Abstract
The secondary metabolite gene clusters of euascomycete fungi are among the largest known clusters of functionally related genes in eukaryotes. Most of these clusters are species specific or genus specific, and little is known about how they are formed during evolution. We used a comparative genomics approach to study the evolutionary origins of a secondary metabolite cluster that synthesizes a polyketide derivative, namely, the fumonisin (FUM) cluster of Fusarium verticillioides, and that of Aspergillus niger another fumonisin (fumonisin B) producing species. We identified homologs in other euascomycetes of the Fusarium verticillioides FUM genes and their flanking genes. We discuss four models for the origin of the FUM cluster in Fusarium verticillioides and argue that two of these are plausible: (i) assembly by relocation of initially scattered genes in a recent Fusarium verticillioides; or (ii) horizontal transfer of the FUM cluster from a distantly related Sordariomycete species. We also propose that the FUM cluster was horizontally transferred into Aspergillus niger, most probably from a Sordariomycete species.
1. Introduction
The order of genes along eukaryotic chromosomes is often assumed to be random, but there is growing evidence that the chromosomal position of some genes is maintained by natural selection [1–3], and that selection can sometimes operate to move genes to new locations [4–6]. Among the eukaryotes, many of the most notable examples of the physical clustering of genes with related functions occur in the filamentous fungi [7–10]. The most striking fungal gene clusters are those involved in the synthesis of secondary metabolites such as sterigmatocystin (a 25-gene cluster in Aspergillus nidulans [11]), fumonisin (a 17-gene cluster in Fusarium verticillioides [12, 13]), and trichothecene (a 12-gene cluster in Fusarium sporotrichioides [14]). Secondary metabolites are organic molecules that are not essential for the normal growth of the fungus but which function in host/pathogen interactions or other forms of communication or warfare between organisms. They are typically modified polyketides, terpenes, alkaloids, or nonribosomal peptides [15]. Synthesis of these molecules requires many successive enzymatic steps, and the genes for these enzymes are almost invariably clustered together at a single genomic location. It has been suggested that physical clustering may allow the genes to be coregulated by means of chromatin modification [15], or that it may facilitate horizontal transfer of intact clusters between species [16].
Since the discovery of secondary metabolite gene clusters, their mechanism of origin and assembly has remained a matter of speculation. The growing number of available euascomycete genome sequences now enables us to both predict new secondary metabolite clusters [17] and take a phylogenomic approach to the evolutionary origins of these clusters. In this study we focus on one of the largest known secondary metabolite clusters “Fumonisin”, a polyketide synthase type cluster. Fumonisins (FB/FC) are mycotoxins produced by some species in the Gibberella fujikuroi species complex, of which F. verticillioides and F. proliferatum are the most studied (Gibberella is a teleomorphic form of the genus Fusarium). The FUM genes are organized as a cluster of 17 genes in both species (including FUM20 and FUM21, two additional genes recently revealed in the cluster [13]), though the location of the cluster differs between the two [12, 18]. The products of the FUM cluster genes include a polyketide synthase, fatty acyl-CoA synthases, and cytochrome P450 monooxygenases. Expression of the genes in the cluster, but not the neighboring genes, is induced under conditions when fumonisin (FB/FC) is synthesized [12]. The ability to synthesize fumonisin (FB/FC) has a patchy phylogenetic distribution across the genus Fusarium, due to the variable presence or absence of the FUM cluster among different isolates [19]. F. graminearum, for instance, does not synthesize fumonisin.
Recently, fumonisin (FB) production has also been reported in Aspergillus niger [20]. The genes in this species are also clustered and homologous to the FUM genes of F. verticillioides [21]. This is surprising given the large evolutionary distance between A. niger and F. verticillioides.
We combined comparative genomics with phylogenetic analysis to investigate whether genes in the FUM cluster have homologs in filamentous fungi that do not synthesize the mycotoxins, and so to study how this cluster was formed.
2. Methods
Our analysis was done using the completely sequenced genomes of the euascomycetes A. nidulans [22], F. graminearum [23], M. grisea [24], and N. crassa [25]. A set of 87,000 expressed sequence tags from F. verticillioides [26] was used for analysis of rates of sequence evolution in this species compared to F. graminearum.
