Abstract
Objective
Brain-derived neurotrophic factor (BDNF) and apolipoprotein E (ApoE) are thought to be implicated in a variety of neuronal processes, including cell growth, resilience to noxious stimuli and synaptic plasticity. A Val to Met substitution at codon 66 in the BDNF protein has been associated with a variety of neuropsychiatric conditions. The ApoE4 allele is considered a risk factor for late-onset Alzheimer’s disease, but its effects on young adults are less clear. We sought to investigate the effects of those two polymorphisms on hemispheric and lateral ventricular volumes of young healthy adults.
Methods
Hemispheric and lateral ventricular volumes of 144 healthy individuals, aged 19–35 years, were measured using high resolution magnetic resonance imaging and data were correlated with BDNF and ApoE genotypes.
Results
There were no correlations between BDNF or ApoE genotype and hemispheric or lateral ventricular volumes.
Conclusion
These findings indicate that it is unlikely that either the BDNF Val66Met or ApoE polymorphisms exert any significant effect on hemispheric or lateral ventricular volume. However, confounding epistatic genetic effects as well as relative insensitivity of the volumetric methods used cannot be ruled out. Further imaging analyses are warranted to better define any genetic influence of the BDNF Val6Met and ApoE polymorphism on brain structure of young healthy adults.
Keywords: apolipoprotein E, brain-derived neurotrophic factor, magnetic resonance imaging, polymorphism, volumetry
Introduction
In the last 15 years, there has been an evolving interest in the field of molecular and biological psychiatry. There is a growing amount of evidence from basic and clinical research to suggest that morphological characteristics of the human nervous system are at least to some degree influenced by genetic factors.
Brain-derived neurotrophic factor (BDNF) is a trophic factor produced mainly in the central nervous system. It is necessary for survival of several neuronal populations in the cortex, hippocampus, striatum, olfactory bulb and cerebellum. It is widely expressed in the mammalian brain with the hippocampus (1) and the prefrontal cortex (2) having the highest levels of expression. BDNF also affects axonal and dendritic outgrowth as well as synaptic plasticity (3) and prevents cellular death in the adult brain (4). Its synthesis and release is controlled by activation of glutamate receptors (5,6). A recently described single nucleotide polymorphism (SNP) in the 5′ prodomain region of the BDNF gene in which valine is replaced by methionine, due to substitution of guanine by adenine (G to A) at nucleotide 196, is an example of a neurotrophin gene allelic variation implicated in brain structure and function. This polymorphism seems to impair intracellular trafficking and packaging of BDNF and its activation-dependent release (7,8). It is relatively common in Caucasian subjects with a frequency of 19–25% (9–13), while its frequency is estimated to be somehow higher in Asian populations (approximately 30%) (5). The most widely investigated structural effect of this polymorphism to date has been its association to a reduced hippocampal volume (14–17) without, though, absolute consensus (14,18). Nemoto et al. (18) found a decreased volume in the dorsal lateral prefrontal cortex, anterior cingulate and temporal and parietal lobes in healthy, aged 20–72, subjects. A negative effect of the Val66Met polymorphism on hippocampal volume was demonstrated in another trial that recruited healthy and depressed subjects, irrespective of the presence of disease (17). There may also be a more general rather than regional effect of the BDNF Val66Met polymorphism. In an analysis of 331 young adolescents, Toro et al. (19) found an average decrease of 28.34 cm3 in Met carriers compared to Val homozygotes.
