Abstract
Adjuvant tamoxifen therapy of breast cancer patients with estrogen receptor-positive tumors reduces the rate of breast cancer recurrence by approximately a half. Tamoxifen is metabolized by several polymorphic enzymes, including cytochrome P450 2D6 (CYP2D6), to more active metabolites. We have reviewed the clinical pharmacology of tamoxifen and evaluated the evidence from clinical epidemiology studies regarding the association between CYP2D6 inhibition and tamoxifen effectiveness. We conclude that the impact of CYP2D6 inhibition on tamoxifen effectiveness is likely to be null or small, at least in the populations studied so far. Understanding the effect of variations in tamoxifen metabolism on breast cancer outcomes, if any, will likely require a broader perspective, including examination of the complete metabolic pathway and subgroups of patients with other markers of potentially poor tamoxifen response.
Keywords: breast cancer, breast cancer recurrence, cytochrome P450 2D6, selective serotonin reuptake inhibitors, tamoxifen
Tamoxifen, a selective estrogen receptor (ER) modulator, was a trailblazer for personalized cancer therapy. In stage I, II and III breast cancer patients with tumors that express the estrogen receptor (ER+) – approximately two out of three patients – tamoxifen reduces, by half, the rate of breast cancer recurrence and the risk of mortality by a quarter [1] . Current editions of the major treatment guidelines all reach the same recommendations [2,3,201] . ER+ premenopausal patients should receive tamoxifen for 5 years; aromatase inhibitors (AIs) are contraindicated outside of clinical trials. ER+ postmenopausal patients should receive AI either as initial therapy or in sequence with tamoxifen. Postmenopausal women with contraindications to AI, or who decline AI, should receive 5 years of tamoxifen therapy. Thus, tamoxifen remains fundamental in adjuvant breast cancer therapy.
Despite its impressive therapeutic effects, and more than 30 years history as a cancer therapy, recurrent disease refractive to endocrine therapy develops in some tamoxifen-treated women and they succumb to their cancer. Women with seemingly identical clinical and prognostic factors at breast cancer diagnosis, and who are treated with the same tamoxifen regimen, can have very different outcomes.
The era of personalized medicine is rapidly evolving [4], and developments related to tamoxifen treatment of breast cancer patients are no exception. A longstanding research field has focused on identifying predictive markers of tamoxifen resistance [5–7] . Most recently, modification of tamoxifen’s effectiveness by functional variants in the enzymes that activate and deactivate the parent drug, or by inhibition of these enzymes by other prescription drugs, has received a lot of attention in the evolution of tamoxifen personalization [8,9].
Cytochrome P450 (CYP) enzymes (CYP2D6, CYP2C19, CYP2C9, CYP3A4 and CYP3A5) metabolize tamoxifen to more active forms [10] , each with its own binding affinity to the ER. The compounds that most efficiently bind to the ER are hydroxylated at tamoxifen’s four-carbon [11]. The strength of ER affinity predicts the anti-tumor response [12] . The major enzymes involved in solubilizing tamoxifen metabolites into excretable forms are uridine 5′-diphospho-glucuronosyltransferases (UGTs; primarily UGT1A8, UGT1A10, UGT2B7 and UGT2B15) [13] and sulfotransferases (primarily SULT1A1) [14]. All of the enzymes in this pathway are polymorphic, and the changes in function related to genotypes contribute to interindividual differences in tamoxifen metabolite concentrations in the serum [14–16].
Despite the complexity of this pathway, most clinical epidemiology studies examining the associations between gene variants and breast cancer outcomes have focused on only one player in this intricate pathway, namely CYP2D6. The current article therefore has two objectives. First, we will evaluate the evidence to date regarding the association between CYP2D6 inhibition and tamoxifen effectiveness, by reviewing the clinical pharmacology of tamoxifen and meta-analyzing the clinical epidemiologic studies. Second, we suggest that understanding the effect of variations in tamoxifen metabolism on breast cancer outcomes, if any, will require a broader perspective than that taken so far.
Methods
Search strategy & selection criteria
For our review of the association between CYP2D6 inhibition and tamoxifen effectiveness, we searched for the terms ‘tamoxifen’ and ‘CYP2D6’ in PubMed. No language restrictions were imposed. All papers published or presented as abstracts through 21 March 2011 regarding the association between CYP2D6 gene variants or drug–drug interactions and the risk of breast cancer recurrence or mortality were reviewed to determine whether their results should be included. Citations included within the selected scientific papers or other reference sources were also used to locate other articles, for example, conference abstracts.