To identify homologs of genes in the FUM clusters, we first used each protein as a query in a BLASTP search against the NCBI nonredundant protein sequence database. Because the F. verticillioides genome is not present in this database, we made a local BLAST database for expressed sequence tags from this species. Sequences giving hits with Expect (E) values of less than 1e − 4 were retained and used for phylogenetic analysis. Each set of proteins was aligned using ClustalW [27] and poorly aligned regions were removed using Gblocks [28]. Maximum likelihood trees were constructed using PHYML [29] with the JTT amino acid substitution matrix and four categories of substitution rates. Bootstrapping was done using the default options in PHYML with 100 replicates per run. The trees were eyeballed, the distant sequences were removed, and the steps above (ClustalW, Gblocks, and PHYML) were repeated on the remaining sequences.
3. Results
In the following we are interested in identifying scenarios that can explain the origins of the current FUM gene cluster. We do this by listing all possible scenarios that might have taken place in evolution, and comparing their plausibility. Building an evolutionary scenario is not straightforward because many of the events took place in extinct species and only a few clues remain in the current organisms. Almost any scenario is formally possible, but what makes one scenario more likely than another is parsimony-consideration of the number of separate events that are required to have taken place in order to account for it. In other words, a scenario involving fewer events is a more likely explanation of the observed data.
3.1. Origins of the FUM Gene Cluster in F. verticillioides
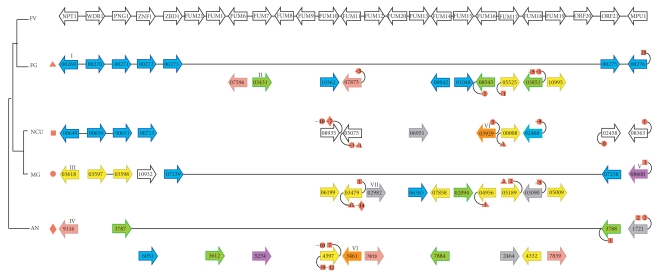
Gene-by-gene phylogenetic analyses were carried out to decipher the evolutionary history of the FUM genes using homologs of these genes in four euascomycetes: Fusarium graminearum, Neurospora crassa, Magnaporthe grisea, and Aspergillus nidulans (we use the word “homologs” for convenience in situations where we are unsure whether genes are orthologs or paralogs). We also constructed individual phylogenies for the genes located on either side of the F. verticillioides FUM cluster. Homology relationships between the genes in or near the F. verticillioides FUM cluster and other euascomycete genes are summarized in Figure 1. We identified probable orthologs of 13 of the 17 FUM genes (Figure 1). These genes are not arranged in clusters in the other genomes.
Figure 1.
Representation of the fumonisin cluster and its flanking genes in F. verticillioides (FV). Columns are homologs of the FUM genes and orthologs of the flanking genes identified by phylogenetic analysis in F. graminearum (FG), N. crassa (NCU), M. grisea (MG), and A. nidulans (AN). Genes in the latter four species are identified by gene numbers from their genome projects. Different colors represent different chromosomes. The long lines in F. graminearum, M. grisea, and A. nidulans show that in those species, there is a site in the genome that corresponds to the FUM cluster location, but no FUM genes are present at that locus. Curved lines and numbers in orange symbols indicate conservation of the neighboring genes around FUM homologs in F. graminearum, N. crassa, M. grisea, and A. nidulans. For each gene show in the figure (the focal genes) we considered the two genes immediately next to it. If these genes have orthologs located <20 genes away from the focal gene's ortholog in another species, a symbol indicates this fact. For example, the numbers −10 (in a circle) and −7 (in a diamond) connected to gene NCU08935 indicate that the gene immediately after NCU08935 (i.e., NCU08936) has an ortholog in M. grisea that is 10 genes away from MG06199 (i.e., MG06189), and an ortholog in A. nidulans that is 7 genes away from AN04397 (i.e., AN04392). Triangles, squares, circles, and diamonds indicate relationships to F. graminearum, N. crassa, M. grisea, and A. nidulans, respectively.
