ABSTRACT
Light control of motility behavior (phototaxis and photophobic responses) in green flagellate algae is mediated by sensory rhodopsins homologous to phototaxis receptors and light-driven ion transporters in prokaryotic organisms. In the phototaxis process, excitation of the algal sensory rhodopsins leads to generation of transmembrane photoreceptor currents. When expressed in animal cells, the algal phototaxis receptors function as light-gated cation channels, which has earned them the name “channelrhodopsins.” Channelrhodopsins have become useful molecular tools for light control of cellular activity. Only four channelrhodopsins, identified in Chlamydomonas reinhardtii and Volvox carteri, have been reported so far. By screening light-induced currents among algal species, we identified that the phylogenetically distant flagellate Mesostigma viride showed photoelectrical responses in vivo with properties suggesting a channelrhodopsin especially promising for optogenetic use. We cloned an M. viride channelrhodopsin, MChR1, and studied its channel activity upon heterologous expression. Action spectra in HEK293 cells match those of the photocurrents observed in M. viride cells. Comparison of the more divergent MChR1 sequence to the previously studied phylogenetically clustered homologs and study of several MChR1 mutants refine our understanding of the sequence determinants of channelrhodopsin function. We found that MChR1 has the most red-shifted and pH-independent spectral sensitivity so far reported, matches or surpasses known channelrhodopsins’ channel kinetics features, and undergoes minimal inactivation upon sustained illumination. This combination of properties makes MChR1 a promising candidate for optogenetic applications.
IMPORTANCE
Channelrhodopsins that function as phototaxis receptors in flagellate algae have recently come into the spotlight as genetically encoded single-molecule optical switches for turning on neuronal firing or other cellular processes, a technique called “optogenetics.” Only one of four currently known channelrhodopsins is widely used in optogenetics, although electrical currents recorded in diverse flagellates suggest the existence of a large variety of such proteins. We applied a strategy for the search for new channelrhodopsins with desirable characteristics by measuring rhodopsin-mediated photocurrents in microalgae, which helped us identify MChR1, a new member of the channelrhodopsin family. MChR1 exhibits several sought-after characteristics and thus expands the available optogenetic toolbox. The divergence of the MChR1 sequence from those of the four known channelrhodopsins contributes to our understanding of diversity in the primary structures of this subfamily of sensory rhodopsins.
Introduction
The genomes of microorganisms from all three domains of life encode retinylidene proteins of shared sequence homology that are called type 1 rhodopsins to distinguish them from the animal visual pigment proteins called type 2 rhodopsins (1). Initially, microbial rhodopsins were discovered in haloarchaea but were subsequently also found in proteobacteria, cyanobacteria, green algae, cryptomonads, dinoflagellates, fungi, and representatives of some other phylogenetic groups. Functionally, microbial rhodopsins act as light-driven ion pumps or light sensors that initiate diverse signal transduction mechanisms (2).
The first eukaryotic microbial rhodopsins with a sensory function were cloned from green flagellate algae, in which they serve as light receptors for photomotility responses (3–6). Four channelrhodopsins have been identified to date, ChR1 and ChR2 from Chlamydomonas reinhardtii (3, 7–9) and VChR1 and VChR2 from Volvox carteri (6, 10). They contain a 7-transmembrane-helix (7TM) domain characteristic of type 1 rhodopsins (1) followed by a conserved but more varied extended C-terminal region. Photoexcitation of algal sensory rhodopsins leads to generation of the photoreceptor currents that depolarize the plasma membrane and trigger a sensory transduction cascade that eventually leads to a change in the flagellar beating pattern (11, 12). Heterologous expression in Xenopus oocytes revealed that their 7TM domains exhibit a unique property of light-gated ion conductance (7, 8), so far not observed in any other protein. To highlight this feature, the name “channelrhodopsins” has been coined to distinguish algal sensory rhodopsins from other microbial rhodopsins (7, 8). When transfected into and expressed in animal cells, e.g., defined subpopulations of rodent brain neurons, channelrhodopsins enable their targeted light activation in tissue culture and in living organisms (13–15). This approach, called “optogenetics,” offers temporal and spatial resolution superior to that of conventional electrical or chemical stimulation. Channelrhodopsins have greatly benefited research into brain circuitry and pathogenic mechanisms of neurological and other disorders, and they are candidates for future gene therapy applications, such as for restoring vision lost to retinal-degeneration diseases. Although today channelrhodopsins as optogenetic tools are most widely used in neurons, the range of their application is rapidly expanding to include glial, muscle, and embryonic stem cells (reviewed in reference 16) and potentially many more cellular systems in which rapid control of the membrane potential and/or the intracellular ion concentration is desirable.
