ABSTRACT
Galactose is important for the survival and virulence of bacteria. In Escherichia coli, galactose is utilized by the Leloir pathway, which is controlled by a complex network. To shed light on the potential functions the galactose network could perform, we performed bioinformatical analysis of reference genome sequences belonging to the Enterobacteriaceae family. We found that several genomes have reduced numbers of components compared to the E. coli galactose system, suggesting that the network can be optimized for different environments. Typically, genes are removed by deletions; however, in Yersinia pestis, the galactose mutarotase (galM) gene is inactivated by a single-base-pair deletion. Lack of GalM activity indicates that the two anomers of d-galactose are used for different purposes, α-d-galactose as a carbon source and β-d-galactose for induction of UDP-galactose synthesis for biosynthetic glycosylation. We demonstrate that activity of the galM gene can be restored by different single-base-pair insertions. During the evolution of Y. pestis to become a vector-transmitted systemic pathogen, many genes were converted to pseudogenes. It is not clear whether pseudogenes are present to maintain meiotrophism or are in the process of elimination. Our results suggest that the galM pseudogene has not been deleted because its reactivation may be beneficial in certain environments.
IMPORTANCE
Evolution of bacteria to populate a new environment necessarily involves reengineering of their molecular network. Members of the Enterobacteriaceae family of bacteria have diverse lifestyles and can function in a wide range of environments. In this study we performed bioinformatical analysis of 34 reference genome sequences belonging to the Enterobacteriaceae family to gain insight into the natural diversity of the d-galactose utilization network. Our bioinformatical analysis shows that in several species, some genes of the network are completely missing or are inactivated by large deletions. The only exception is the galactose mutarotase (galM) gene of Yersinia pestis, which is converted to a pseudogene by a single-base-pair deletion. In this paper, we discuss the possible consequences of galM inactivation on network function. We suggest that galM was converted to a pseudogene rather than being deleted in evolution because its reactivation can be beneficial in certain environments.
Introduction
Evolution of bacteria to populate a new environment necessarily involves reengineering of their molecular network. Changes can affect the elements (e.g., genes) of the network as well as the interactions in the network. Redundant genes or those that antagonize successful colonization in the new environment can be inactivated by point mutations or removed by deletions (1). Also, genes required for adaptation to a new niche can be acquired by horizontal transfer (e.g., pathogenicity islands) (2). A major determinant of network function is network logic, which depends on the interactions between network elements (3). Recent studies demonstrated that network logic can be easily engineered by mutations in regulatory sequences (4, 5). Rearrangements in metabolic pathways were analyzed in Yersinia pestis, which diverged recently from Yersinia pseudotuberculosis, a gastrointestinal pathogen (6). Analysis of the Yersinia pestis metabolic network suggested excellent agreement between the possible metabolic reaction pathways and the known nutritional needs of Y. pestis cells (1). It is generally assumed that cellular responses to the natural levels of perturbations in the concentration of critical chemical compounds are optimized. Along this logic, capabilities of regulatory networks would reflect the potential nature of environments and the environmental changes cells may face (7).
Members of the Enterobacteriaceae family of bacteria have diverse lifestyles and can function in a wide range of environments. In this study we performed bioinformatical analysis of 34 reference genome sequences belonging to the Enterobacteriaceae family to gain insight into the natural diversity of the d-galactose utilization network. The rationale of using the d-galactose network is that d-galactose metabolism can be an important factor in virulence in different bacteria (8–10). For example, in Erwinia amylovora, the causal agent of fire blight of rosaceous plants, galactose metabolism affects capsule synthesis and virulence (9). In Y. pestis, both the d-galactose transport and metabolism operons are induced when the temperature is shifted from 26°C to 37°C, which corresponds to the temperature change in the transition from the flea to the mammalian host (11). Also, incorporation of β-d-galactose into the lipopolysaccharide (LPS) of Y. pestis inside the transmitting vector (flea) may be important (12). The d-galactose network of Escherichia coli is well characterized (13) and suitable to be used as a reference for the comparative analysis.
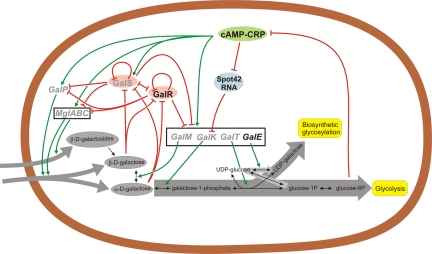
Escherichia coli utilizes d-galactose by the amphibolic Leloir pathway (Fig. 1). The metabolic steps of d-galactose utilization are catalyzed by the GalK (galactokinase), GalT (galactose 1-phosphate uridylyltransferase), and GalE (UDP-galactose-4-epimerase) proteins. In E. coli, the genes encoding these proteins belong to the same operon, together with the galM gene, which encodes a mutarotase that allows the intracellular interconversion of the two anomers of d-galactose (14), α-d-galactose and β-d-galactose. Only α-d-galactose serves as a substrate of galactokinase, the first enzyme of the Leloir pathway. d-Galactose can be transported by two d-galactose-induced transport systems, the low-affinity GalP and the high-affinity Mgl systems (15). Transport and utilization of d-galactose is regulated by the Gal repressor (GalR) and the Gal isorepressor (GalS). These repressors bind the same set of operators in the regulatory regions of the genes belonging to the gal regulon in the absence of d-galactose. d-Galactose binding to GalR (similar to that to GalS) allosterically inhibits its operator binding (13, 16, 17). A recent study demonstrated that both of these anomers of d-galactose are effective in the binding and inactivation of GalR (18).