A region of F. graminearum chromosome 1 (genes FG00269–F00276; Figure 1) contains orthologs of the five genes to the left of the FUM cluster (F. verticillioides NPT1, WDR1, PNG1, ZNF1, and ZBD1) immediately adjacent to orthologs of the two genes to the right of the cluster (F. verticillioides ORF21 and MPU1), with nothing in between them in F. graminearum. Similarly, in M. grisea the ZBD1 ortholog is beside the ORF21 ortholog, and in A. nidulans the PNG1 ortholog is beside the ORF21 ortholog. Thus, chromosomal sites orthologous to the F. verticillioides FUM cluster-flanking regions exist adjacent to one another in F. graminearum, M. grisea, and A. nidulans, but no orthologs of the FUM genes themselves are found at these sites (Figure 1). Instead, homologs of the FUM genes are scattered on different chromosomes of the genomes of these fumonisin-nonproducing species. In F. graminearum, for instance, the 10 homologs of FUM genes are dispersed across four chromosomes and none of them is located close to another or to the FUM-flanking genes. Thus the FUM cluster genes appear to have been inserted into a pre-existing genomic locus between ZBD1 and ORF21.
Further, we examined the genomic contexts around each of the FUM cluster homologs in other species. For example, the F. verticillioides FUM cluster gene FUM11 is homologous to FG07875 in F. graminearum and to MG03479 in M. grisea (Figure 1). We will refer to FG07875 and MG03479 as focal genes. When we examine the regions around these focal genes in the F. graminearum and M. grisea genomes, we find that some of the neighboring genes near them are also orthologs of each other. On one side FG07874 (1 gene away from the focal gene) is an ortholog of MG03474 (5 genes away), and on the other side FG07864 (11 genes away from the focal gene) is an ortholog of MG03480 (1 gene away from the focal gene). In Figure 1, only the focal genes are shown but these similarities of context are indicated by the small numbers at the ends of the curved lines attached to the symbols for the focal genes FG07875 and MG03479.
Overall, the homologs of five FUM cluster genes (FUM10, FUM11, FUM16, FUM17, and FUM18) show some degree of local synteny conservation among the four species that do not contain FUM clusters. In other words, each of these genes is in a conserved location among some of the four species, and these locations are not close to one another.
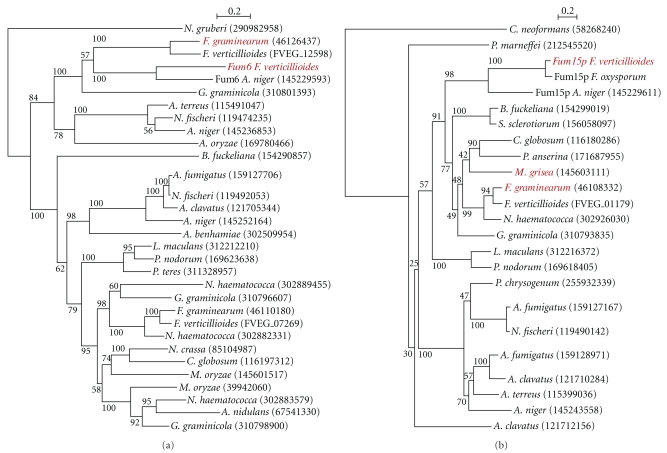
The phylogenetic trees obtained from individual FUM genes are shown in Figure 2 (for FUM6 and FUM15) and supplemental 1 available on line at doi:10.4061/2011/423821 (for the other genes). The trees present a diversity of topologies, such that no one sentence story can explain the origin of the FUM cluster. One cannot expect all the FUM gene trees to have identical topologies—especially given the possibility that different genes have been subject to very different evolutionary constraints—but even still the diversity of topologies is surprising. To interpret these trees, we consider four possible scenarios for the origin of the cluster (Figure 3), and what tree topologies they would predict. To evaluate these scenarios, we concentrate on the FUM genes that are present in both F. verticillioides and A. niger (FUM1, FUM6, FUM7, FUM8, FUM9, FUM10, FUM13, FUM14, FUM15, and FUM19). Below, we discuss these four scenarios.