Several intrinsic properties of the four known channelrhodopsins limit their application as optogenetic tools (reviewed in references 17 and 18). The most widely used, ChR2, has maximal spectral sensitivity at 470 nm (8, 19), but excitation at longer wavelengths is preferable to minimize light scattering and absorption by biological tissues. VChR1 is a red-shifted channelrhodopsin variant (10), but it has slower current kinetics than ChR2, compromising its performance at moderate-to-high stimulation frequencies. Another limiting property is that photocurrents generated by all channelrhodopsins in response to a pulse of continuous light decrease to a plateau level, a process called “inactivation.” In the most commonly used channelrhodopsin, ChR2, this decrease from the peak amplitude can be as large as 80% (8, 20, 21), which correspondingly decreases the light-induced membrane depolarization, requiring more intense or longer light pulses to induce a desired biological action. This limitation is further aggravated by the low unitary conductance of channelrhodopsins, which is less than that of common ion channels, as estimated by whole-cell current noise analysis (22, 23). Taking into account the rapidly growing demand for optogenetic tools, it is highly desirable to improve these characteristics of channelrhodopsins and to expand the toolbox of variants available for a variety of experimental systems. The challenge here is that the mechanism of ion conductance by channelrhodopsins remains poorly understood, in particular because their crystal structures are not yet available.
Molecular engineering of the known channelrhodopsins by site-specific mutagenesis and chimera construction have been applied to produce variants with altered properties and to gain insights into the structure-function relationships within the channelrhodopsin molecule (see, e.g., references 21 and 23 to 29). An alternative approach is to search for natural channelrhodopsin variants in algae. To screen algal species for candidates of new channelrhodopsins with desirable characteristics, we used the photoelectrophysiological population assay for recording rhodopsin-mediated photocurrents (30). We found that photocurrents in the scaly green flagellate Mesostigma viride showed faster kinetics and more red-shifted spectral sensitivity than those of the earlier-studied organisms. M. viride is an alga that attracted considerable interest from evolutionary biologists, because early ultrastructural studies (31) and later molecular phylogenetic analyses (32) revealed that this unicellular organism is closer to land plants (Streptophyta) than to other green algae (Chlorophyta). We cloned a channelopsin-encoding nucleotide sequence from this alga and studied its channel activity by expression in HEK293 cells. Here we show that this new channelrhodopsin variant, MChR1, refines our understanding of what structural features are essential for channelrhodopsin function. Furthemore, it has a more red-shifted spectral sensitivity at neutral pH than the previously available channelrhodopsin with the most red-shifted spectral sensitivity, VChR1, and surpasses VChR1 by having faster current kinetics and smaller inactivation, all of which makes MChR1 a promising candidate for potential optogenetic applications.
RESULTS
Rhodopsin-mediated photoelectric currents in M. viride cells.
Photoreceptor currents in native M. viride cells were measured in cell suspensions under unilateral laser flash excitation. The response was similar to those observed previously in other phototactic flagellates with intrachloroplast eyespots (for a review, see reference 33). However, it was significantly faster, with a peak time of <0.5 ms and a half-rise time of 100 µs (Fig. 1A).
FIG 1 .
(A) Photoelectric response in a suspension of native cells of Mesostigma viride. Unilateral excitation with a 6-ns laser pulse at 530 nm. (B) Fluence-response dependence of the photoreceptor current. Excitation was with a 520 ± 20-nm light from a photoflash. (C) Action spectrum of the photoreceptor current in native M. viride cells. Solid line, Gaussian fit of the main peak; maximum, 531 nm; half-bandwidth, ~93 nm (for details, see the text). rel. u., relative units.
The contribution of the late (delayed) photoreceptor current to the recorded signal, discovered earlier in other green algae (34), is much smaller in M. viride (Fig. 1A). The second component was also not found in the fluence response dependence (Fig. 1B), which was well fitted with a single exponential function that saturated at about the same light intensities as those of previously studied early photoreceptor currents in other flagellate algae. However, the curve in M. viride, in contrast to those in other flagellates (35, 36), was well fitted without introducing an additional low-saturation component, which means that the amplified current component (33) did not contribute to the signals. One possible explanation is that localization of this component in M. viride cells is unfavorable for registration in the unilateral mode of our suspension method (for a more detailed discussion of geometric foundations of the method, see reference 37). Alternatively, this component could be relatively small, another argument for which is the observation that photomotility responses in M. viride were not highly sensitive to light (reference 38 and our unpublished observations). Analysis of signal transduction in M. viride is beyond the scope of this report, in which we focus on the early photoreceptor current that directly reflects channelrhodopsin function in native cells.
The action spectrum for the photoreceptor current in suspensions of M. viride cells is shown in Fig. 1C. The spectral sensitivity was estimated by calculating the mean value of the current response between 50 µs and 2 ms. The first 50 µs was omitted, because during this time, a fast photosynthetic signal developed (30). To completely eliminate the contribution of this signal, the traces were further corrected by subtracting the responses evoked by a 680-nm laser flash. This wavelength is ineffective for generation of channelrhodopsin-mediated currents or motility responses in flagellates but is absorbed by the photosynthetic pigments. The spectrum was corrected for photon density, taking into account the logarithmic fluence response dependence (Fig. 1B). The spectral maximum at 531 nm was determined by Gaussian fit of the main peak (the solid line in Fig. 1C). The 90-nm half-bandwidth of the spectrum is typical of retinal proteins, indicating that a single rhodopsin species is responsible for the fast light-induced currents in M. viride. Significant light sensitivity of the electric response is observed even beyond 620 nm. This long-wavelength action spectrum and the fast kinetics of the photoelectric current made this organism a promising candidate for cloning a channelrhodopsin likely to be useful for optogenetic applications.