Apolipoprotein E (ApoE) is a 299 amino acid polypeptide which plays a significant role in lipid metabolism and circulation. Human ApoE has three major isoforms, namely E2, E3 and E4 coded for by respective alleles. Isoform E3 is the most common one in most populations representing likely a mutation of the phylogenetically older E4 isoform. E2 is the least common of all alleles (20,21). In their study, including a German sample, Corbo et al.(22) reported frequencies of 7.7, 77.8 and 14.5% for E2, E3 and E4, respectively, for the German sample. Signalling through binding of ApoE to its receptors mediates several processes such as synaptic plasticity, neuronal survival and neurite outgrowth (23,24). There is a plethora of data to suggest linkage of the Apo E4 allele with Alzheimer’s disease, although it is neither a necessary nor a sufficient presupposition. The E2 isoform seems, however, to exert a protective effect. The mechanisms through which the E4 isoform increases the risk for Alzheimer’s disease remain to be further clarified but proposed theories include differential domain interaction with its receptors (25), instability to protein unfolding (26) and impaired clearance of the amyloid-beta polypeptide (27). Neuroimaging studies have demonstrated diminished hippocampal volumes and increased hippocampal atrophy rates in Alzheimer’s patients. There is, however, ongoing research in other, nondemented, populations to investigate whether this gene may have any effect at an earlier age and before any clinical symptoms, by conferring a neural substrate for susceptibility to neurodegeneration at a later age. Hippocampal volume, rate of volume loss and cortical thickness were lower, higher and decreased, respectively in ApoE4 carriers in studies that recruited nondemented middle and older age probands (28–30) Earlier studies had also found a decreased hippocampal volume asymmetry (31) and diminished right hippocampal volume in elderly nondemented subjects (32). Regionally, reduced grey matter (GM) density was detected by Wishart et al. (33) in nondemented adults, carriers of the ApoE4 gene. Reduced cortical thickness in adults (34) and thickness of the entorhinal cortex in children and young adolescents (35) has also been linked to ApoE4 carrier status. Mondadori et al. (36), however, in a sample of 34 young, healthy subjects found no difference in whole brain or hippocampal volumes among the three alleles.
Ventricular volume measurement may be an objective and easily reproducible and obtainable variable to assess for neuropathological changes. On magnetic resonance images (MRI) there is a robust contrast between the ventricular spinal fluid and surrounding tissue in volumetric T1-weighted images. It has been a widely implemented tool in the field of neurodegeneration (37–41). Furthermore, hemispheric atrophy rates as defined by ventricular enlargement correlate significantly with cognitive changes and disease progression, at least in Alzheimer’s disease (39). This may, to a certain degree, be explained by the fact that parts of the lateral ventricles line specific medial temporal lobe structures that are affected during the disease course (42,43). Although ventricular volumetry has not been used in a young population, it has been implemented in normal elderly controls. As BDNF may exert trophic effects on multiple brain regions, we sought to evaluate this easily quantifiable and sensitive method in our population. We employed the same method for the ApoE4 polymorphism to define whether there are volumetric differences between E4 and noncarriers.
Materials and methods
Subjects
From February 2007 to August 2008, 144 young, healthy individuals, aged 19–35 years, were recruited and underwent a neuropsychological evaluation, BDNF genotyping and volumetric brain MRI at the University of Erlangen-Nuremberg as part of the GENES study. This study was conducted in accordance with the declaration of Helsinki and approved by the local Ethics Committee. All probands provided written informed consent after deemed eligible for the study. Exclusion criteria were the following: age younger than 18 or older than 35 years, inability to provide appropriate consent, concurrent or past medical, neurological or psychiatric conditions, history of head trauma with loss of consciousness, substance abuse, positive family history of a neurological or psychiatric disease in first degree relatives, birth complications, any contraindications for having MRI, history of heart or brain surgery and pregnancy or lactation. There were 51 men (35.4%) and 93 women (64.6%) with a mean age of 24.79 years (range 19–35) and a mean number of 17.03 years of education (range 12–23). All subjects were genotyped for the BDNF gene which yielded a percentage of 28.5% for the Val66Met polymorphism close to percentages previously reported. We also genotyped all subjects for the ApoE4 allele yielding a percentage of approximately 14%, similar to previous studies.
Structural neuroimaging and image analysis
Brain MRIs were analysed for 144 subjects. Axial FLAIR images with a slice thickness of 5 mm were obtained to exclude any intracranial pathology and reviewed by the neuroradiology department at the University of Erlangen-Nuremberg. Volumetric sagittal T1-weighted MPRAGE images were acquired in a Siemens Sonata 1.5 T scanner (Siemens, Erlangen, Germany) using the following parameters: TR/TE = 2030/3.93 ms, field of view = 280 × 280 mm, slices = 176, slice thickness = 1.0 mm, pixel size = 1×1 mm2 (voxel size= 1× 1×1 mm3) and matrix size = 256 × 256.
The T1-weighted images were first converted from DICOM format to ANALYZE using a MAT-LAB (MathWorks, Natick, MA, USA, Version 7.6) code. We then applied hierarchical attribute matching mechanism for elastic registration (HAMMER) (44) to generate labels of different structures. HAMMER is an elastic registration procedure to warp a template brain image to an individual. With the provided brain template, HAMMER locates more than 90 brain regions including the ventricles and subcortical structures.