FIG 1 .
Regulation of proteins involved in d-galactose transport and utilization. Vast grey arrows indicate the major metabolic flows. This figure is based on the E. coli d-galactose network (13) but for simplicity shows only the gene products (proteins and sRNA) of the 12 genes involved in the bioinformatical analysis. Products of the four genes which are present in all 34 genomes studied are typed in black, and the others which are missing from one or more genomes are shown in grey. Red lines indicate inhibitions, while green lines indicate enhancements. Proteins which often belong to the same operon are shown in black boxes.
In the Leloir pathway, only the galE gene product is involved in making substrates for biosynthetic glycosylation reactions, while all of the Gal enzymes are needed for catabolism of d-galactose. Therefore, when d-galactose is not available or not preferred as a carbon source, expression of the gal operon genes is discoordinated, resulting mostly in GalE synthesis (19–21). Enterobacteria evolved different mechanisms to allow such discoordination. These include using small RNA (sRNA)-mediated translational regulation (22), differentially regulated promoters and natural polarity (19–21, 23), and physical separation of the galE gene from the rest of the gal operon genes (23).
In this paper, we focus primarily on the role of galactose mutarotase in the d-galactose utilization network. We show that the function of the galM pseudogene of Y. pestis can be restored by single-base-pair insertions in different ways. Our analysis predicts that the two d-galactose anomers, α- and β-d-galactose, play different roles in Y. pestis. Only α-d-galactose can be used as a carbon source, while β-d-galactose can induce the production of UDP-galactose, a compound used in biosynthetic glycosylation reactions.
RESULTS
Identification of elements of the d-galactose network in enterobacteria.
Because metabolic enzyme sequences are highly conserved across species, metabolic networks can be successfully reconstructed primarily based on genome sequence data (24). Therefore, we used the BLAST program (25) to identify DNA and protein sequences in reference genome sequences of strains belonging to the Enterobacteriaceae family, which are similar to components of the d-galactose network of E. coli K-12 MG1655 (Fig. 2). We found only four genes, galR (encoding the galactose repressor), galE (encoding the UDP-galactose-4-epimerase), crp (encoding the cyclic AMP [cAMP] receptor protein [CRP]), and spf (encoding the Spot42 sRNA), which are present in all the genomes studied. All 12 genes studied (galETKM, mglBAC, galR, galS, galP, crp, and spf) are present in 19 of the 34 genomes analyzed. However, in six of these strains, the galE gene is not part of the gal operon. Six strains have the high-affinity Mgl transport system but not the low-affinity GalP symporter, suggesting that the galactose utilization networks of these strains are optimized for low-galactose environments (Dickeya dadantii, Dickeya zeae, Sodalis glossinidius, Yersinia enterocolitica, Y. pestis, and Y. pseudotuberculosis strains). As opposed to those strains, Edwardsiella ictaluri, Edwardsiella tarda, Erwinia pyrifoliae, Shigella boydii, and Shigella dysenteriae have only the low-affinity transporter, which requires high levels of extracellular d-galactose for proper function. The three Pectobacterium strains and Proteus mirabilis lack both transport systems. Genes responsible for galactose metabolism (galE, galK, galT, galM) can be found in different arrangements in the strains analyzed; however, there are two strains in which we found inactive components. In Sodalis glossinidius, only the galE gene is intact, suggesting that in these cells, UDP-galactose required for biosynthetic purposes is produced from UDP-glucose. In Y. pestis, the galM gene contains a single-nucleotide deletion, resulting in a frameshift.
FIG 2 .
Presence of genes responsible for d-galactose transport (galP, mglBAC), utilization (galETKM), and regulation of these two processes (galR, galS, crp, spf) in genomes of strains belonging to the Enterobacteriaceae family (GenBank accession numbers are shown on the left, and more details are provided in Materials and Methods). Filled boxes indicate the presence of intact genes, based on bioinformatical analysis. Empty boxes indicate genes disrupted by extensive deletions or insertions. The galM pseudogene of Y. pestis, which is inactivated by a single-base-pair deletion, is marked by an asterisk. Arrowheads indicate the direction of transcription for putative operons and ORFs separated by less than 1 kbp.
Our bioinformatical analysis shows that in most of the cases, genes are completely missing or are inactivated by large deletions. The only exception is the galM gene of Y. pestis, which is converted to a pseudogene. The same pseudogene was found in all the Y. pestis genome sequences (e.g., GenBank accession no. NC_010159.1, NC_009381.1, NC_008149.1, NC_008150.1, NC_005810.1, NC_003143.1, and NC_004088.1).
Functional restoration of the galM pseudogene of Y. pestis.