Meta-analyses
We created four separate meta-analytical models. The first two focused on population-based studies associating concurrent use of weak or strong CYP2D6 inhibitors (selective serotonin reuptake inhibitors [SSRIs]) and breast cancer recurrence or breast cancer-specific mortality in tamoxifen-treated women. The third and fourth models focused on population-based studies associating CYP2D6 inherited mutations and breast cancer recurrence or breast cancer-specific mortality in tamoxifen-treated women. The first of these compared recurrence risks of homozygote and heterozygote carriers of decreased-function alleles with homozygote carriers of the corresponding full-function allele, and the second compared recurrence risks of homozygote carriers of decreased-function alleles with homozygote or heterozygote (only Xu et al. [17]) carriers of the corresponding full-function allele. For these analyses, we searched all scientific papers or conference abstracts to obtain study-specific effect estimates for the association of inheritance of at least one variant allele of CYP2D6 (either *4 or *10 ) with breast cancer recurrence or mortality. When studies presented associations for heterozygote and homozygote variant alleles separately, we estimated an inverse variance weighted average of these two associations, which was then used in the first of the genetic meta-analytic models.
Statistical analysis
We used random-effects meta-analytic models to generate summary effect estimates. In all cases, estimates from fixed-effects models were similar. We constructed funnel plots to evaluate publication and other sources of bias in the meta-analyses. These plots, which are available from the authors, showed no evidence of publication bias. All analyses were performed using STATA software, version 11.0 (StataCorp LP, College Station, TX, USA). All statistical tests were two-sided.
Results
Pharmacological evidence: CYP2D6 inhibition & the profile of tamoxifen metabolite concentrations
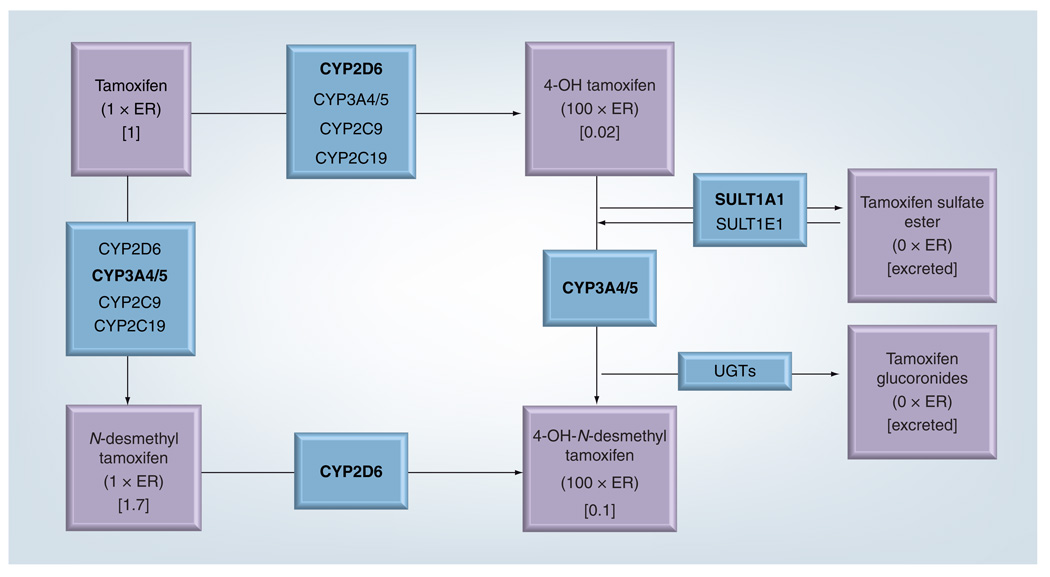
Tamoxifen is metabolized mostly in the liver, where it primarily undergoes 4-hydroxylation [18,19] and N-demethylation reactions (FIGURE 1) [20]. CYP enzymes metabolize tamoxifen and N-desmethyl-tamoxifen to 4-hydroxytamoxifen and 4-hydroxy-N-desmethyl tamoxifen (sometimes called endoxifen), respectively [21] . As noted previously, these metabolites bind the ER approximately 100-fold more readily than their respective parent molecules, so they are important modulators of the ER pathway [11]. Since 4-hydroxy-N-desmethyl-tamoxifen is present at a threefold to fivefold higher concentration in the serum than 4-hydroxytamoxifen, it is a key tamoxifen metabolite.
Figure 1. Major metabolic pathways for tamoxifen.
Bold type denotes the enzyme(s) primarily involved in each step.
C: Plasma concentration of the metabolite, relative to tamoxifen’s concentration, after 4 months of tamoxifen therapy at 20 mg/day; CYP: Cytochrome P450; N × ER: Binding affinity to estrogen receptor relative to tamoxifen itself; UGT: Uridine 5′-diphospho-glucuronosyltransferases.
