factor VIIIa forms. While the mean number of binding sites appeared somewhat greater for the factor VIIIa forms compared with the unactivated cofactors, we observed no significant differences as a result of mutation at E113. High factor VIII/VIIIa concentrations (>8 nM) resulted in apparent decreases in the observed maximal binding capacity (<15% of maximal values), and this effect was more pronounced for the E113A proteins. The reason(s) for this observation is (are) not known.
Figure 5.
Equilibrium binding of labeled factor VIII forms on activated platelets. The results shown represent specific binding of factor VIII (Panel A) and factor VIIIa (Panel B) to activated platelets as described in Methods, where open and closed circles represent specific binding of factor VIII wild type and factor VIII E113A, respectively.
Table 2.
Binding constants for factor VIII reagents to activated platelets
| Ligand | Number of Binding Sites (per platelet) | Apparent Kd (nM) |
|---|---|---|
| wild type FVIII | 627 ± 48 | 1.68 ± 0.43 |
| E113A FVIII | 750 ± 46 (p>0.05)* | 0.86 ± 0.23 (p> 0.05) |
| wild type FVIIIa | 963 ± 75 | 2.51 ± 0.55 |
| E113A FVIIIa | 1083 ± 59 (p>0.05) | 1.20 ± 0.25 (p<0.05) |
Parameter values were estimated by nonlinear least square regression of the data shown in Figures 5A and 5B.
Statistical significance of the difference of the values between wild type and E113A was calculated by Student s t-test.
Discussion
We have recently mapped a Ca2+-binding site within residues 110–126 of the A1 domain of factor VIII (11). This region was identified following site-directed mutagenesis of candidate acidic residues within this segment, expression and purification of the recombinant factor VIII forms, and functional assay assessing affinity for Ca2+. Results from that study identified several acidic residues that contribute to Ca2+ coordination, and showed that occupancy of this site was necessary for maximal factor VIII specific activity. Mn2+ also binds near this region yielding a similar effect to that seen with Ca2+. While the mechanism for metal ion-induced increase in activity is not fully understood, we proposed it resulted from stabilizing the conformation in and around the A1-C1 interface, thereby facilitating functional alignment of factor VIIIa A domains with factor IXa in the intrinsic factor Xase.
The glutamate residue at position 113 (E113), which is conserved in all known species of factor VIII, does not appear to directly contribute to coordinating the calcium or manganese ions (11). However that study determined that substitution at position 113 with alanine increased the affinity for either ion by ~2-fold. One possible explanation for this effect is the negative charge at residue 113 may tend to compete for the divalent cation, weakening its binding at the desired position. Thus elimination of this charge may facilitate binding of the divalent cation to its appropriate coordination site. This hypothesis is supported by the mutation, E113D that yields a hemophilic phenotype (mild or severe) as indicated in the Hemophilia A database (12). Consistent with the database, we observed a >50% reduced activity with factor VIII E113D relative to wild type factor VIII. This result is not predicted given the conservation of charge with the Glu to Asp substitution. One possible explanation for this phenotype is that the smaller Asp residue may position its carboxylic group somewhat further removed from the site for Ca2+ coordination, in effect drawing the ion away from its requisite binding pocket. Since occupancy of this site is required for maximal cofactor activity (11), this competition may destabilize the coordination site leading to incorrect and/or weakened binding of the metal ion.
The mechanism for the increase in cofactor activity observed for E113A factor VIII is not likely related to its enhanced affinity for Ca2+ (or Mn2+), since saturating levels of Ca2+were included in reaction buffers. While this factor VIII retained a ~2-fold greater specific activity following activation by thrombin, the apparent rate of its activation and subsequent rate of decay of activity appeared similar to the wild type protein, suggesting that the mutation did not affect either interaction with the activating enzyme or dissociation of the A2 subunit from factor VIIIa. In addition, wild type and E113A showed equivalent stability of factor VIII activity at 50°C, inter-factor VIII heavy chain-light chain affinity, binding affinity for von Willebrand factor and effects on the fluorescence anisotropy of fluorescein-Phe-Phe-Arg-factor IXa in the presence of synthetic vesicles (data not shown). Furthermore, when we measured the functional affinity for factor IXa and Km for factor X, as judged by factor Xa generation assays in the presence of synthetic phospholipids, we found virtually no differences. However, when we performed factor Xa generation assays in the presence of platelets, E113A exhibited a 3–4 fold increase in factor IXa binding affinity compared with wild type.