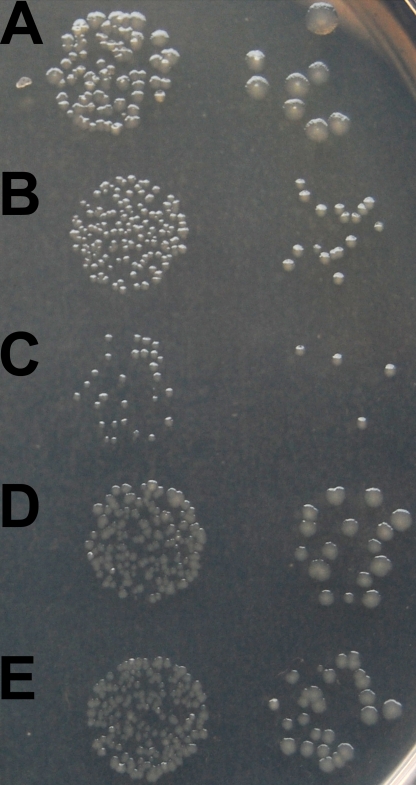
Comparison of the Y. pestis and Y. pseudotuberculosis genomes indicated that the transition from the enteropathogen form to the vector-transmitted systemic pathogen form resulted in more than 300 pseudogenes (26, 27). However, it remains to be answered whether pseudogenes are present to maintain meiotrophism or are in the process of elimination. The Y. pestis galM pseudogene differs from the Y. pseudotuberculosis YPIII galM gene only by a single-base-pair deletion (at nucleotide position 117 of the open reading frame [ORF]) and a same-sense mutation (at nucleotide position 180). Therefore, this pseudogene can likely be reactivated by insertion of a single base pair. The GalM protein that would be encoded by the “repaired” Y. pestis galM gene is identical to the Y. pseudotuberculosis YPIII GalM protein (GenBank accession no. YP_001721673.1). Interestingly, the Yersinia pseudotuberculosis IP 32953 galM gene encodes a different amino acid at the position of the required insertion (D instead of N), suggesting that the Y. pestis pseudogene can be restored in multiple ways. To test this hypothesis we synthesized the Y. pestis galM pseudogene and its different “restored” versions and inserted them into a low-copy-number plasmid. The plasmid-borne genes were tested whether they can complement a chromosomal galM deletion in E. coli. GalM function was tested by studying growth on intracellularly produced β-d-galactose. Cells were grown in the presence of phenyl-β-d-galactopyranoside as the sole carbon source. Hydrolysis of phenyl-β-d-galactopyranoside by β-galactosidase generates phenol and β-d-galactose. Phenol is excreted into the culture medium and does not accumulate in the cell. Cells were grown overnight in LB medium and then washed in M63 minimal medium. Washed cells were plated on M63 minimal medium supplemented with 2 mM phenyl-β-d-galactopyranoside, 0.0004% vitamin B1, 0.5 mM IPTG (isopropyl-thiogalactoside; to induce β-galactosidase production), and 15 µg/ml tetracycline. We found that growth strongly depends on the presence of functional galM. Cells containing the Y. pestis galM pseudogene showed very slow growth, similar to that of the E. coli ΔgalM cells harboring the empty plasmid. This slow growth reflects the low level of nonenzymatic mutarotation of the d-galactose anomers. However, two “restored” versions of GalM (N39, D39) conferred similar growth to wild-type E. coli MG1655 cells (Fig. 3). The difference in the size of colonies reflects a minimum doubling time of 135 to 150 min for the galM+ and about 1,000 min for the ΔgalM strains, as measured in liquid cultures. GalM version Q39 grew similarly to galM+, I39 and T39 showed intermediate growth, while K39, R39, and E39 were not growing substantially faster than ΔgalM.
FIG 3 .
Complementation of a chromosomal galM mutation (ΔgalM) in E. coli by two restored versions of the Y. pestis galM pseudogene carried on a low-copy-number plasmid. Cells containing a functional copy of galM grow faster on intracellularly produced β-d-galactose as the sole carbon source. Hydrolysis of phenyl-β-d-galactopyranoside by β-galactosidase generates phenol and β-d-galactose. Phenol is excreted into the culture medium and does not accumulate in the cell. MG1655ΔgalM cells containing plasmid pLG338E carrying the Y. pestis galM pseudogene (C) and its two restored versions, N39 (D) and D39 (E), were plated on M63 minimal medium containing 2 mM phenyl-β-d-galactopyranoside and also 0.5 mM IPTG to induce β-galactosidase production. E. coli MG1655/pLG338E (A) and E. coli MG1655ΔgalM/pLG338E (B) were plated as controls for comparison.
Effect of galM and galP deletion on utilization of extracellular d-galactose in vivo.
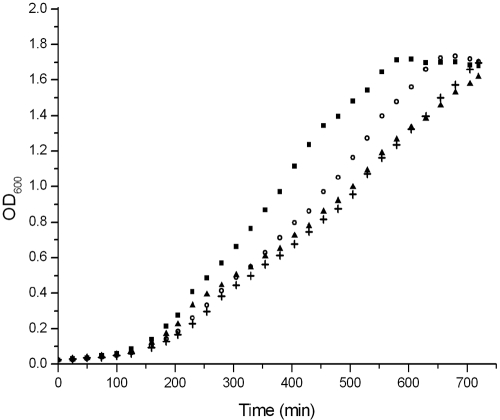
Y. pestis strains have only one of the d-galactose-induced transport systems. The three genes of the high-affinity Mgl (β-methylgalactoside) transport system are part of a single operon, similar to E. coli (mglBAC). However, Y. pestis strains lack the low-affinity GalP transporter. To simulate the function of the reduced network lacking both the low-affinity transporter and the galactose mutarotase, we created a ΔgalM ΔgalP double mutant of E. coli MG1655. We compared the growth of this double mutant with wild-type and single mutant strains (Fig. 4) in minimal d-galactose medium. We found that deletion of galM increases the time needed to reach stationary phase by about 100 minutes, while deletion of galP increases it by about 200 minutes, compared to the time needed by the wild-type strain. As expected, in the logarithmic phase, the ΔgalM mutant shows a growth rate similar to that of the wild-type strain (14). However, the double mutant shows growth similar to that of the ΔgalP strain, suggesting that growth is limited by the lack of the low-affinity transporter and not by the low rate of spontaneous d-galactose mutarotation. Because intracellular mutarotation of d-galactose is inefficient in the absence of GalM (14), our results suggest that besides β-d-galactose, the Mgl transport system can efficiently transport α-d-galactose as well; therefore, the presence of mutarotase does not substantially increase the rate of d-galactose utilization. Also, because β-d-galactose inactivates GalR similarly to α-d-galactose (18, 28), the unused β-d-galactose pool in the cell may result in higher derepression of the gal regulon genes; therefore, it allows higher rates of transport and utilization, which can compensate for the lack of β-d-galactose utilization in the double mutant.