M. viride opsin sequence.
The opsin sequence cloned from M. viride (593 residues) consists of a 7TM domain and an extended C-terminal domain, as is characteristic of other known channelrhodopsins. Most of the residues known to form the retinal binding pocket in bacteriorhodopsin (BR) are conserved. However, the overall homology of this sequence to other channelrhodopsins is lower than between any two of them: its 7TM domain shows 38% identity and 52% similarity with ChR1, 35% identity and 51% similarity with ChR2, 36% identity and 51% similarity with VChR1, and 37% identity and 53% similarity with VChR2. The genomes of C. reinhardtii and V. carteri are completely sequenced, and each contains two channelopsin genes. Complete genomic information is not available for M. viride, and there is a possibility that its genome also contains more than one channelopsin. The M. viride protein cannot be definitively distinguished as channelrhodopsin 1 or 2 on the basis of sequence homology since it is only slightly closer to the two channelrhodopsin 1 sequences than to the channelrhodopsin 2 sequences (Fig. 2A). Also, this sequence lacks two structural features that differentiate ChR1/VChR1 from ChR2/VChR2. First, there is no Glu residue corresponding to Glu87 (ChR1 numbering), which is implicated by mutagenesis as required for the pH-dependent spectral shift characteristic of ChR1/VChR1 (21). Also, the positions of Tyr226 (ChR1) and Asn187 (ChR2), conserved in VChR1 and VChR2, respectively, are two of the molecular determinants of spectral sensitivity, desensitization and kinetics, by which ChR1 differs from ChR2 (24). In the M. viride protein, this site is occupied by a Trp residue.
FIG 2 .
(A) Phylogenetic tree of the channelrhodopsin family constructed by the neighbor-joining method; (B) alignment of channelrhodopsin sequences (regions of predicted helices B and C of the 7TM domain). The conserved Glu residues are shown in green, the conserved His residue at the proton donor position in BR (Asp96) is shown in blue, other conserved residues of the retinal binding pocket are shown in yellow, and important residues that are not conserved in MChR1 are shown in red.
In the two known channelrhodopsin pairs, the more red-shifted one is defined as channelrhodopsin 1. The spectral sensitivity of M. viride channelrhodopsin is more red-shifted than that of any previously known species, including the most red-shifted VChR1 (see below). Therefore, we named it MChR1.
Unexpectedly, several residues in the 7TM region previously considered defining features of the channelrhodopsin family based on the four known homologs are not conserved in MChR1, as described below under “Mutagenesis studies.” The C-terminal domain of MChR1 shows even lower homology than the 7TM domain to the C-terminal domains of other channelrhodopsins.
Channel activity in HEK293 cells.
The light-gated channel activity of Mesostigma rhodopsin was tested in HEK293 cells (Fig. 3). It has been reported that the C domain of channelrhodopsins did not influence their function in heterologous systems (7, 8), so here we studied only the 7TM domain of MChR1 (331 residues). As with the other known channelrhodopsins, the photocurrent generated by MChR1 under sustained illumination decayed from a peak to a plateau, a phenomenon called inactivation (Fig. 3A). The relatively small inactivation of MChR1-generated currents showed that this protein is functionally closer to ChR1 than to ChR2, as the former exhibits a lower degree of inactivation (21). The amplitude and the sign of the photocurrent depended on the holding potential, as is typical of other channelrhodopsins, with the reversal potential close to 0 (Fig. 3B). The mean peak value of MChR1 photocurrents at the saturating light intensity and a −60-mV holding potential was 276 ± 97 pA (mean ± standard error of the mean [SEM]; n = 12), which is comparable to that reported for VChR1 in neurons (10) and to our own measurements of VChR1 in HEK cells. The plateau amplitude of MChR1 photoresponses saturated at lower light intensities than the peak value (Fig. 4), as in other channelrhodopsins (20, 39), and the degree of inactivation increased with the light intensity. When successive pulses were applied with a variable dark interval, the amplitude of the peak recovered, with a time constant of ~3.5 s at pH 7.4, which was faster than that in other channelrhodopsins (20, 39, 40). However, the level to which the amplitude recovered was only about 90% of that elicited by the first flash, even after minutes of dark adaptation. This suggests that in addition to this short-term process, another, long-term recovery might take place. Interestingly, the same behavior of channelrhodopsin-mediated electrical responses was observed earlier in native algal cells (34).
FIG 3 .
(A) Photocurrents in HEK293 cells expressing the 7TM domain of MChR1 recorded by the whole-cell patch clamp method at different holding potentials changed in 20-mV steps from −100 mV (the bottom trace) to +40 mV (the top trace). (B) Current voltage dependence of the peak photocurrent. Data are mean values ± SEM of results from four successive scans.
FIG 4 .