Comparison of factor Xa generation rates on the synthetic phospholipid vesicle and platelet membrane surfaces revealed significantly reduced reaction rates on the latter surface. This observation likely reflects limiting sites on the platelet surface for factor Xase assembly with more sites available when the platelet is activated compared to non-activated (see Ref. (20) for review). This limitation is not the case with the synthetic membrane surface where a Vmax concentration of vesicles allows for essentially complete utilization of the input factors VIIIa and IXa. Hence this surface does not appear to allow for subtle discrimination of factor IXa affinity exhibited by the E113A and wild type factor VIIIa forms. Thus we speculate that under conditions leading to low concentrations of factor Xase, that is, complex formation on the platelet surface, as well as in a one-stage clotting assay due to low levels of factor IXa formed, factor Xase formation is facilitated by the relatively higher inter-protein affinity demonstrated by the E113A mutant. An alternate explanation, that of a plasma-derived contaminant that is also present in the platelet preparation, responsible for enhancing the activity of E113A factor VIIIa in the clotting and platelet assays is not likely since dilution of wild type and E113A factor VIII in hemophilic plasma yielded similar results in factor Xa generation assays using synthetic vesicles (data not shown).
One mechanism for this observed increase in factor IXa affinity shown for E113A factor VIIIa may result from the observed increased affinity of this factor VIIIa form for the platelet surface, since the inter-protein affinity value is a membrane-dependent parameter. The reason for the enhanced affinity of E113A factor VIII for platelets is not currently known. Clearly, the E113A mutation occurs in a site (A1 domain) far removed from regions of factor VIII involved in membrane binding. While significant evidence implicates residues within the C2 domain as interacting with the membrane surface (21, 22), the recent x-ray structure of the factor Vai A1/A3-C1-C2 dimer (23) suggests C1 may also contact the membrane surface. Interestingly, the E113A mutation occurs in a region of A1 involved in Ca2+ coordination that appears to juxtapose the C1 domain (24) and occupancy of this coordination site modulates the structure of factor VIII to yield the active conformation (9). Thus one potential effect of the mutation could be an alteration in conformation in and around this Ca2+ site that is transduced into C1, subsequently affecting a membrane-binding site.
Extensive studies on factor Xase assembly and function on the (activated) platelet surface have defined unique coordinate effects related to component binding affinity and number of sites (20). Thus the E113A mutation may possess either enhanced interaction for a specific membrane structure and/or membrane-bound factors IXa and/or factor X. One or more of these properties may be responsible for the ~2-fold increase in specific activity measured for E113A factor VIII in the one-stage clotting assay compared to the equivalence of this mutant with wild type factor VIII when assayed in the factor Xa generation assay using vesicles. The reason(s) for this activity disparity is (are) not known, but likely reflect differences in the assays used to measure cofactor activity. Assay disparities between the plasma-based and factor Xa generation (chromogenic) assays, especially as noted for B-domainless factor VIII forms, are a source of controversy (25). Indeed, we observed that factor VIII wild type yielded a specific activity value in the one-stage assay that was ~50% that observed in the chromogenic assay. On the other hand, specific activity values obtained for E113A showed minimal assay-dependent differences, suggesting that the E113A mutation substantially reduced discrepancies in the two activity assays.