FIG 4 .
Effect of galM and galP deletion on growth on d-galactose as a carbon source. We compared the growth of the ΔgalM ΔgalP double mutant (+) with those of the wild-type (▪), ΔgalM (○), and ΔgalP (▴) strains in minimal d-galactose medium. Cells were grown overnight in LB medium and diluted in M63 minimal medium supplemented with 0.0004% vitamin B1 and 0.3% d-galactose. OD600, optical density at 600 nm.
Prediction of regulatory links in the Y. pestis galactose network by sequence analysis.
By searching for genes similar to the regulator genes of the E. coli d-galactose system, we could confirm the presence of galR, crp (encoding the cAMP receptor protein), and spf (encoding the Spot42 small RNA). The amino acid sequences of the GalR proteins in Y. pestis strains are about 80% identical to those of the E. coli GalR protein. The DNA recognition helices are identical, suggesting that the same sequences are recognized by the Y. pestis and E. coli GalR proteins. However, the galS repressor gene was not found in Y. pestis strains. In order to predict regulatory links in the Y. pestis d-galactose network, we compared the regulatory regions of the Y. pestis galETK, mglBAC, and galR genes with the corresponding regions in E. coli. We found that the binding sites of the regulatory proteins in the control region of the galETK operon are arranged in a way similar to that found in E. coli. There are two GalR binding sites (operators); however, the spacing between the two operators is 1 bp shorter than that in E. coli. Comparison of the promoter elements suggested that the P1galE promoter is functional but that the P2galE promoter is significantly weaker than that in E. coli. In Y. pestis strains, the sequence found at the position of the extended −10 element of the E. coli P2galE promoter (TTTGTTATGCT) is AATGGCGTGCT, which is not similar to the consensus −10 element.
Based on the sequence similarities, the galR gene is assumed to be autoregulated in the same way as that in E. coli (13). The regulatory region and promoter of the mglBAC operon in Y. pestis strains are also highly similar to those of E. coli; however, the 5′ untranslated region of the mglBAC mRNA (about 250 bp in both species) shows no similarity. The sequence found in Y. pestis is conserved in Y. pestis and Y. pseudotuberculosis strains. A similar 5′ untranslated region of the mglBAC mRNA is found in Yersinia enterocolitica, but we could not find any other similar sequences in the database.
Transcription of the Y. pestis galETK operon is initiated from a single promoter.
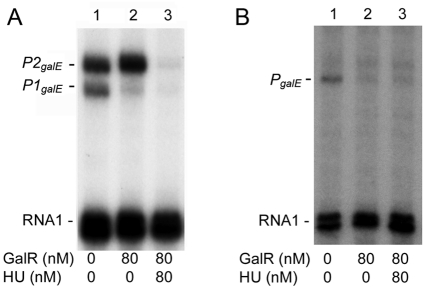
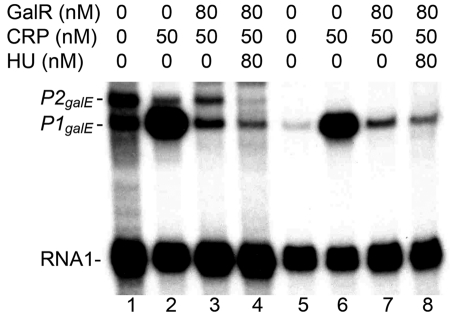
In order to test the predictions of sequence analysis, we studied the regulation of the Y. pestis galETK operon in a purified system. We performed in vitro transcription assays using E. coli RNA polymerase (σ70), purified Y. pestis GalR (GalRYP), and plasmid templates containing the regulatory region of the Y. pestis galETK operon. We used a similar plasmid construct containing the corresponding regulatory region from E. coli (pSA850) (29) for comparison (Fig. 5). We found that the GalRYP protein can regulate the E. coli galETKM operon the same way as E. coli GalR. In the presence of GalRYP, the level of P2galE transcription is increased, while P1galE transcription is repressed. When both GalRYP and HU (from E. coli) are present, both promoters are repressed by DNA looping (30) (Fig. 5). However, unlike in E. coli, transcription of the Y. pestis galETK operon is initiated only from a single site, corresponding to the start site of the P1galE promoter in E. coli. GalRYP repressed this promoter equally in both the presence and absence of HU. Similar to the P1galE promoter of E. coli, cAMP-CRP strongly activated the Y. pestis gal promoter, and GalR reduced the effect of cAMP-CRP (Fig. 6).
FIG 5 .
Transcription regulation of the Y. pestis galETK operon. In vitro transcription assays were performed on plasmid templates containing the regulatory region of the gal operon from E. coli (A) and from Y. pestis (B). Lane 2 shows the effect of GalRYP, while lane 3 shows the combined effect of GalRYP and HU. Protein concentrations are indicated below the lanes. The RNA1 transcript, which does not vary with the GalR concentration, was used as an internal control between lanes.