(A) Photocurrents in HEK293 cells expressing the 7TM domain of MChR1 recorded by the whole-cell patch clamp method. Excitation was at 530 nm for 2 s, with a 30-s dark interval; relative light intensities, starting from the bottom trace, were 100%, 20%, 7%, 3%, and 0.5%. (B) Dependence of the amplitudes of the peak and plateau currents (filled squares and filled circles, respectively) and the initial slope of photocurrent (open squares) on the light intensity. The initial slope of the photocurrent was normalized to the peak value at the maximal intensity.
Estimation of the absorption properties of a receptor responsible for a photobiological response is usually done by action spectroscopy. For the action spectrum to faithfully reflect the absorption spectrum of the pigment, the response needs to be measured far from saturation, ideally in the linear range of the intensity response curve. Also, the action spectrum should be measured in response to a short stimulus to avoid secondary photochemistry, which may produce a spectrally dependent mixture of intermediates, affecting the action spectrum. We measured the initial slope of the photocurrent (assessed from the mean amplitude of the signal recorded during the first 50 ms of illumination with low-intensity light) to evaluate spectral properties of the ground state of MChR1 in HEK cells. This parameter showed a close-to-linear dependence on light, even at intensities when both the peak and plateau levels saturated (Fig. 4B), and hence intensities could easily be corrected for equal photon densities.
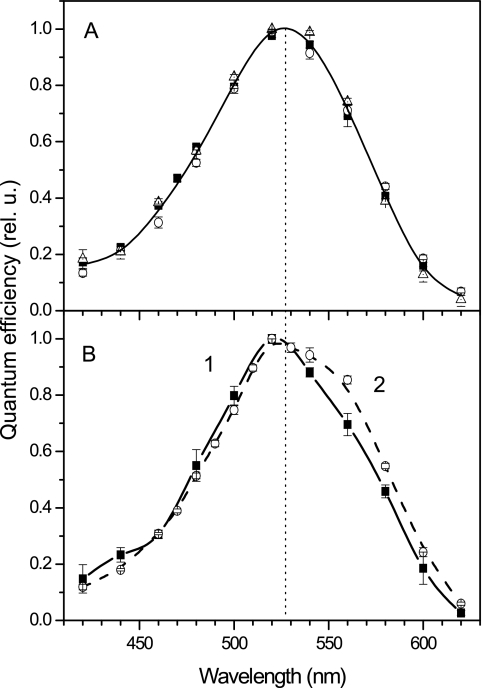
The action spectra of MChR1 light-gated channel activity in HEK cells measured as outlined above at three values of external pH are shown in Fig. 5A. To reveal the position of their maxima, the main peaks of the spectra were fitted with a Gaussian function (as shown in Fig. 1C for the spectrum in native M. viride cells). At all three pH values, the spectral maximum was at 528 nm, which corresponded within a 3-nm accuracy to the maximum of the spectral efficiency of the pigment in native M. viride cells (Fig. 1C).
FIG 5 .
Low-light-intensity action spectra of photoelectric responses in HEK293 cells. (A) MChR1 at the external pHs of 9.0 (filled squares), 7.4 (open circles), and 5.3 (open triangles). As these data were very close, a single B-spline solid line was drawn through the average values at each wavelength. (B) VChR1 at external pHs of 7.4 (filled squares) and 5.3 (open circles). Data for each spectrum are mean values ± SEM of results from 6 to 10 successive scans in opposite directions obtained on 2 to 3 cells.
The long-wavelength spectral sensitivity found in MChR1 is one of the properties highly desirable for optogenetic applications. The most red-shifted channelrhodopsin variant reported earlier was VChR1 from V. carteri (10). We measured the action spectra of the light-gated channel activity of VChR1 under the same experimental conditions as we used for MChR1 and found, using the same fitting algorithm, that the VChR1 spectrum peaked at 520 nm at neutral pH (Fig. 6B, solid line), which was ~8 nm less than that measured in MChR1 (Fig. 6A). The 520-nm peak of the VChR1 spectrum corresponded to the absorption maximum of the isolated pigment measured at this pH (6). We found that acidification of the external medium led to the appearance of a red-shifted shoulder in the VChR1 action spectrum (Fig. 6B, dashed line). The maximal absorption of purified VChR1 was reported to shift to 540 nm at low pH, indicating conversion of the pigment to a protonated form (6). Our results suggest that only partial conversion occurred in HEK cells or that the conductance of the protonated state was much smaller than that of the 520-nm form.
FIG 6 .
Current kinetics (A) and dependence of the rate of the fast-decay component on external pH (mean values ± SEM of results from 6 to 10 cells for each pH value) (B) of MChR1 and VChR1 (filled squares and open circles, respectively) expressed in HEK293 cells. Excitation was at 530 nm for 2 s. In panel A, time zero corresponds to the end of the light pulse.
The main difference between the two red-shifted channelrhodopsins was that the current decay after switching off the light was much faster in MChR1 than in VChR1. In both cases, the decay kinetics was better fitted with two exponentials. At neutral pH, the time constants of both components were smaller in MChR1 than in VChR1 (Fig. 6A). In addition, the relative amplitude of the slow component was smaller in MChR1, further increasing the rate of the overall decay. Due to the small contribution of the slow component to the decay kinetics in MChR1, here we analyzed only its fast component. Its rate strongly increased upon acidification of the external medium from pH 9.0 to pH 5.3 (Fig. 6B, filled symbols), whereas in VChR1 it only slightly increased (Fig. 6B, open symbols). However, the rate of the slow component in VChR1 showed a stronger dependence on pH than the fast component (data not shown).