To make HAMMER robust to different acquisition protocols, a distribution has been provided that assumes images have been skull-stripped and segmented into (GM), white matter (WM) and ventricular cerebrospinal fluid (CSF) (Fig. 1). We used skull strip (http://idealab.ucdavis.edu/index.php) followed by brain extraction tool (BET) (45) for skull-stripping and FSL FAST (46), a clustering algorithm, for segmentation to GM, WM and CSF. For subjects with poor skull-stripping outcome, manual skull-stripping/correction was applied. We used the free available software MRIcro (Chris Rorden, University of South Carolina, USA) (47) to check the quality of skull-stripping results. The majority of images did not need manual correction. At the end, we used a MATLAB code to calculate the volumes of different structures using the produced label image.
Fig. 1.
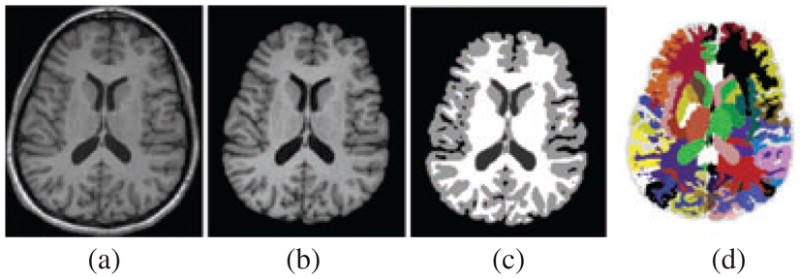
Segmentation of the brain using HAMMER in a representative T1-weighted axial image. (a) Original image. (b) Skull-removal outcome using skull strip (http://idealab.ucdavis.edu/index.php) followed by BET. (c) Outcome of segmentation into WM, GM and CSF using FSL FAST. (d) Colour-coded anatomical segmentation generated by HAMMER.
Genotyping
Peripheral blood was collected from the study subjects and leucocyte DNA was isolated and used for ApoE and BDNF genotyping at the Department of Psychiatry and Psychotherapy of the University Hospital of Erlangen, Germany. The methodology is described in detail in the paper by Richter-Schmidinger et al. (48)
Statistical analysis
We used the SPSS software (Statistical Package for the Social Sciences, Chicago, IL, USA) for Windows, Version 17.0. p Values of less than 0.05 were considered statistically significant. Normal distribution of data was checked using the Kolmogorov–Smirnov test. Differences regarding demographic variables and volumetric parameters among the different genotypic groups were tested using t-test for independent samples under consideration of the Levene test.
Results
To detect any statistically significant differences, we divided our population into different groups according to the presence of one or two A alleles compared to the G allele in the BDNF gene. We performed the same analysis dichotomising our groups in ApoE4 carriers and noncarriers.
When comparing A allele carriers versus noncarriers, there were 52 carriers and 92 noncarriers with a mean age of 24.12 versus 25.17 years and standard deviation (SD) 2.57 versus 3.71 years.
No statistical significance was attained for any of the four volumetric measures using a p value of 0.05 as a cut-off (Table 1). Comparing G allele homozygotes with heterozygotes, there were 92 versus 41 probands with mean ages of 25.17 ± 3.71 and 24.17 ± 2.65 years, respectively. Mean right hemispheric, left hemispheric, right lateral ventricular and left lateral ventricular volumes were 520.22 ± 55.92, 527.72 ± 57.46, 10.06 ± 5.5 and 8.67 ± 5.62 cm3 in the former group versus 504.34 ± 48.76, 512.81 ± 49.4, 9.95 ± 4.56 and 8.3 ± 4.34 cm3 in the latter.
Table 1.
Summary of demographic, genotypic and volumetric data of the study sample
| BDNF Val homozygotes (n = 92), mean (SD) | BDNF Met carriers (n = 52), mean (SD) | ApoE E4 noncarriers (n = 124), mean (SD) | ApoE E4 carriers (n = 20), mean (SD) | |
|---|---|---|---|---|
| Demographic characteristics | ||||
| Age (years) | 25.17 (3.71) | 24.12 (2.57) | 24.9 (3.31) | 24.1 (3.74) |
| Volumetric parameters | ||||
| RHV (cm3) | 520.22 (55.92) | 509 (50.62) | 516.25 (54.63) | 515.65 (52.5) |
| LHV (cm3) | 527.92 (57.46) | 517.4 (51.52) | 524 (56.03) | 523.95 (52.95) |
| RLVV (cm3) | 10.06 (5.5) | 10.1 (4.48) | 10 (5.27) | 10.57 (4.37) |
| LLVV (cm3) | 8.67 (5.62) | 8.31 (4.06) | 8.55 (5.3) | 8.49 (3.73) |
RHV, right hemispheric volume; LHV, left hemispheric volume; RLVV, right lateral ventricular volume; LLVV, left lateral ventricular volume.