FIG 6 .
Combined effect of GalR and CRP on transcription of the Y. pestis gal operon promoter (lanes 5 to 8). Regulation of the E. coli gal operon promoters is shown for comparison (lanes 1 to 4). Protein concentrations are indicated on top. The RNA1 transcript, which does not vary with the GalR, HU, or CRP concentrations, was used as an internal control between lanes.
DISCUSSION
Functional diversity of the d-galactose network in enterobacteria.
The functional role of different genes in the d-galactose utilization network of E. coli has been studied extensively. Therefore, based on the set of genes present in a certain reduced network, it is possible to formulate predictions about the functional consequences of reduction. Functional diversity of the reduced networks affects both transport and metabolism. Some networks contain only the low-affinity transporter (5), while others have only the high-affinity transport system (6), suggesting optimal performance in high-galactose and low-galactose environments, respectively. In four cases, we found that both of these transporters are missing; therefore, the function of the galactose system is limited to endogenous inducer synthesis (31) and metabolism of intracellular d-galactose obtained from galactose-containing compounds. Galactose metabolism is affected in two cases, as follows. In the S. glossinidius network, only the galE gene of the gal operon is functional, indicating that this network is incapable of amphibolic utilization of d-galactose. In Y. pestis, the last gene of the gal operon, galM, is inactivated by a single-base-pair deletion.
d-Galactose utilization in Y. pestis.
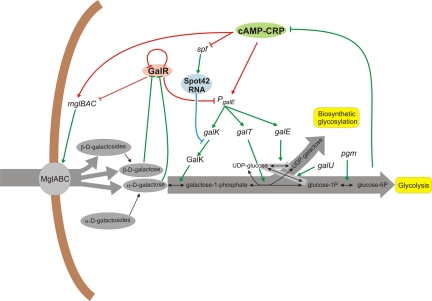
To understand the consequences of galM inactivation, we identified elements and interactions in the d-galactose network of Y. pestis by combining experimental results with the results of bioinformatical analyses (Fig. 7). The Y. pestis network is less complex than the E. coli d-galactose network. Both the number of elements and the number of interaction links are reduced. The Y. pestis d-galactose system is regulated by a single d-galactose-responsive regulator, which is interchangeable with E. coli GalR in in vitro experiments. d-Galactose transport of Y. pestis strains is also simplified, having only the high-affinity Mgl transport system, which can transport d-glucose, d-galactose, and β-methylgalactosides in E. coli.
FIG 7 .
Predicted d-galactose network in Y. pestis. Vast grey arrows show major metabolic flows (Leloir pathway). Grey ellipses designate intracellular d-galactose and galactoside pools containing α- and β-d-galactose anomers. Red lines indicate transcriptional regulation. The blue line indicates Spot42 RNA-mediated translational control. Other enhancements and inhibitions are indicated by green lines.
Our results suggest that the Y. pestis d-galactose network, unlike that in E. coli, can utilize α-d-galactose but not β-d-galactose. Based on the comparative network analysis presented in this work, we can formulate predictions about the potential d-galactose-related environments and the function of the d-galactose network in such environments. Our results show that the simplified network found in Y. pestis is less competitive in utilizing large extracellular d-galactose pools; however, it can use d-galactose when present at a constant low level due to the presence of the high-affinity Mgl transport system. This suggests that niches occupied by Y. pestis (e.g., arthropod vector, macrophages, and human blood) (32) are generally poor in d-galactose. Also, from the absence of galS we can conclude that extracellular d-galactose levels are less dynamic in these niches than in the niches occupied by E. coli (33).
The d-galactose system is also involved in the utilization of intracellular d-galactose obtained from the degradation of oligosaccharides (e.g., lactose and melibiose) (14). Because galM is inactivated in the Y. pestis network, intracellular degradation of α-d-galactose- and β-d-galactose-containing compounds could have different effects. The system can utilize intracellular α-d-galactose as a carbon source and also to provide building blocks for biosynthetic glycosylation. For example, unlike E. coli, Y. pestis has a galactan transport and utilization system which can degrade extracellular galactan into smaller oligomers that are transported and processed to α-d-galactose oligomers inside the cell (34). The Mgl transport system can transport β-d-galactosides (e.g., methyl-β-d-galactoside and d-glycerol-β-d-galactoside) (35). However, β-d-galactose obtained from intracellular degradation of such compounds could induce the gal regulon genes but would not be utilized by the system. Induction of the galETK operon by a nonmetabolized inducer in the absence of α-d-galactose can serve biosynthetic purposes because GalE can catalyze the production of UDP-galactose (from UDP-glucose), which is required for biosynthetic glycosylation (Fig. 7). Such metabolic flow control in the amphibolic d-galactose pathway is common in enterobacteria. Synthesis of d-galactose-containing polysaccharides (e.g., in LPS) is often required for pathogenesis (8–10), and the need for UDP-galactose is independent of d-galactose availability. Different mechanisms have been reported so far, which take advantage of the fact that only the galE gene product is involved in making substrates for biosynthetic glycosylation reactions, and all of the gal enzymes are needed for catabolism of the sugar d-galactose. A common mechanism in enterobacteria is discoordinated expression of the gal operon genes. This can be achieved by using a small regulatory RNA (Spot42) which does not affect translation of GalE and GalT but blocks GalK production (22) or by transcribing the gal operon from two different promoters, one of which transcribes mostly the first gene of the operon, galE, because of natural polarity (19–21, 23). In certain pathogenic enterobacteria, the galE gene is physically separated from the other gal operon genes, and its expression is independent of intracellular d-galactose levels (9). Intracellular synthesis of a nonmetabolized inducer constitutes a novel mechanism for regulation of the amphibolic d-galactose pathway. The observation that utilization of intracellular β-d-galactose by the Y. pestis network can be turned on by single-base-pair insertions in galM suggests that the galM pseudogene has not been deleted in evolution because its reactivation is beneficial in certain environments.