In view of possible optogenetic applications, we compared the responses of MChR1 and VChR1 to high-frequency stimulation. At a 25-Hz stimulus frequency, the photocurrent generated by MChR1 was modulated to at least 50% amplitude, whereas that generated by VChR1 showed practically no modulation (Fig. 7A). The much higher degree of amplitude modulation in MChR1 than in VChR1 derives not only from the faster current decay but also from significantly less inactivation of the photocurrent by a series of light pulses (Fig. 7A). Consequently, at the level of 50% amplitude modulation, MChR1 demonstrated an ~15-fold-better frequency response than VChR1 (Fig. 7B). This probably reflects the adaptation of unicellular M. viride for a rotation frequency higher than that of colonial V. carteri.
FIG 7 .
(A) Photoresponses generated by MChR1 and VChR1 expressed in HEK293 cells upon 25-Hz stimulation with 530-nm light; (B) dependence of the amplitude of modulation on the stimulus frequency for MChR1 (filled squares) and VChR1 (open circles). Data are the mean values ± SEM of results from 8 successive frequency changes in the opposite directions.
Mutagenesis studies.
As shown above, wild-type MChR1 exhibited typical light-gated channel activity upon heterologous expression, although not all structural features found in other channelrhodopsins are conserved in its primary sequence. To test for the functional importance of these features, we generated and studied three groups of MChR1 point mutants.
(i) V102E and A116E mutants.
All other channelrhodopsins contain an array of five Glu residues in the predicted second helix, but only three of these are conserved in MChR1 (Fig. 2B). It might be expected that introducing the missing glutamates (corresponding to Glu83 and Glu97 in the ChR2 sequence) in the MChR1 sequence would enhance its channel activity. However, both mutations resulted in a dramatic decrease in the whole-cell current amplitude; the plateau current at the saturating light intensity was 28.3 ± 8.0 pA (mean ± SEM; n = 6) in the A116E mutant and 0.3 ± 0.3 pA (mean ± SEM; n = 8) in the V102E mutant. The intensity of fluorescence of the enhanced yellow fluorescent protein (EYFP) in cells that expressed the mutant sequences did not change compared to that of the wild type. Inhibition of the channel function of the MChR1 molecule was also revealed by a significant slowing down of the current kinetics (see Fig. 8 for the A116E mutant; data for the V102E mutant are not shown). In the V102E A116E double mutant (glutamate was introduced at both positions), the photocurrents were below the detection limit of our measurements.
FIG 8 .
Photocurrents generated by the A116E and C147A mutants and wild-type MChR1 (WT) in HEK293 cells. Traces were normalized at the plateau level to reveal the differences in the decay kinetics. The increased level of the noise reflects a decrease in the absolute current amplitudes. Excitation was at 530 nm for 0.5 s. Time zero corresponds to the end of the light pulse.
(ii) A153/H/R mutant.
All known channelrhodopsins so far have a His residue at the position of the proton donor in BR (Asp96), and its replacement with Arg resulted in an increase in the stationary current amplitude in ChR2 (15). When Ala found in MChR1 in this position was replaced with His or Arg, the current was completely suppressed without decreasing the EYFP fluorescence in the membrane.
(iii) C147A and D175A mutants.
It has been suggested that residues Cys128 and Asp156 in ChR2 form a hydrogen bond between helices C and D that is important for channel gating (41, 42). Its disruption leads to a dramatic increase in the lifetime of the channel’s open state (25, 43). The corresponding residues (C147 and D175) are conserved in MChR1. Replacing either of these residues with Ala led to a large decrease in the EYFP fluorescence, indicating a decrease in protein expression levels. The current amplitude was also dramatically decreased; the plateau amplitude was only 0.7 ± 0.5 pA (mean ± SEM; n = 9) in the C147A mutant and 7.3 ± 7.3 pA (mean ± SEM; n = 3) in the D175A mutant. In agreement with the ChR2 result, analysis of the current kinetics showed similar significant increases in the decay time (see Fig. 8 for the D175A mutant; data for the C147A mutant are not shown).
DISCUSSION
Our results show that screening flagellate species by measuring rhodopsin-mediated photoelectric currents in vivo is an efficient strategy for selecting potential source organisms for cloning new channelrhodopsins with characteristics desirable for optogenetic applications. Such screening cannot be performed by measuring phototaxis itself, i.e., the behavioral response. First of all, if an alga exhibits phototaxis, it is not necessarily mediated by a channelrhodopsin; a well-known example is Euglena, which uses a flavin-binding adenylyl cyclase as a receptor for light control of motility (44). Second, even in those species in which channelrhodopsins do act as receptors for phototaxis, a change in the flagellar beating pattern occurs as the final result of a not yet fully understood amplification cascade (for reviews, see references 45 to 47). Consequently, the strength of phototaxis in a particular species might reflect the efficiency of amplification, rather than the properties of the cell’s channelrhodopsins. The latter can be probed in algal cells only by measuring photoelectric responses.