We then studied G allele homozygotes versus A allele homozygotes. There were 92 G allele homozygotes versus 11 A allele homozygotes with mean ages 25.17 ± 3.71 and 23.91 ± 2.34 years, respectively. Right hemispheric, left hemispheric, right lateral ventricular and left lateral ventricular volumes were 520.22 ± 55.92, 527.72 ± 57.46, 10.06 ± 5.51 and 8.67 ± 5.62 cm3 in the former versus 526.42 ± 55.94, 534.52 ± 58.02, 10.65 ± 4.33 and 8.35 ± 2.92 cm3 in the latter group, respectively, without reaching statistical significance for any of the volumetric measures. When studying heterozygotes versus A allele homozygotes, there were 41 heterozygotes versus 41 A allele homozygotes with mean ages 24.17 ± 2.65 and 23.91 ± 2.34 years, respectively. Right hemispheric, left hemispheric, right lateral ventricular and left lateral ventricular volumes in the two groups were 504.34 ± 48.76, 512.81 ± 49.4, 9.95 ± 4.56 and 8.3 ± 4.34 cm3 versus 526.42 ± 55.94, 534.53 ± 58.02, 10.65 ± 4.33 and 8.35 ± 2.92 cm3. None of the differences was statistically significant.
With regard to ApoE genotype, there were 124 non-E4 carriers versus 20 E4 carriers with mean ages 24.9 ± 3.31 versus 24.1 ± 3.74 years. Again no statistical significance was attained for any volumetric parameter (Table 1).
Discussion
In our study, there was no association of the BDNF or ApoE genotype with any of the volumetric parameters that were evaluated, both hemispheric and lateral ventricular volumes. It is of note that we used a comparatively large and homogeneous subject material of young, healthy adults chosen based on stringent exclusion criteria. Although we did not use any specific neuropsychological test batteries to further stratify our sample, we believe that based on the questionnaire used, we could reasonably avoid any major confounding factors in the recruitment process. Thus, the variability in brain volumes can be postulated to emanate from their genetic background and much less likely from external factors such as substance abuse or medical illness. There is evidence that brain weight starts diminishing in the third decade of life. The age range used, though some-how arbitrary, achieved a satisfactory compromise of rapid recruitment, due to the study environment and avoidance of the possibility of any underlying neurodegenerative process or simply ageing effect, as stated above. Furthermore, all probands were scanned using the same MR instrument avoiding any variability in imaging protocols or device quality. Despite those strengths we could not detect any volumetric differences among the genotypes tested for. The literature concerning brain volumetric parameters comparing Met to Val carriers has yielded inconsistent results so far, as discussed earlier. Although some reports found a decreased volume of the hippocampal formation in Met carriers (15), they used either a small sample or a more wide spectrum of ages in the subjects they recruited. It is thus possible that the BDNF Val66Met polymorphism may exert differential effects on brain ageing and not during the phase of neuronal growth and stabilisation. However, there is some evidence that the Met allele may have negative effects on the volume of partial brain structures such as the prefrontal cortex or the cingulate gyrus. In our study, the volume of the lateral ventricles did not differ significantly between any of the genotypic combinations tested for, meaning that most likely neither the hippocampal formation nor any of the structures lining the lateral ventricles differ in their size. Volumes of the two hemispheres were similar among all genotypic groups tested, negating a global trophic effect of BDNF alleles. However, the tools used in the study were not designed to detect any partial volumetric asymmetries and thus, any such, may have been diluted in the overall sample. Further-more, differences in the imaging study methodology may also account for the variability in the results published in the literature, including our study.