MATERIALS AND METHODS
Bacterial strains.
Genome sequences of 34 strains were involved in the bioinformatical analysis. The strains used are the following: Citrobacter koseri ATCC BAA-895, Citrobacter rodentium ICC168, Cronobacter sakazakii ATCC BAA-894, Cronobacter turicensis z3032, Dickeya dadantii 3937, Dickeya zeae Ech 1591, Edwardsiella ictaluri 93-146, Edwardsiella tarda EIB202, Enterobacter cloacae subsp. cloacae ATCC 13047, Enterobacter sp. strain 638, Erwinia amylovora CFBP1430, Erwinia billingiae Eb661, Erwinia pyrifoliae Ep1/96, Erwinia tasmaniensis Et1/99, Escherichia coli MG1655, Escherichia fergusonii ATCC 35469, Klebsiella pneumoniae 342, Klebsiella variicola At-22, Pantoea ananatis LMG 20103, Pantoea vagans C9-1, Pectobacterium atrosepticum SCRI1043, Pectobacterium carotovorum PC1, Pectobacterium wasabiae WPP163, Proteus mirabilis HI4320, Salmonella enterica serovar Typhimurium strain LT2, Serratia proteamaculans 568, Shigella boydii CDC3083-94, Shigella dysenteriae Sd197, Shigella flexneri 5 strain 8401, Shigella sonnei Ss046, Sodalis glossinidius strain “morsitans,” Yersinia enterocolitica subsp. enterocolitica 8081, Yersinia pestis KIM10, and Yersinia pseudotuberculosis IP32953.
E. coli MG1655ΔgalM87::Kanr was described by Bouffard et al. (14). MG1655ΔgalP::cmr and MG1655ΔgalM::KanrΔgalP::cmr are derivatives of E. coli MG1655 and MG1655ΔgalM87::Kanr, respectively. The chloramphenicol resistance cassette was inserted into the galP gene by recombineering (36). The cat gene from plasmid pRFB122 (37) was amplified using the primers GalPupcat (5′-CTCACCTATCTAATTCACAATAAAAAATAACCATATTGGAGGGCATCAAAATGAGACGTTGATCGGCACGTAAGA-3′) and GalPdowncat (5′-CCTCCGCGATGGGAGGAAGCTTGGGGAGATTAATCGTGAGCGCCTATTTCTTACGCCCCGCCCTGCCACTCATCGCAG-3′). Recombineering was performed according to the protocol described by Datsenko and Wanner (38).
DNA manipulation methods.
Bacterial growth conditions and plasmid manipulations followed the protocols described by Sambrook and Russell (39). Transformations were performed with chemically competent XL1-Blue cells (Stratagene). Restriction endonucleases and DNA oligonucleotide primers were purchased from Invitrogen, PCR (GeneAmp XL) and sequencing (ABI Prism) kits from Applied Biosystems, and DNA purification kits from Qiagen. DNA sequencing reactions were analyzed in a PerkinElmer/Applied Biosystems (model 373 A) automated sequencer.
Plasmid construction.
Plasmid pSA850YP was made by inserting the Y. pestis galETK regulatory region between the EcoRI and PstI sites of plasmid pSA850 (29). Because of safety considerations, the DNA fragment corresponding to the chromosomal region 3350221 to 3350532 of Y. pestis KIM (GenBank accession no. NC_004088.1) was amplified from Yersinia pseudotuberculosis ATCC 23207, which contains an identical sequence. The primers used for amplification were YPP1 (5′-AAAAGAATTCGCGCACCACAAACAGGACATTCC) and YPP2 (5′-AAAACTGCAGCAATTGCACACAGGTATGGCTACC). The sequence of the Y. pestis galETK regulatory region in plasmid pSA850YP was verified.
The plasmid pSEM1026YP for the expression of Y. pestis GalR was created by amplifying the galR gene from Yersinia pseudotuberculosis ATCC 23207 using primers YPGNCO (5′-AAAACCATGGCCACTATAAAGGATGTTGCCAAGCT) and YPGCTBI (5′-AAAAGGATCCATCAGTGTCATCCCGTAGGCTTGGC) and inserting it between the NcoI and BamHI sites of plasmid pSEM1026 (40). The cloned Y. pseudotuberculosis galR gene is 99% identical to the sequence of Y. pestis galR, while the amino acid sequences of the encoded proteins are identical.