Figure 2.
Maximum likelihood trees for FUM6, 15, and their homologs. (a) FUM6; (b) FUM15. In each tree, genes that appear in Figure 1 are named in red. The species name and the NCBI ID are provided on each branch. Bootstrap percentages are shown for all nodes. Trees were constructed from amino acid sequences as described in Section 2 using PHYML after alignment with ClustalW.
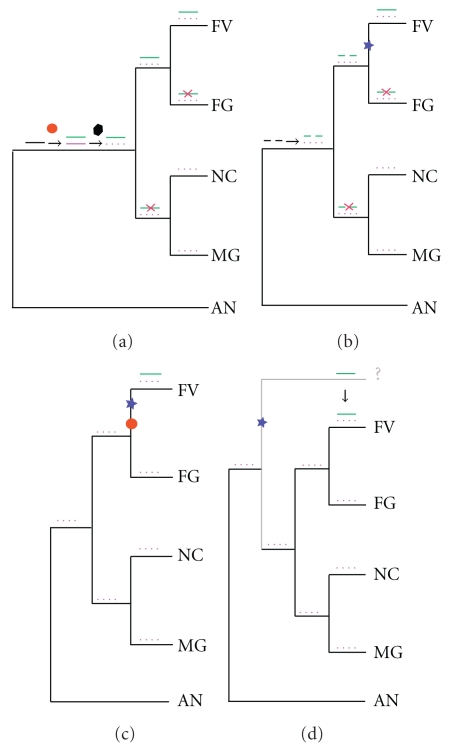
Figure 3.
The four most likely scenarios giving rise to the current fumonisin cluster in F. verticillioides. The species represented on all four trees are: F. verticillioides (FV), F. graminearum (FG), N. crassa (NCU), M. grisea (MG), and A. nidulans (AN). A red circle represents a duplication event. A blue star represents an assembly and clustering event, while a disassembly is shown using a black square. Red crosses indicate a loss event. When the FUM genes, or their ancestral genes, are clustered they are represented by a short line. The line is dashed if the genes are not clustered. Two colors are assigned to the duplicated genes and green genes are ancestors of the FUM genes (or the current FUM genes), while pink genes are the paralogs of FUM genes (ones found in all other Sordariomycetes). (a) represents the vertical transfer where the ancestor processed a version of the FUM cluster. (b) The ancestor in this scenario contained the ancestral genes of the FUM cluster (scattered). (c) represents a recent event of duplication and assembly of the FUM cluster in an ancestor of F. verticillioides. Finally, (d) represents the horizontal gene transfer scenario.
Scenario (vertical inheritance of an ancestral cluster) —
This scenario is illustrated schematically in Figure 3(a). According to it, a cluster existed in the common ancestor of Sordariomycetes (i.e., F. verticillioides, F. graminearum, N. crassa, and M. grisea). This cluster became duplicated in this ancestor, and then one copy disintegrated, dispersing its genes around the genome. F. verticillioides retained both the cluster and the scattered genes, whereas the other Sordariomycete species retained only the scattered genes.
Support for this scenario comes from some trees (Fum6, Fum10, Fum15, and Fum19) which show that the FUM genes have duplicates in F. verticillioides, but are single copy in non-producing fumonisin species. A duplication in the common ancestor of Sordariomycetes is suggested by the trees for some genes (Fum6, Fum13, and Fum15), whereas an older duplication in the common ancestor of Sordariomycetes plus Eurotiomycetes (not as shown in Figure 3(a)) is suggested by the trees for other genes (Fum7, Fum8, Fum10, and Fum19).
Scenario (ancient duplications of scattered genes, followed by recent assembly of a cluster in F. verticillioides) —
This scenario is illustrated in Figure 3(b). Scenarios 1 and 2 both require that through evolution numerous independent events of loss have occurred in very distantly related species (for illustration purposes all the losses in Figures 3(a) and 3(b) are placed on the F. graminearum branch and in the common ancestor of N. crassa and M. graminearum, but other combinations or losses on other branches are also possible).