In regards to ApoE polymorphisms and its effects on volumetric changes, it is known that the entorhinal cortex as well as the medial temporal and orbitofrontal lobe (49) are among the regions more severely affected in Alzheimer’s disease, probably in part due to their cytoarchitectural homology (50). Furthermore, cognitive and MRI measure changes are evident in ApoE4 carriers long before the onset of dementia (51). Young healthy adults with the E4 allele exhibit altered cerebral activation pattern both at rest and during performance of cognitive tasks (52). Children and adolescents carrying the E4 allele were found in one study to have thinner entorhinal cortex with E2 carriers having the greatest thickness (35). In general, volumetric studies involving the hippocampal formation have yielded rather equivocal results with a trend towards lower volumes in E4 carriers that did not reach always statistical significance (32,36,53–55). Differences in image analysis methods (56), sample size and homogeneity may account for some variability in results. However, plain volumetry may not be a sufficiently sensitive method to detect subtle regional changes and localised volume loss may be washed out in more generalised measurements. It is thus not surprising that ventricular volumes, which are influenced by medial lobe structures, among other periventricular formations, did not differ statistically between E4 and non carriers. Another explanation may be the homogeneity of our sample, which recruited a relatively young population. The E4 status may result either in an endophenotype even at young age, i.e. difficult to detect with standard volumetric methods or may confer a greater susceptibility to age-related volume loss that may be evident at an older age than the one tested in our study.
To our knowledge, this is the first study to investigate the effects of the Val66Met BDNF polymorphism and ApoE genotype on whole brain and lateral ventricular volume in a large, homogeneous sample of young and healthy adults. Especially, lateral ventricular volumetry is employed for the first time in a young and nondemented population. Our study did not detect any differences among Met and non carriers, as well as among E4 and non carriers, in any of the volumetric parameters, despite the use of a large, highly homogeneous sample. However, it should also be viewed in light of its limitations. The lack of association between the Val66Met polymorphism as well as the E4 genotype speaks against any direct or significant global trophic effect. Similarly, lateral ventricular volumes did not differ between the study groups negating any significant difference in periventricular structures, primarily medial temporal lobe and hippocampus, contrary to some previous studies that had detected a volumetric difference in the hippocampal formation. However, no study has thus far evaluated the lateral ventricular volumes in young subjects and our results are difficult to generalise. Furthermore, the study was not designed to measure regional or lobar volumes. The lack of a standardised volumetric methodology may confer some variability as well. Moreover, interactions between genes, such as the RE1 silencing transcription factor gene, which decreases BDNF secretion (57), may modify the final effect on hemispheric and lateral ventricular volumes.
In conclusion, this study did not detect any trophic influence of either the BDNF Val66Met polymorphism or the E4 allele on hemispheric or lateral ventricular volumes, using a robust and highly accurate method. To our knowledge, this is the first study to evaluate the above structural parameters in young adults, whereas only a few have focused on children and adolescents and most on a wider age range. It is also the first to study lateral ventricular volume in a nondemented population. It should be noted that, due to great variability in the inclusion criteria and volumetric parameters, there are significant controversies in the current literature. The lack of an association between genetic status and the volumes measured in our study is not surprising. It is likely that multiple gene interplay, known and unknown, confers an individual trophic effect that is not evident with gross volumetry and may be more regional and even restricted to specific cortical layers. Nevertheless, the field of structural neuroimaging and molecular psychiatry remains a fascinating and promising one in our attempt to unravel the mysteries of brain development and function.
Acknowledgments
The authors would like to thank Dr M. Doelken (Department of Neuroradiology, Friedrich-Alexander Universität Erlangen-Nürnberg, Germany) for reviewing magnetic resonance scans. Dr Mitsias was supported by NINDS grant R01 NS 070922.