The galM pseudogene of Y. pestis (ATCC BAA-1504) was PCR amplified from a genomic DNA preparation (purchased from ATCC) using the primers YP_Mup_XhoI (5′-TTTTCTCGAGGTTCGCACCACCGTTGCGCAAGAATAC-3′) and YP_Mdn_Acc65I (5′-TTTTGGTACCATCATTCATAACGTCATCATTCATAAC-3′). The resulting DNA fragment was digested with XhoI and Acc65I and inserted between the XhoI and Acc65I sites of the low-copy-number plasmid pLG338E, which was derived from pLG338 (41) by eliminating the EcoRI site. The open reading frame of galM was restored by 8 different base pair insertions, using PCR mutagenesis with four designed primers and the previously used YP_Mdn_Acc65I primer. The designed primers are Yps_D (5′-TTTTGAATTCACCAAATTGCAGAATAAAAGCGGTATGACCGTCACCTTTATGGATTGGGGGGCAACCTGGTTATCGGCCATAT-3′), Yps_EKQ (5′-TTTTGAATTCACCAAATTGCAGAATAAAAGCGGTATGACCGTCACCTTTATGVAATGGGGGGCAACCTGGTTATCGGCCATAT-3′), Yps_N (5′-TTTTGAATTCACCAAATTGCAGAATAAAAGCGGTATGACCGTCACCTTTATGAACTGGGGGGCAACCTGGTTATCGGCCATAT-3′), and Yps_ITR (5′-TTTTGAATTCACCAAATTGCAGAATAAAAGCGGTATGACCGTCACCTTTATGABATGGGGGGCAACCTGGTTATCGGCCATATTACCGCTGAAAAAT-3′). The amplified fragments were used to replace the EcoRI-Acc65I fragment in the pLG338 plasmid containing the galM pseudogene. The DNA sequence of the inserted fragment was verified in all the constructs created.
Protein purification.
Expression and purification of the hexahistidine-tagged Y. pestis GalR followed the protocol described before for E. coli GalR purification (40). HU protein was purified according to the method described by Aki et al. (42). CRP was purified as described by Ryu et al. (43).
In vitro transcription.
Transcription reactions were performed as described previously (16). The reaction mixture (50 µl) contained 20 mM Tris acetate at pH 7.8, 10 mM magnesium acetate, 200 mM potassium glutamate, 100 µM cAMP, and 2 nM supercoiled plasmid DNA template. GalR and CRP concentrations are as indicated in Fig. 5 and 6. HU was used at an 80 nM concentration when present. Twenty nanomolar RNA polymerase (USB) was added before incubation of the reactions at 37°C for 5 minutes. Transcription was started by the addition of 1.0 mM ATP, 0.1 mM GTP, 0.1 mM CTP, 0.01 mM UTP, and 5 µCi of [α-32P]UTP (3,000 Ci/mmol). Reactions were terminated after 10 minutes by addition of an equal volume of transcription loading buffer (0.025% bromophenol blue, 0.025% xylene cyanol, 0.01 M EDTA, and 90% deionized formamide). After heating at 90°C for 3 minutes, the samples were loaded onto 7% polyacrylamide-urea DNA sequencing gels. RNA bands were quantified using the ImageQuant PhosphorImager (Molecular Dynamics, CA). Band intensities were corrected by the background and normalized to the RNA1 band intensities of the corresponding lanes.
ACKNOWLEDGMENTS
We thank our colleagues for various input on the project, particularly Thomas Soares and Mofang Liu for purification of HU and CRP and Takácsné Botond Judit for technical assistance.
This research was supported by the Hungarian Scientific Research Fund (OTKA) grant PD75496 to S.S., by the Danish Council for Independent Research∣Natural Sciences, by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, United States, and by the Danish National Research Foundation.
Footnotes
Citation Csiszovszki Z, Krishna S, Orosz L, Adhya S, Semsey S. 2011. Structure and function of the d-galactose network in enterobacteria. mBio 2(4):e00053-11. doi:10.1128/mBio.00053-11.
REFERENCES
- 1. Navid A, Almaas E. 2009. Genome-scale reconstruction of the metabolic network in Yersinia pestis, strain 91001. Mol. Biosyst. 5:368–375 [DOI] [PubMed] [Google Scholar]
- 2. Hacker J, Kaper JB. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641–679 [DOI] [PubMed] [Google Scholar]
- 3. Sneppen K, Krishna S, Semsey S. 2010. Simplified models of biological networks. Annu. Rev. Biophys. 39:43–59 [DOI] [PubMed] [Google Scholar]
- 4. Hunziker A, Tuboly C, Horváth P, Krishna S, Semsey S. 2010. Genetic flexibility of regulatory networks. Proc. Natl. Acad. Sci. U. S. A. 107:12998–13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mayo AE, Setty Y, Shavit S, Zaslaver A, Alon U. 2006. Plasticity of the cis-regulatory input function of a gene. PLoS Biol. 4:e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Achtman M, et al. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 96:14043–14048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dekel E, Mangan S, Alon U. 2005. Environmental selection of the feed-forward loop circuit in gene-regulation networks. Phys. Biol. 2:81–88 [DOI] [PubMed] [Google Scholar]
- 8. Maskell DJ, Szabo MJ, Deadman ME, Moxon ER. 1992. The gal locus from Haemophilus influenzae: cloning, sequencing and the use of gal mutants to study lipopolysaccharide. Mol. Microbiol. 6:3051–3063 [DOI] [PubMed] [Google Scholar]
- 9. Metzger M, Bellemann P, Bugert P, Geider K. 1994. Genetics of galactose metabolism of Erwinia amylovora and its influence on polysaccharide synthesis and virulence of the fire blight pathogen. J. Bacteriol. 176:450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robertson BD, Frosch M, van Putten JP. 1993. The role of galE in the biosynthesis and function of gonococcal lipopolysaccharide. Mol. Microbiol. 8:891–901 [DOI] [PubMed] [Google Scholar]
- 11. Motin VL, et al. 2004. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J. Bacteriol. 186:6298–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knirel YA, et al. 2006. Structural features and structural variability of the lipopolysaccharide of Yersinia pestis, the cause of plague. J. Endotoxin Res. 12:3–9 [DOI] [PubMed] [Google Scholar]