One problem with Scenarios 1 and 2 is that, according to the phylogenies of Fum7, Fum10, Fum15, and Fum19, the duplicates retained in all the fumonisin-nonproducing Sordariomycetes are coincidently always the same copy (as illustrated by the parallel losses of multiple green genes, but not pink ones, in Figures 3(a) and 3(b)). This can be visualized in the trees by the fact that the homologs of the FUM genes in fumonisin-non-producing species are orthologs of each other, showing a more or less typical species phylogeny. If the second copy in F. graminearum, M. grisea, or N. crassa (the one represented in green dots in Figures 3(a) and 3(b) had been retained, this copy would be closer to the FUM gene than to genes in the fumonisin-non-producing species, which is not what we observe. Together these observations make both Scenarios 1 and 2 very unlikely.
Scenario (FUM gene duplication and cluster assembly specifically on the branch leading to F. verticillioides and the GFSC) —
The specificity of the FUM cluster to the Gibberella fujikuroi species complex (GFSC, which includes F. verticillioides, F. oxysporum, and F. proliferatum) points towards a complex-specific cluster. This scenario is shown in Figure 3(c). It proposes that the FUM cluster was built in an ancestor of F. verticillioides after its speciation from F. graminearum. Most of the gene trees do not support this model, because with the exception of two genes (Fum10 and Fum19) no homolog of a FUM gene has remained in F. verticillioides. Although the trees for Fum6 and Fum15 show homologs in F. verticillioides, the duplications in these cases greatly precede the origin of the GFSC clade.
The numbers of steps required for the different scenarios in Figures 3(a), 3(b), and 3(c) make it more parsimonious to argue that the FUM cluster became assembled in an ancestor of F. verticillioides (Figure 3(c)) than to argue that either the cluster or the individual genes underwent early duplication and then got lost multiple times (Figures 3(a) and 3(b)). Additionally, as explained above, Figures 3(a) and 3(b) would also imply that the same copy of the duplicates in the fumonisin-non-producing species were independently retained (a minimum of two independent retentions of the same copy are required: one on the F. graminearum lineage, and one in the common ancestor of N. crassa and M. grisea, pink genes in Figures 3(a) and 3(b)).
The hypothesis of assembly requires that each gene transposed once, from an ancestral location, to its current location in F. verticillioides. The genes must have been sequentially relocated, with selection for each step.
Scenario (origin of the F. verticillioides fum cluster by horizontal transfer from a distantly related fungus) —
This scenario is shown in Figure 3(d). Under this scenario the donor could be a Sordariomycete (as suggested by the trees for Fum6, Fum13, and Fum15), or a more distant species that is an outgroup to both Sordariomycetes and Eurotiomycetes (as suggested by Fum7, Fum9, Fum10, and Fum19). Although we cannot identify a specific donor, we cannot rule out this possibility. Indeed this scenario reduces the number of events leading to the current trees. Under this scenario, we would not have multiple losses, nor the unexpected retention of the same copy in many of the trees (represented in pink in Figure 3(b); Fum7, Fum10, Fum15, and Fum19). This scenario may explain the unexpected phylogenetic positioning of some FUM gene outside the expected class of species. For example, in the case of Fum7, Fum 9, Fum 10, Fum13, Fum15, and Fum19 a horizontal gene transfer of the FUM genes into F. verticillioides would explain such topologies.
Figure 3(d) illustrates how the horizontal transfer scenario, like the recent assembly hypothesis (Figure 3(c)), reduces the number of independent events necessary to explain the FUM cluster. However, the horizontal transfer scenario does not posit any mechanism for the assembly of the cluster; it just shifts the question to how the cluster became assembled in the donor species.
3.2. The Fumonisin Cluster in A. niger Results from Horizontal Gene Transfer
A. niger has been shown to produce fumonisin [13] and contains clustered homologs of many of the F. verticillioides FUM genes [21] (Figure 2). Our phylogenetic analysis illuminates the origin of this cluster in A. niger and how it relates to the cluster in F. verticillioides (phylogenetic trees in Figure 2 and Supplemental Figure 1).
The first trend evident from this phylogenetic analysis is that genes from the FUM cluster in F. verticillioides, F. oxysporum, and A. niger define clades supported by high bootstrap values (>90%, Figure 2), to the exclusion of homologous genes from Sordariomycetes and Eurotiomycetes.