MChR1 is a channelrhodopsin that we cloned from the flagellate alga M. viride, identified as a candidate species by the above approach. The MChR1 sequence significantly expands channelrhodopsin diversity, as it deviates from other known channelrhodopsins in several important aspects, thereby refining the sequence criteria for light-gated channel activity. Our results showed that two out of five glutamate residues in the putative helix B that are conserved in the other four channelrhodopsins and that have been suggested to contribute to forming a water-containing channel (48, 49) are not essential for the channel function, at least in some channelrhodopsins. Similarly, although the position of the proton donor (Asp96 in BR) is confirmed to be sensitive, the presence of His at this site cannot be considered an essential feature of the channelrhodopsin family. On the other hand, formation of the predicted hydrogen bond between putative helices C and D (Cys128 and Asp156 according to ChR2 numbering) is required for proper gating even in such a distant member of the channelrhodopsin family as MChR1. Also, the role of Glu87 in pH-dependent color tuning in channelrhodopsins (21) is indirectly confirmed by the lack of such tuning in MChR1, in which no Glu residue is found at this position.
Analysis of MChR1 properties upon expression in HEK cells showed that (i) it exhibits the most red-shifted absorption among all studied channelrhodopsins; (ii) the spectral sensitivity of its channel activity is pH independent; (iii) its current kinetics is significantly faster than that of the other known red-shifted channelrhodopsin, VChR1; and (iv) it shows lower inactivation than VChR1 when stimulated with light of the same wavelength. These four qualities are highly desirable for a prospective optogenetic tool, especially when high-frequency stimulation is required.
MATERIALS AND METHODS
Strain and growth conditions.
Mesostigma viride strain CCMP2046 (also known as NIES 296) was obtained from the Provasoli-Guillard National Center for the Culture of Marine Phytoplankton. Cells were grown at 25°C in modified Jaworski medium [the concentration of Ca(NO3)2 was increased to 0.17 mM; 0.5 mM KNO3, 0.15 µM ZnSO4, and 0.04 µM CoCl2 were added] under a 12-h light (~3,000 lx)–12-h dark cycle.
Measurements of rhodopsin-mediated photocurrents in M. viride cells.
Currents were measured with the population assay described in reference 30. Two platinum wires immersed in a cell suspension pick up an electrical current generated in response to a unilateral excitation flash from a Vibrant HE 355II tunable laser (Opotek Inc., Carlsbad, CA), set at desired wavelengths. The signal was amplified by a low-noise current amplifier (model 428; Keithley Instruments, Cleveland, OH) and digitized by a Digidata 1322A digitizer, supported by pCLAMP 10 software (both from Molecular Devices, Union City, CA).
Cloning and expression of M. viride channelopsin.
Total RNA was extracted from 500 ml of 1-week-old culture of M. viride using Trizol reagent (Invitrogen, Carlsbad, CA). Synthesis of 3′ and 5′ rapid amplification of cDNA ends (RACE)-ready first-strand cDNAs and 3′ and 5′ RACE PCR were carried out using the SMARTer RACE cDNA amplification kit (Clontech Laboratories, Takara Bio Company, Mountain View, CA) using primers designed according to the opsin sequence fragment found in the Taxonomically Broad EST Database (http://amoebidia.bcm.umontreal.ca/pepdb/searches/welcome.php). The overlapping RACE fragments were combined by fusion PCR, cloned into the pCR2.1-TOPO vector (Invitrogen), and fully sequenced. The 7TM domain (encoding residues 1 to 331) was inserted between BamHI and NotI sites to replace the VChR1 sequence in the pcDNA3.1/VChR1-EYFP mammalian expression vector provided by K. Deisseroth (Stanford University). This vector was also used for expression of VChR1 in HEK cells. The vector map and sequence are available at the EveryVector website. The presence of a fluorescent tag is not expected to affect channelrhodopsin properties, as has been shown by quantitative comparison of photocurrents generated by YFP-, mCherry-, and myc-tagged ChR2 (50). Point mutations were introduced using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). HEK293 cells were transfected using the TransPass COS/293 transfection reagent (New England Biolabs, Ipswich, MA). All-trans-retinal (stock solution in ethanol) was added to a final concentration of 2.5 µM.
Whole-cell patch clamp recording.
Measurements were performed 24 to 48 h after transfection with an Axopatch 200B amplifier (Molecular Devices, Union City, CA). The signals were digitized with a Digidata 1440A digitizer using pCLAMP 10 software (both from Molecular Devices). Patch pipettes with resistances of 2 to 5 MΩ were fabricated from borosilicate glass and filled with the following solution (in mM): KCl (126), MgSO4 (2), CaCl2 (0.5), EGTA (5), and HEPES (25, pH 7.2). The bath solution contained (in mM) NaCl (150), CaCl2 (1.8), KCl (4), MgCl2 (1), glucose (5), and HEPES (10, pH 7.4), unless otherwise indicated. For experiments at pH 9, Tris was used in the bath solution instead of HEPES. Unless otherwise indicated, the holding potential was −60 mV. Light excitation was provided by a polychrome IV light source (TILL Photonics GmbH, Grafelfing, Germany) pulsed with a mechanical shutter (Uniblitz model LS6; Vincent Associates, Rochester, NY). The light intensity was attenuated with the built-in polychrome system or with neutral density filters.