References
- 1.Hofer M, Pagliusi SR, Hohn A, Leibrock J, Barde YA. Regional distribution of brain-derived neurotrophic factor mRNA in the adult mouse brain. EMBO J. 1990;9:2459–2464. doi: 10.1002/j.1460-2075.1990.tb07423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baquet ZC, Bickford PC, Jones KR. Brain-derived neurotrophic factor is required for the establishment of the proper number of dopaminergic neurons in the substantia nigra pars compacta. J Neurosci. 2005;25:6251–6259. doi: 10.1523/JNEUROSCI.4601-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorski JA, Zeiler SR, Tamowski S, Jones KR. Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J Neurosci. 2003;23:6856–6865. doi: 10.1523/JNEUROSCI.23-17-06856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morse JK, Wiegand SJ, Anderson K, et al. Brain-derived neurotrophic factor (BDNF) prevents the degeneration of medial septal cholinergic neurons following fimbria transection. J Neurosci. 1993;13:4146–4156. doi: 10.1523/JNEUROSCI.13-10-04146.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipsky RH, Marini AM. Brain-derived neurotrophic factor in neuronal survival and behavior-related plasticity. Ann N Y Acad Sci. 2007;1122:130–143. doi: 10.1196/annals.1403.009. [DOI] [PubMed] [Google Scholar]
- 6.Tian F, Marini AM, Lipsky RH. NMDA receptor activation induces differential epigenetic modification of BDNF promoters in hippocampal neurons. Amino Acids. 2009 doi: 10.1007/s00726-009-0315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24:4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egan MF, Kojima M, Callicott JH, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 9.Sen S, Nesse RM, Stoltenberg SF, et al. A BDNF coding variant is associated with the NEO personality inventory domain neuroticism, a risk factor for depression. Neuropsy-chopharmacology. 2003;28:397–401. doi: 10.1038/sj.npp.1300053. [DOI] [PubMed] [Google Scholar]
- 10.Sklar P, Gabriel SB, McInnis MG, et al. Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. Brain-derived neutrophic factor. Mol Psychiatry. 2002;7:579–593. doi: 10.1038/sj.mp.4001058. [DOI] [PubMed] [Google Scholar]
- 11.Pezawas L, Verchinski BA, Mattay VS, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neves-Pereira M, Mundo E, Muglia P, King N, Macciardi F, Kennedy JL. The brain-derived neurotrophic factor gene confers susceptibility to bipolar disorder: evidence from a family-based association study. Am J Hum Genet. 2002;71:651–655. doi: 10.1086/342288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agartz I, Sedvall GC, Terenius L, Kulle B, Frigessi A, Hall H, Jonsson EG. BDNF gene variants and brain morphology in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:513–523. doi: 10.1002/ajmg.b.30338. [DOI] [PubMed] [Google Scholar]
- 15.Bueller JA, Aftab M, Sen S, Gomez-Hassan D, Burmeister M, Zubieta JK. BDNF Val66Met allele is associated with reduced hippocampal volume in healthy subjects. Biol Psychiatry. 2006;59:812–815. doi: 10.1016/j.biopsych.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 16.Szeszko PR, Lipsky R, Mentschel C, et al. Brain-derived neurotrophic factor val66met polymorphism and volume of the hippocampal formation. Mol Psychiatry. 2005;10:631–636. doi: 10.1038/sj.mp.4001656. [DOI] [PubMed] [Google Scholar]
- 17.Frodl T, Schule C, Schmitt G, et al. Association of the brain-derived neurotrophic factor Val66Met polymorphism with reduced hippocampal volumes in major depression. Arch Gen Psychiatry. 2007;64:410–416. doi: 10.1001/archpsyc.64.4.410. [DOI] [PubMed] [Google Scholar]
- 18.Nemoto K, Ohnishi T, Mori T, Moriguchi Y, Hashimoto R, Asada T, Kunugi H. The Val66Met polymorphism of the brain-derived neurotrophic factor gene affects age-related brain morphology. Neurosci Lett. 2006;397:25–29. doi: 10.1016/j.neulet.2005.11.067. [DOI] [PubMed] [Google Scholar]
- 19.Toro R, Chupin M, Garnero L, et al. Brain volumes and Val66Met polymorphism of the BDNF gene: local or global effects? Brain Struct Funct. 2009;213:501–509. doi: 10.1007/s00429-009-0203-y. [DOI] [PubMed] [Google Scholar]