Because we extended our analysis to many species, we were faced with the problem of low bootstrap values for many of the FUM genes trees. The two trees shown in Figure 2(b) (FUM6 and FUM15) are the ones with the high bootstrap support for relevant branches, and a reasonably correct species phylogeny. Our analysis shows that both these genes in A. niger clearly group with genes from the Sordariomycetes, rather than with genes in the more closely related (Eurotiomycetes) species including other A. niger genes. Bootstrap values for grouping the A. niger FUM genes with the Sordariomycete homologs are 98–100% (Figure 2).
The disagreement of this result with the expected A. niger species relationships, are suggestive of horizontal gene transfer between A. niger and an ancestor existing prior to the divergence of F. verticillioides and F. oxysporum. More importantly, it is more likely that the transfer occurred from Sordariomycetes to A. niger (or an ancestor of this species), rather than the opposite. Indeed, the opposite would result in the FUM genes (from F. verticillioides, F. oxysporum, and A. niger) clustering in the Eurotiomycetes subphylum as opposed to the Sordariomycetes as seen in Figure 2. For the FUM6 and FUM15 trees, we used the likelihood ratio test (LRT) to test whether the topologies shown (Figure 2) have significantly higher likelihoods than alternative trees where the A. niger was placed in the Eurotiomycetes and constrained to form a monophyletic group. In both cases the topology shown in Figure 2 is significantly more likely than the tree expected if genes were inherited vertically (P < .001 for each).
Because the clusters in A. niger and in F. verticillioides share only 11 of the 17 known FUM genes (including FUM21), these two types of cluster have probably had a long history of independent evolution, although they certainly share a common ancestor. We conclude that the cluster in A. niger originated by horizontal transfer from an ancestor of F. verticillioides and F. oxysporum.
4. Discussion
In the literature three scenarios for the creation of a gene cluster have been described: horizontal of an existing cluster from one genome to another [30, 31]; the duplication of an ancestral cluster [31]; the de novo creation of a cluster from initially scattered genes that become relocated into one locus [4]. We find that the FUM genes are apparent duplicates of conserved genes in Sordariomycetes (Figures 1 and 2). We think that two of the scenarios we discussed could plausibly account for the observed data. First, the FUM cluster could be the result of horizontal cluster transfer into an ancestor of F. verticillioides (Scenario 4). This scenario is similar to our observation of the ACE1 cluster in A. clavatus [31], and the more recent observation of the horizontal gene transfer of the sterigmatocystin cluster [32]. Secondly, the FUM cluster may have been assembled after recent gene duplication in an ancestor of F. verticillioides (Scenario 3). The latter scenario resembles our previous observations on the DAL gene cluster of S. cerevisiae [4], though it should be noted that the DAL genes code for a catabolic pathway (degradation of allantoin, a secondary nitrogen source), whereas the FUM genes are part of an anabolic pathway (secondary metabolite biosynthesis).
On the other hand we propose that the fumonisin cluster in A. niger was acquired via horizontal gene transfer. It has been shown in recent years that horizontal gene transfer between filamentous fungi is more common than was originally thought. Many independent genes can transfer between distantly related species such as that observed between and ancestor of A. oryzae and Sordariomycetes [33]; also an entire secondary metabolite cluster has been shown to have horizontally transferred between a relative of M. grisea into an ancestor of A. clavatus [31]. Here again this finding adds to the repertoire of horizontally transferred genes between fungal species and shows that this exchange mechanism is not so uncommon after all. Moreover it shows how an entire cluster can transfer between distantly related species and remain functional in the new species. In addition, the differences between the A. niger and F. verticillioides Fum clusters highlights how a cluster can diverge by adding, removing, or reshuffling the genes.
Our lack of knowledge about what benefit the metabolite confers on the organism hampers our understanding of the selective purpose of this clustering. However, we can be almost certain that the reason behind the clustering is not simply to synthesize the metabolite, which is possible with scattered genes. It is more likely that the selective force involves selection for a tight coregulation of gene expression, perhaps mediated by a LaeA-type universal regulator.