There is significant interest in using recombinant DNA technology to develop superior factor VIII therapeutics that may improve secretion, reduce antigenicity and immunogenicity, prolong circulatory half-life, and resist inactivation due to subunit dissociation and/or proteolysis (see Ref. (26) for review). The E113A factor VIII reagent described in this report offers a further addition to this repertoire; that of possessing increased specific activity due to a point mutation within a Ca2+ binding site that yields enhanced factor Xase formation on physiological membrane surfaces.
Acknowledgements
We thank Pete Lollar and John Healey for generous provision of the factor VIII construct HSQ-MSAB-NotI-RENeo, Lisa Regan and Bayer Corporation for the gifts of recombinant factor VIII, and Jan Freas and Qian Zhou for excellent technical assistance.
This work was supported by grants HL38199 and HL76213 (to P.J.F.) and HL70683, HL64943, HL46213 and HL74124 (to P.N.W.) from the National Institutes of Health.
Footnotes
The abbreviations used are: EDTA, ethylenediamine tetraacetic acid; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PS, phosphatidylserine; HEPES, N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid; MES, 4-morpholinethanesulfonic acid; BSA, bovine serum albumin; BHK, baby hamster kidney; SDS-PAGE, sodium dodecyl sulfate -polyacrylamide gel electrophoresis; ELISA, enzyme-linked immunoadsorbent assay.
References
- 1.Davie EW. Biochemical and molecular aspects of the coagulation cascade. Thrombosis & Haemostasis. 1995;74:1–6. [PubMed] [Google Scholar]
- 2.Lollar P. Structure and function of Factor VIII. Advances in Experimental Medicine & Biology. 1995;386:3–17. doi: 10.1007/978-1-4613-0331-2_1. [DOI] [PubMed] [Google Scholar]
- 3.Fass DN, Knutson GJ, Katzmann JA. Monoclonal antibodies to porcine factor VIII coagulant and their use in the isolation of active coagulant protein. Blood. 1982;59:594–600. [PubMed] [Google Scholar]
- 4.Fay PJ, Anderson MT, Chavin SI, Marder VJ. The size of human factor VIII heterodimers and the effects produced by thrombin. Biochimica et Biophysica Acta. 1986;871:268–278. doi: 10.1016/0167-4838(86)90208-6. [DOI] [PubMed] [Google Scholar]
- 5.Fay PJ. Reconstitution of human factor VIII from isolated subunits. Archives of Biochemistry & Biophysics. 1988;262:525–531. doi: 10.1016/0003-9861(88)90404-3. [DOI] [PubMed] [Google Scholar]
- 6.Nordfang O, Ezban M. Generation of active coagulation factor VIII from isolated subunits. Journal of Biological Chemistry. 1988;263:1115–1118. [PubMed] [Google Scholar]
- 7.Fay PJ, Smudzin TM. Intersubunit fluorescence energy transfer in human factor VIII. Journal of Biological Chemistry. 1989;264:14005–14010. [PubMed] [Google Scholar]
- 8.Wakabayashi H, Koszelak ME, Mastri M, Fay PJ. Metal ion-independent association of factor VIII subunits and the roles of calcium and copper ions for cofactor activity and inter-subunit affinity. Biochemistry. 2001;40:10293–10300. doi: 10.1021/bi010353q. [DOI] [PubMed] [Google Scholar]
- 9.Wakabayashi H, Schmidt KM, Fay PJ. Ca(2+) binding to both the heavy and light chains of factor VIII is required for cofactor activity. Biochemistry. 2002;41:8485–8492. doi: 10.1021/bi025589o. [DOI] [PubMed] [Google Scholar]
- 10.Wakabayashi H, Zhen Z, Schmidt KM, Fay PJ. Mn2+ binding to factor VIII subunits and its effect on cofactor activity. Biochemistry. 2003;42:145–153. doi: 10.1021/bi026430e. [DOI] [PubMed] [Google Scholar]