Nucleotide sequence accession number.
The nucleotide sequence of the opsin sequence cloned from M. viride was deposited in GenBank under accession no. JF922293.
ACKNOWLEDGMENTS
We are grateful to K. Deisseroth (Stanford University) for the pcDNA3.1/VChR1-EYFP mammalian expression vector, V. Jayaraman (UT Houston Health Science Center) for the HEK293 cell line and advice on whole-cell patch clamp recording, and K. D. Ridge (UT Houston Health Science Center) for his help with HEK cell cultivation.
The project described was supported by award no. RC1AG035779 from the National Institute on Aging, grant R37GM027750 from the National Institute of General Medical Sciences, and endowed chair AU-0009 from the Robert A. Welch Foundation.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging, the National Institute of General Medical Sciences, or the National Institutes of Health.
Footnotes
Citation Govorunova EG, Spudich EN, Lane CE, Sineshchekov OA, Spudich JL. 2011. New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride. mBio 2(3):e00115-11. doi:10.1128/mBio.00115-11.
REFERENCES
- 1. Spudich JL, Yang CS, Jung KH, Spudich EN. 2000. Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16:365–392 [DOI] [PubMed] [Google Scholar]
- 2. Spudich JL, Jung KH. 2005. Microbial rhodopsins: phylogenetic and functional diversity, p. 1–23 In Briggs WR, Spudich JL, Handbook of photosensory receptors. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 3. Sineshchekov OA, Jung KH, Spudich JL. 2002. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A. 99:8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Govorunova EG, Jung KH, Sineshchekov OA, Spudich JL. 2004. Chlamydomonas sensory rhodopsins A and B: cellular content and role in photophobic responses. Biophys. J. 86:2342–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berthold P, et al. 2008. Channelrhodopsin-1 initiates phototaxis and photophobic responses in Chlamydomonas by immediate light-induced depolarization. Plant Cell 20:1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kianianmomeni A, Stehfest K, Nematollahi G, Hegemann P, Hallmann A. 2009. Channelrhodopsins of Volvox carteri are photochromic proteins that are specifically expressed in somatic cells under control of light, temperature, and the sex inducer. Plant Physiol. 151:347–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nagel G, et al. 2002. Channelrhodopsin-1: a light-gated proton channel in green algae. Science 296:2395–2398 [DOI] [PubMed] [Google Scholar]
- 8. Nagel G, et al. 2003. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U. S. A. 100:13940–13945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suzuki T, et al. 2003. Archaeal-type rhodopsins in Chlamydomonas: model structure and intracellular localization. Biochem. Biophys. Res. Commun. 301:711–717 [DOI] [PubMed] [Google Scholar]
- 10. Zhang F, et al. 2008. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat. Neurosci. 11:631–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Litvin FF, Sineshchekov OA, Sineshchekov VA. 1978. Photoreceptor electric potential in the phototaxis of the alga Haematococcus pluvialis. Nature 271:476–478 [DOI] [PubMed] [Google Scholar]
- 12. Harz H, Hegemann P. 1991. Rhodopsin-regulated calcium currents in Chlamydomonas. Nature 351:489–491 [Google Scholar]
- 13. Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. 2005. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8:1263–1268 [DOI] [PubMed] [Google Scholar]
- 14. Li X, et al. 2005. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. U. S. A. 102:17816–17821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nagel G, et al. 2005. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 15:2279–2284 [DOI] [PubMed] [Google Scholar]
- 16. Deisseroth K. 2011. Optogenetics. Nat. Methods 8:26–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin JY. 2011. A user’s guide to channelrhodopsin variants: features, limitations and future developments. Exp. Physiol. 96:19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hegemann P, Möglich A. 2011. Channelrhodopsin engineering and exploration of new optogenetic tools. Nat. Methods 8:39–42 [DOI] [PubMed] [Google Scholar]
- 19. Bamann C, Kirsch T, Nagel G, Bamberg E. 2008. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J. Mol. Biol. 375:686–694 [DOI] [PubMed] [Google Scholar]
- 20. Ishizuka T, Kakuda M, Araki R, Yawo H. 2006. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci. Res. 54:85–94 [DOI] [PubMed] [Google Scholar]
- 21. Tsunoda SP, Hegemann P. 2009. Glu 87 of channelrhodopsin-1 causes pH-dependent color tuning and fast photocurrent inactivation. Photochem. Photobiol. 85:564–569 [DOI] [PubMed] [Google Scholar]