Competition, either between one fungal species and another, or between a fungus and a host species, is likely to result in strong selection on the secondary metabolite repertoire of filamentous fungal species. This arms race between organisms pressurizes the organism to create new chemical weapons, which are the products of new secondary metabolite gene clusters. It is relatively easy to envisage that neofunctionalization after gene duplication, or partial cluster duplication as appears to have happened in the origins of the Ace1 cluster [31], could result in the production of a new secondary metabolite and so could be selectively advantageous. It is harder to understand why relocating genes, as has happened in the FUM cluster, can be evolutionarily advantageous. One possibility is that the mere act of relocating a gene can have the consequence of changing the end product of a pathway, because the expression of all the genes in a cluster is coordinated. For example, imagine that we have two secondary metabolite biosynthesis pathways, 1 and 2. If a cytochrome P450 oxidoreductase gene that originally functioned in pathway 1 is suddenly relocated so that it becomes coexpressed with the genes in pathway 2 (and no longer co-expressed with pathway 1), it is possible that its product could begin to act on one of the intermediate molecules in pathway 2. The result would be that the products of pathways 1 and 2 are both changed. Alternative possibilities include that there is selection for tighter regulation (e.g., if an intermediate molecule in the pathway is toxic), or that there is epistatic selection for tight linkage between interacting alleles [34].
Supplementary Material
Maximum likelihood trees for the common fumonisin proteins between F. verticillioides and A. niger and their homologs. The species name and the NCBI ID are provided on each branch. Bootstrap percentages are shown for all nodes. Trees were constructed from amino acid sequences as described in Methods using PHYML after alignment with ClustalW.
Acknowledgment
The authors would like to thank the reviewers for their valuable comments and suggestions.
References
- 1.Hurst LD, Pál C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nature Reviews Genetics. 2004;5(4):299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 2.Hurst LD, Williams EJB, Pál C. Natural selection promotes the conservation of linkage of co-expressed genes. Trends in Genetics. 2002;18(12):604–606. doi: 10.1016/s0168-9525(02)02813-5. [DOI] [PubMed] [Google Scholar]
- 3.Singer GAC, Lloyd AT, Huminiecki LB, Wolfe KH. Clusters of co-expressed genes in mammalian genomes are conserved by natural selection. Molecular Biology and Evolution. 2005;22(3):767–775. doi: 10.1093/molbev/msi062. [DOI] [PubMed] [Google Scholar]
- 4.Wong S, Wolfe KH. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nature Genetics. 2005;37(7):777–782. doi: 10.1038/ng1584. [DOI] [PubMed] [Google Scholar]
- 5.Field B, Osbourn AE. Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science. 2008;320(5875):543–547. doi: 10.1126/science.1154990. [DOI] [PubMed] [Google Scholar]
- 6.Qi X, Bakht S, Leggett M, Maxwell C, Melton R, Osbourn A. A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(21):8233–8238. doi: 10.1073/pnas.0401301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giles NH, Case ME, Baum J. Gene organization and regulation in the qa (quinic acid) gene cluster of Neurospora crassa. Microbiological Reviews. 1985;49(3):338–358. doi: 10.1128/mr.49.3.338-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hull EP, Green PM, Arst HN, Scazzocchio C. Cloning and physical characterization of the L-proline catabolism gene cluster of Aspergillus nidulans. Molecular Microbiology. 1989;3(4):553–559. doi: 10.1111/j.1365-2958.1989.tb00201.x. [DOI] [PubMed] [Google Scholar]
- 9.Keller NP, Hohn TM. Metabolic pathway gene clusters in filamentous fungi. Fungal Genetics and Biology. 1997;21(1):17–29. [PubMed] [Google Scholar]
- 10.Cary JW, Chang P-K, Bhatnagar D. Clustered metabolic pathway genes in filamentous fungi. In: Khachatourians GG, Arora DK, editors. Applied Mycology and Biotechnology. Amsterdam, The Netherlands: Elsevier; 2001. pp. 165–198. [Google Scholar]
- 11.Brown DW, Yu JH, Kelkar HS, et al. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(4):1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proctor RH, Brown DW, Plattner RD, Desjardins AE. Co-expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genetics and Biology. 2003;38(2):237–249. doi: 10.1016/s1087-1845(02)00525-x. [DOI] [PubMed] [Google Scholar]