- 13. Semsey S, et al. 2007. Signal integration in the galactose network of Escherichia coli. Mol. Microbiol. 65:465–476 [DOI] [PubMed] [Google Scholar]
- 14. Bouffard GG, Rudd KE, Adhya SL. 1994. Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J. Mol. Biol. 244:269–278 [DOI] [PubMed] [Google Scholar]
- 15. Wilson DB. 1974. Source of energy for the Escherichia coli galactose transport systems induced by galactose. J. Bacteriol. 120:866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geanacopoulos M, Adhya S. 1997. Functional characterization of roles of GalR and GalS as regulators of the gal regulon. J. Bacteriol. 179:228–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weickert MJ, Adhya S. 1993. Control of transcription of gal repressor and isorepressor genes in Escherichia coli. J. Bacteriol. 175:251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee SJ, Lewis DE, Adhya S. 2008. Induction of the galactose enzymes in Escherichia coli is independent of the C-1-hydroxyl optical configuration of the inducer d-galactose. J. Bacteriol. 190:7932–7938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adhya S. 2003. Suboperonic regulatory signals. Sci. STKE 2003:pe22. [DOI] [PubMed] [Google Scholar]
- 20. Lee HJ, Jeon HJ, Ji SC, Yun SH, Lim HM. 2008. Establishment of an mRNA gradient depends on the promoter: an investigation of polarity in gene expression. J. Mol. Biol. 378:318–327 [DOI] [PubMed] [Google Scholar]
- 21. Semsey S, Virnik K, Adhya S. 2006. Three-stage regulation of the amphibolic gal operon: from repressosome to GalR-free DNA. J. Mol. Biol. 358:355–363 [DOI] [PubMed] [Google Scholar]
- 22. Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. 2002. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 16:1696–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ullmann A, Joseph E, Danchin A. 1979. Cyclic AMP as a modulator of polarity in polycistronic transcriptional units. Proc. Natl. Acad. Sci. U. S. A. 76:3194–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Herrgård MJ, Covert MW, Palsson BØ. 2004. Reconstruction of microbial transcriptional regulatory networks. Curr. Opin. Biotechnol. 15:70–77 [DOI] [PubMed] [Google Scholar]
- 25. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 26. Wren BW. 2003. The Yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 1:55–64 [DOI] [PubMed] [Google Scholar]
- 27. Lerat E, Ochman H. 2005. Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res. 33:3125–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brown MP, Shaikh N, Brenowitz M, Brand L. 1994. The allosteric interaction between d-galactose and the Escherichia coli galactose repressor protein. J. Biol. Chem. 269:12600–12605 [PubMed] [Google Scholar]
- 29. Lewis DE, Adhya S. 2004. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Mol. Microbiol. 54:692–701 [DOI] [PubMed] [Google Scholar]
- 30. Choy HE, Adhya S. 1992. Control of gal transcription through DNA looping: inhibition of the initial transcribing complex. Proc. Natl. Acad. Sci. U. S. A. 89:11264–11268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Death A, Ferenci T. 1994. Between feast and famine: endogenous inducer synthesis in the adaptation of Escherichia coli to growth with limiting carbohydrates. J. Bacteriol. 176:5101–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oyston PC, Isherwood KE. 2005. The many and varied niches occupied by Yersinia pestis as an arthropod-vectored zoonotic pathogen. Antonie Van Leeuwenhoek 87:171–177 [DOI] [PubMed] [Google Scholar]
- 33. Semsey S, et al. 2009. Dominant negative autoregulation limits steady-state repression levels in gene networks. J. Bacteriol. 191:4487–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Delangle A, et al. 2007. Characterization of the Erwinia chrysanthemi Gan locus, involved in galactan catabolism. J. Bacteriol. 189:7053–7061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilson DB. 1976. Properties of the entry and exit reactions of the beta-methyl galactoside transport system in Escherichia coli. J. Bacteriol. 126:1156–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Court DL, Sawitzke JA, Thomason LC. 2002. Genetic engineering using homologous recombination. Annu. Rev. Genet. 36:361–388 [DOI] [PubMed] [Google Scholar]
- 37. Fekete RA, Chattoraj DK. 2005. A cis-acting sequence involved in chromosome segregation in Escherichia coli. Mol. Microbiol. 55:175–183 [DOI] [PubMed] [Google Scholar]
- 38. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40. Semsey S, Geanacopoulos M, Lewis DE, Adhya S. 2002. Operator-bound GalR dimers close DNA loops by direct interaction: tetramerization and inducer binding. EMBO J. 21:4349–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stoker NG, Fairweather NF, Spratt BG. 1982. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene 18:335–341 [DOI] [PubMed] [Google Scholar]
- 42. Aki T, Choy HE, Adhya S. 1996. Histone-like protein HU as a specific transcriptional regulator: co-factor role in repression of gal transcription by GAL repressor. Genes Cells 1:179–188 [DOI] [PubMed] [Google Scholar]
- 43. Ryu S, Kim J, Adhya S, Garges S. 1993. Pivotal role of amino acid at position 138 in the allosteric hinge reorientation of cAMP receptor protein. Proc. Natl. Acad. Sci. U. S. A. 90:75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]