- 20.Gerdes LU, Gerdes C, Hansen PS, Klausen IC, Faergeman O, Dyerberg J. The apolipoprotein E polymorphism in Greenland Inuit in its global perspective. Hum Genet. 1996;98:546–550. doi: 10.1007/s004390050257. [DOI] [PubMed] [Google Scholar]
- 21.Gerdes LU, Klausen IC, Sihm I, Faergeman O. Apolipoprotein E polymorphism in a Danish population compared to findings in 45 other study populations around the world. Genet Epidemiol. 1992;9:155–167. doi: 10.1002/gepi.1370090302. [DOI] [PubMed] [Google Scholar]
- 22.Corbo RM, Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a ‘thrifty’ allele? Ann Hum Genet. 1999;63:301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- 23.Beffert U, Nematollah Farsian F, Masiulis I, Hammer RE, Yoon SO, Giehl KM, Herz J. ApoE receptor 2 controls neuronal survival in the adult brain. Curr Biol. 2006;16:2446–2452. doi: 10.1016/j.cub.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 24.Beffert U, Stolt PC, Herz J. Functions of lipoprotein receptors in neurons. J Lipid Res. 2004;45:403–409. doi: 10.1194/jlr.R300017-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Dong LM, Weisgraber KH. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J Biol Chem. 1996;271:19053–19057. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- 26.Morrow JA, Hatters DM, LU B, Hochtl P, Oberg KA, Rupp B, Weisgraber KH. Apolipoprotein E4 forms a molten globule. A potential basis for its association with disease. J Biol Chem. 2002;277:50380–50385. doi: 10.1074/jbc.M204898200. [DOI] [PubMed] [Google Scholar]
- 27.Jiang Q, Lee CY, Mandrekar S, et al. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58:681–693. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burggren AC, Zeineh MM, Ekstrom AD, Braskie MN, Thompson PM, Small GW, Bookheimer SY. Reduced cortical thickness in hippocampal subregions among cognitively normal apolipoprotein E e4 carriers. Neuroimage. 2008;41:1177–1183. doi: 10.1016/j.neuroimage.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jak AJ, Houston WS, Nagel BJ, Corey-Bloom J, Bondi MW. Differential cross-sectional and longitudinal impact of APOE genotype on hippocampal volumes in nonde-mented older adults. Dement Geriatr Cogn Disord. 2007;23:382–389. doi: 10.1159/000101340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lind J, Larsson A, Persson J, et al. Reduced hippocampal volume in non-demented carriers of the apolipoprotein E epsilon4: relation to chronological age and recognition memory. Neurosci Lett. 2006;396:23–27. doi: 10.1016/j.neulet.2005.11.070. [DOI] [PubMed] [Google Scholar]
- 31.Soininen H, Partanen K, Pitkanen A, et al. Decreased hippocampal volume asymmetry on MRIs in nondemented elderly subjects carrying the apolipoprotein E epsilon 4 allele. Neurology. 1995;45:391–392. doi: 10.1212/wnl.45.2.391. [DOI] [PubMed] [Google Scholar]
- 32.Tohgi H, Takahashi S, Kato E, et al. Reduced size of right hippocampus in 39- to 80-year-old normal subjects carrying the apolipoprotein E epsilon4 allele. Neurosci Lett. 1997;236:21–24. doi: 10.1016/s0304-3940(97)00743-x. [DOI] [PubMed] [Google Scholar]
- 33.Wishart HA, Saykin AJ, McAllister TW, et al. Regional brain atrophy in cognitively intact adults with a single APOE epsilon4 allele. Neurology. 2006;67:1221–1224. doi: 10.1212/01.wnl.0000238079.00472.3a. [DOI] [PubMed] [Google Scholar]
- 34.Espeseth T, Westlye LT, Fjell AM, Walhovd KB, Rootwelt H, Reinvang I. Accelerated age-related cortical thinning in healthy carriers of apolipoprotein E epsilon 4. Neurobiol Aging. 2008;29:329–340. doi: 10.1016/j.neurobiolaging.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 35.Shaw P, Lerch JP, Pruessner JC, et al. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol. 2007;6:494–500. doi: 10.1016/S1474-4422(07)70106-0. [DOI] [PubMed] [Google Scholar]
- 36.Mondadori CR, de Quervain DJ, Buchmann A, et al. Better memory and neural efficiency in young apolipoprotein E epsilon4 carriers. Cereb Cortex. 2007;17:1934–1947. doi: 10.1093/cercor/bhl103. [DOI] [PubMed] [Google Scholar]