- 13.Brown DW, Butchko RAE, Busman M, Proctor RH. The Fusarium verticillioides FUM gene cluster encodes a Zn(II)2Cys6 protein that affects FUM gene expression and fumonisin production. Eukaryotic Cell. 2007;6(7):1210–1218. doi: 10.1128/EC.00400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown DW, Dyer RB, McCormick SP, Kendra DF, Plattner RD. Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genetics and Biology. 2004;41(4):454–462. doi: 10.1016/j.fgb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism—from biochemistry to genomics. Nature Reviews Microbiology. 2005;3(12):937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 16.Rosewich UL, Kistler HC. Role of horizontal gene transfer in the evolution of fungi. Annual Review of Phytopathology. 2000;38:325–363. doi: 10.1146/annurev.phyto.38.1.325. [DOI] [PubMed] [Google Scholar]
- 17.Khaldi N, Seifuddin FT, Turner G, et al. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genetics and Biology. 2010;47(9):736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waalwijk C, Van Der Lee T, De Vries I, Hesselink T, Arts J, Kema GHJ. Synteny in toxigenic Fusarium species: the fumonisin gene cluster and the mating type region as examples. European Journal of Plant Pathology. 2004;110(5-6):533–544. [Google Scholar]
- 19.Proctor RH, Plattner RD, Brown DW, Seo JA, Lee YW. Discontinuous distribution of fumonisin biosynthetic genes in the Gibberella fujikuroi species complex. Mycological Research. 2004;108(7):815–822. doi: 10.1017/s0953756204000577. [DOI] [PubMed] [Google Scholar]
- 20.Frisvad JC, Smedsgaard J, Samson RA, Larsen TO, Thrane U. Fumonisin B production by Aspergillus niger. Journal of Agricultural and Food Chemistry. 2007;55(23):9727–9732. doi: 10.1021/jf0718906. [DOI] [PubMed] [Google Scholar]
- 21.Baker SE. Aspergillus niger genomics: past, present and into the future. Medical Mycology. 2006;44(1):17–21. doi: 10.1080/13693780600921037. [DOI] [PubMed] [Google Scholar]
- 22.Galagan JE, Calvo SE, Cuomo C, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438(7071):1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 23.Cuomo CA, Güldener U, Xu J-R, et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science. 2007;317(5843):1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- 24.Dean RA, Talbot NJ, Ebbole DJ, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434(7036):980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 25.Galagan JE, Calvo SE, Borkovich KA, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422(6934):859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 26.Brown DW, Cheung F, Proctor RH, et al. Comparative analysis of 87,000 expressed sequence tags from the fumonisin-producing fungus Fusarium verticillioides. Fungal Genetics and Biology. 2005;42(10):848–861. doi: 10.1016/j.fgb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 29.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 30.Patron NJ, Waller RF, Cozijnsen AJ, et al. Origin and distribution of epipolythiodioxopiperazine (ETP) gene clusters in filamentous ascomycetes. BMC Evolutionary Biology. 2007;7, article 174 doi: 10.1186/1471-2148-7-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khaldi N, Collemare J, Lebrun MH, Wolfe KH. Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biology. 2008;9(1, article R18) doi: 10.1186/gb-2008-9-1-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slot JC, Rokas A. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Current Biology. 2011;21(2):134–139. doi: 10.1016/j.cub.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 33.Khaldi N, Wolfe KH. Elusive origins of the extra genes in Aspergillus oryzae. PLoS ONE. 2008;3(8, article e3036) doi: 10.1371/journal.pone.0003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nei M. Modification of linkage intensity by natural selection. Genetics. 1967;57(3):625–641. doi: 10.1093/genetics/57.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood trees for the common fumonisin proteins between F. verticillioides and A. niger and their homologs. The species name and the NCBI ID are provided on each branch. Bootstrap percentages are shown for all nodes. Trees were constructed from amino acid sequences as described in Methods using PHYML after alignment with ClustalW.