- 11.Wakabayashi H, Freas J, Zhou Q, Fay PJ. Residues 110–126 in the A1 domain of factor VIII contain a Ca2+ binding site required for cofactor activity. Journal of Biological Chemistry. 2004;279:12677–12684. doi: 10.1074/jbc.M311042200. [DOI] [PubMed] [Google Scholar]
- 12.Kemball-Cook G, Tuddenham EG, Wacey AI. The factor VIII Structure and Mutation Resource Site: HAMSTeRS version 4. Nucleic Acids Research. 1998;26:216–219. doi: 10.1093/nar/26.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mimms LT, Zampighi G, Nozaki Y, Tanford C, Reynolds JA. Phospholipid vesicle formation and transmembrane protein incorporation using octyl glucoside. Biochemistry. 1981;20:833–840. doi: 10.1021/bi00507a028. [DOI] [PubMed] [Google Scholar]
- 14.Ahmad SS, Scandura JM, Walsh PN. Structural and functional characterization of platelet receptor-mediated factor VIII binding. Journal of Biological Chemistry. 2000;275:13071–13081. doi: 10.1074/jbc.275.17.13071. [DOI] [PubMed] [Google Scholar]
- 15.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins PV, Freas J, Schmidt KM, Zhou Q, Fay PJ. Mutations associated with hemophilia A in the 558–565 loop of the factor VIIIa A2 subunit alter the catalytic activity of the factor Xase complex. Blood. 2002;100:501–508. doi: 10.1182/blood-2001-12-0361. [DOI] [PubMed] [Google Scholar]
- 17.Casillas G, Simonetti C, Pavlovsky A. Artificial substrate for the assay of factors V and VIII. Coagulation. 1971;4:107–111. [Google Scholar]
- 18.Lollar P, Fay PJ, Fass DN. Factor VIII and factor VIIIa. Methods in Enzymology. 1993;222:128–143. doi: 10.1016/0076-6879(93)22010-d. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad SS, Rawala-Sheikh R, Walsh PN. Comparative interactions of factor IX and factor IXa with human platelets. Journal of Biological Chemistry. 1989;264:3244–3251. [PubMed] [Google Scholar]
- 20.Ahmad SS, London FS, Walsh PN. The assembly of the factor X-activating complex on activated human platelets. Journal of Thrombosis & Haemostasis. 2003;1:48–59. doi: 10.1046/j.1538-7836.2003.00020.x. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert GE, Kaufman RJ, Arena AA, Miao H, Pipe SW. Four hydrophobic amino acids of the factor VIII C2 domain are constituents of both the membrane-binding and von Willebrand factor-binding motifs. Journal of Biological Chemistry. 2002;277:6374–6381. doi: 10.1074/jbc.M104732200. [DOI] [PubMed] [Google Scholar]
- 22.Pratt KP, Shen BW, Takeshima K, Davie EW, Fujikawa K, Stoddard BL. Structure of the C2 domain of human factor VIII at 1.5 A resolution. Nature. 1999;402:439–442. doi: 10.1038/46601. [DOI] [PubMed] [Google Scholar]
- 23.Adams TE, Hockin MF, Mann KG, Everse SJ. The crystal structure of activated protein C-inactivated bovine factor Va: Implications for cofactor function. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8918–8923. doi: 10.1073/pnas.0403072101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pemberton S, Lindley P, Zaitsev V, Card G, Tuddenham EG, Kemball-Cook G. A molecular model for the triplicated A domains of human factor VIII based on the crystal structure of human ceruloplasmin. Blood. 1997;89:2413–2421. [PubMed] [Google Scholar]
- 25.Lollar P. The factor VIII assay problem: neither rhyme nor reason. J. Thromb. Haemost. 2003;1:2275–2279. doi: 10.1046/j.1538-7836.2003.00484.x. [DOI] [PubMed] [Google Scholar]
- 26.Saenko EL, Ananyeva NM, Shima M, Hauser CAE, Pipe SW. The future of recombinant coagulation factors. Journal of Thrombosis and Haemostasis. 2003;1:922–930. doi: 10.1046/j.1538-7836.2003.00196.x. [DOI] [PubMed] [Google Scholar]