- 22. Feldbauer K, et al. 2009. Channelrhodopsin-2 is a leaky proton pump. Proc. Natl. Acad. Sci. U. S. A. 106:12317–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin JY, Lin MZ, Steinbach P, Tsien RY. 2009. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys. J. 96:1803–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang H, et al. 2009. Molecular determinants differentiating photocurrent properties of two channelrhodopsins from Chlamydomonas. J. Biol. Chem. 284:5685–5696 [DOI] [PubMed] [Google Scholar]
- 25. Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. 2009. Bi-stable neural state switches. Nat. Neurosci. 12:229–234 [DOI] [PubMed] [Google Scholar]
- 26. Gunaydin LA, et al. 2010. Ultrafast optogenetic control. Nat. Neurosci. 13:387–392 [DOI] [PubMed] [Google Scholar]
- 27. Wen L, et al. 2010. Opto-current-clamp actuation of cortical neurons using a strategically designed channelrhodopsin. PLoS One 5:e12893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kleinlogel S, et al. 2011. Ultra light-sensitive and fast neuronal activation with the Ca2+-permeable channelrhodopsin CatCh. Nat. Neurosci. 14:513–518 [DOI] [PubMed] [Google Scholar]
- 29. Berndt A, et al. 2011. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl. Acad. Sci. U. S. A. 108:7595–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sineshchekov OA, Govorunova EG, Der A, Keszthelyi L, Nultsch W. 1992. Photoelectric responses in phototactic flagellated algae measured in cell suspension. J. Photochem. Photobiol. B Biol. 13:119–134 [Google Scholar]
- 31. Melkonian M. 1983. Functional and phylogenetic aspects of the basal apparatus in algal cells. J. Submicrosc. Cytol. 15:121–125 [Google Scholar]
- 32. Rodríguez-Ezpeleta N, Philippe H, Brinkmann H, Becker B, Melkonian M. 2007. Phylogenetic analyses of nuclear, mitochondrial, and plastid multigene data sets support the placement of Mesostigma in the Streptophyta. Mol. Biol. Evol. 24:723–731 [DOI] [PubMed] [Google Scholar]
- 33. Sineshchekov OA, Spudich JL. 2005. Sensory rhodopsin signaling in green flagellate algae, p. 25–42 In Briggs WR, Spudich JL, Handbook of photosensory receptors. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 34. Sineshchekov OA, Litvin FF, Keszthelyi L. 1990. Two components of photoreceptor potential in phototaxis of the flagellated green alga Haematococcus pluvialis. Biophys. J. 57:33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sineshchekov OA. 1991. Photoreception in unicellular flagellates: bioelectric phenomena in phototaxis, p. 523–532 In Douglas RD, Light in biology and medicine, vol. II Plenum Press, New York, NY. [Google Scholar]
- 36. Sineshchekov OA, Govorunova EG. 1999. Rhodopsin-mediated photosensing in green flagellated algae. Trends Plant Sci. 4:58–63 [DOI] [PubMed] [Google Scholar]
- 37. Sineshchekov OA, Govorunova EG. 2001. Electrical events in photomovements of green flagellated algae, p. 245–280 In Hader DP, Lebert M, Comprehensive series in photosciences, vol. 1 Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 38. Matsunaga S, Watanabe S, Sakaushi S, Miyamura S, Hori T. 2003. Screening effect diverts the swimming directions from diaphototactic to positive phototactic in a disk-shaped green flagellate Mesostigma viride. Photochem. Photobiol. 77:324–332 [DOI] [PubMed] [Google Scholar]
- 39. Ernst OP, et al. 2008. Photoactivation of channelrhodopsin. J. Biol. Chem. 283:1637–1643 [DOI] [PubMed] [Google Scholar]
- 40. Hegemann P, Ehlenbeck S, Gradmann D. 2005. Multiple photocycles of channelrhodopsin. Biophys. J. 89:3911–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Radu I, et al. 2009. Conformational changes of channelrhodopsin-2. J. Am. Chem. Soc. 131:7313–7319 [DOI] [PubMed] [Google Scholar]
- 42. Nack M, et al. 2010. The DC gate in channelrhodopsin-2: crucial hydrogen bonding interaction between C128 and D156. Photochem. Photobiol. Sci. 9:194–198 [DOI] [PubMed] [Google Scholar]
- 43. Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. 2010. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry 49:267–278 [DOI] [PubMed] [Google Scholar]
- 44. Iseki M, et al. 2002. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature 415:1047–1051 [DOI] [PubMed] [Google Scholar]
- 45. Witman GB. 1993. Chlamydomonas phototaxis. Trends Cell Biol. 3:403–408 [DOI] [PubMed] [Google Scholar]
- 46. Kreimer G. 1994. Cell biology of phototaxis in flagellate algae. Int. Rev. Cytol. 148:229–310 [Google Scholar]
- 47. Sineshchekov OA, Govorunova EG, Spudich JL. 2009. Photosensory functions of channelrhodopsins in native algal cells. Photochem. Photobiol. 85:556–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hegemann P. 2008. Algal sensory photoreceptors. Annu. Rev. Plant Biol. 59:167–189 [DOI] [PubMed] [Google Scholar]
- 49. Sugiyama Y, et al. 2009. Photocurrent attenuation by a single polar-to-nonpolar point mutation of channelrhodopsin-2. Photochem. Photobiol. Sci. 8:328–336 [DOI] [PubMed] [Google Scholar]
- 50. Nikolic K, et al. 2009. Photocycles of channelrhodopsin-2. Photochem. Photobiol. 85:400–411 [DOI] [PubMed] [Google Scholar]