- 37.Bradley KM, Bydder GM, Budge MM, Hajnal JV, White SJ, Ripley BD, Smith AD. Serial brain MRI at 3–6 month intervals as a surrogate marker for Alzheimer’s disease. Br J Radiol. 2002;75:506–513. doi: 10.1259/bjr.75.894.750506. [DOI] [PubMed] [Google Scholar]
- 38.Carmichael OT, Kuller LH, Lopez OL, et al. Ventricular volume and dementia progression in the Cardiovascular Health Study. Neurobiol Aging. 2007;28:389–397. doi: 10.1016/j.neurobiolaging.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jack CR, Jr, Shiung MM, Gunter JL, et al. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson PM, Hayashi KM, De Zubicaray GI, et al. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 41.Wang D, Chalk JB, Rose SE, et al. MR image-based measurement of rates of change in volumes of brain structures. Part II: application to a study of Alzheimer’s disease and normal aging. Magn Reson Imaging. 2002;20:41–48. doi: 10.1016/s0730-725x(02)00472-1. [DOI] [PubMed] [Google Scholar]
- 42.Ferrarini L, Palm WM, Olofsen H, van Buchem MA, Reiber JH, Admiraal-Behloul F. Shape differences of the brain ventricles in Alzheimer’s disease. Neuroimage. 2006;32:1060–1069. doi: 10.1016/j.neuroimage.2006.05.048. [DOI] [PubMed] [Google Scholar]
- 43.Giesel FL, Hahn HK, Thomann PA, et al. Temporal horn index and volume of medial temporal lobe atrophy using a new semiautomated method for rapid and precise assessment. AJNR Am J Neuroradiol. 2006;27:1454–1458. [PMC free article] [PubMed] [Google Scholar]
- 44.Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 45.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 47.Rorden C, Brett M. Stereotaxic display of brain lesions. Behav Neurol. 2000;12:191–200. doi: 10.1155/2000/421719. [DOI] [PubMed] [Google Scholar]
- 48.Richter-Schmidinger T, Alexopoulos P, Horn M, Maus S, et al. Influence of Brain-derived neurotrophic factor and Apolipoprotein E genetic variants on hippocampal volume and memory performance in healthy young adults. J Neural Transm. 2011;118:249–257. doi: 10.1007/s00702-010-0539-8. [DOI] [PubMed] [Google Scholar]
- 49.Van Hoesen GW, Parvizi J, Chu CC. Orbitofrontal cortex pathology in Alzheimer’s disease. Cereb Cortex. 2000;10:243–251. doi: 10.1093/cercor/10.3.243. [DOI] [PubMed] [Google Scholar]
- 50.Ongur D, Ferry AT, Price JL. Architectonic subdivision of the human orbital and medial prefrontal cortex. J Comp Neurol. 2003;460:425–449. doi: 10.1002/cne.10609. [DOI] [PubMed] [Google Scholar]
- 51.Debette S, Wolf PA, Beiser A, et al. Association of parental dementia with cognitive and brain MRI measures in middle-aged adults. Neurology. 2009;73:2071–2078. doi: 10.1212/WNL.0b013e3181c67833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scarmeas N, Habeck CG, Hilton J, Anderson KE, Flynn J, Park A, Stern Y. APOE related alterations in cerebral activation even at college age. J Neurol Neurosurg Psychiatry. 2005;76:1440–1444. doi: 10.1136/jnnp.2004.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jack CR, Jr, Petersen RC, XU YC, et al. Hippocam-pal atrophy and apolipoprotein E genotype are independently associated with Alzheimer’s disease. Ann Neurol. 1998;43:303–310. doi: 10.1002/ana.410430307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reiman EM, Uecker A, Caselli RJ, et al. Hippocampal volumes in cognitively normal persons at genetic risk for Alzheimer’s disease. Ann Neurol. 1998;44:288–291. doi: 10.1002/ana.410440226. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt H, Schmidt R, Fazekas F, Semmler J, Kapeller P, Reinhart B, Kostner GM. Apolipoprotein E e4 allele in the normal elderly: neuropsychologic and brain MRI correlates. Clin Genet. 1996;50:293–299. doi: 10.1111/j.1399-0004.1996.tb02377.x. [DOI] [PubMed] [Google Scholar]
- 56.Geuze E, Vermetten E, Bremner JD. MR-based in vivo hippocampal volumetrics: 1. Review of methodologies currently employed. Mol Psychiatry. 2005;10:147–159. doi: 10.1038/sj.mp.4001580. [DOI] [PubMed] [Google Scholar]
- 57.Miyajima F, Quinn JP, Horan M, Pickles A, Ollier WE, Pendleton N, Payton A. Additive effect of BDNF and REST polymorphisms is associated with improved general cognitive ability. Genes Brain Behav. 2008;7:714–719. doi: 10.1111/j.1601-183X.2008.00409.x. [DOI] [PubMed] [Google Scholar]