One phenotypic categorization of a person’s capacity to metabolize tamoxifen depends on the ratio of the steady-state concentration of the administered drug, or its N-demethylated metabolite, to the steady-state concentration of its 4-hydroxylated metabolites [14,22] . Depending on this ratio, patients can be classified according to their ability to metabolize tamoxifen as ‘poor metabolizers’, ‘intermediate metabolizers’, ‘extensive metabolizers’ or ‘ultra-rapid metabolizers’.
As depicted in FIGURE 1, CYP2D6 catalyzes activation of N-desmethyltamoxifen to 4-hydroxy-N-desmethyltamoxifen [23] and accounts for approximately 45% of the activation of tamoxifen to 4-hydroxytamoxifen [24] . More than 90 polymorphic variants of CYP2D6 have been identified, some of which reduce or eliminate CYP2D6 activity [202] and therefore affect the ability to metabolize tamoxifen [14,15,23,24]. As a second approach to phenotypic categorization, ‘poor metabolizers’ can be categorized as those with two nonfunctional CYP2D6 alleles, ‘intermediate metabolizers’ as those with one functional allele, ‘extensive metabolizers’ as those with two normal alleles, and ‘ultra-rapid metabolizers’ as those with multiple allelic copies of functional alleles and thus excess metabolic capacity. In vivo evidence supports correlated categorization of phenotype by these two approaches [22] . Further complicating the phenotypic categorization is the potential for drug– drug interactions to affect the concentration profile of tamoxifen metabolites [15,23,25]. While the affect of this drug–drug interaction would be apparent using the first strategy for phenotypic characterization, it would not be incorporated into the second strategy, which relies only on genotype to infer phenotype.
Regardless of the strategy used to categorize phenotype, it is clear that interindividual differences in the serum concentration of tamoxifen metabolites – either due to inhibition of the enzyme active site by CYP2D6-inhibiting drugs or to the inheritance of variant alleles in the genes coding for the metabolizing enzymes – could modulate the effectiveness of tamoxifen treatment. While the biologic rationale for this idea seems compelling, it is counterbalanced by further consideration of the mechanism of tamoxifen’s action. As mentioned at the outset, tamoxifen is a selective ER modulator – it acts in concert with its metabolites by competing with estrogen for binding to the tumor’s ERs. As noted by Jordan almost 30 years ago:
“…the metabolic activation of tamoxifen is an advantage rather than a requirement for anti-estrogenic activity. The action of tamoxifen in vivo is the net result of the individual actions of the parent compound and its metabolites competing for the occupation of receptors within target tissues and tumors” [26].
When administered at the standard dose of 20 mg/day, tamoxifen and its metabolites overwhelm estrogen in this competition, thereby occupying almost all of the available receptor binding sites and depriving the tumor of growth stimulation mediated by the estrogen–ER complex. The ER-binding activity of the drug and its metabolites on average outweighs the activity of estradiol by more than 5000 to one in post-menopausal women and by more than 500 to one in premenopausal women [8]. A fewfold reduction in the concentration of one of the active metabolites would be expected to have little effect on the competition.
Tamoxifen effectiveness in the presence of a potential drug–drug interaction: the example of SSRIs inhibiting CYP2D6 function
Although tamoxifen is generally well tolerated, its anti-estrogenic and estrogenic actions sometimes produce mild-to-severe side effects, including hot flashes and vasomotor symptoms, induction or exacerbation of depression (sometimes also a pre-existing condition or a consequence of breast cancer diagnosis), venous thromboembolism and endometrial cancer [27–34]. SSRIs can provide effective clinical control of the depressive and vasomotor side effects and are therefore sometimes prescribed to women undergoing tamoxifen treatment. Both SSRIs and tamoxifen are primarily metabolized by CYP2D6 [21,35,36]. The net result can be competitive inhibition or direct inhibition of tamoxifen metabolism, resulting in a reduced plasma concentration of 4-hydroxy-N-desmethyltamoxifen [15,23] . Therefore, the safety of concurrent use of the two medications has come under scrutiny [37,38].
Selective serotonin reuptake inhibitors inhibit CYP2D6 to varying degrees [36]. Paroxetine and fluoxetine are the strongest inhibitors, with paroxetine irreversibly inhibiting CYP2D6 activity, whereas fluvoxamine and citalopram are weaker inhibitors. Reduced plasma concentrations of 4-hydroxy-N-desmethyltamoxifen have been reported in women who used paroxetine or fluoxetine concomitantly with tamoxifen, intermediate concentrations among women treated with the weaker CYP2D6 inhibitors sertraline and citalopram, and little effect among those using the selective serotonin norepinephrine reuptake inhibitor (SSNRI) venlafaxine – a weak CYP2D6 inhibitor [15,23,39,40].
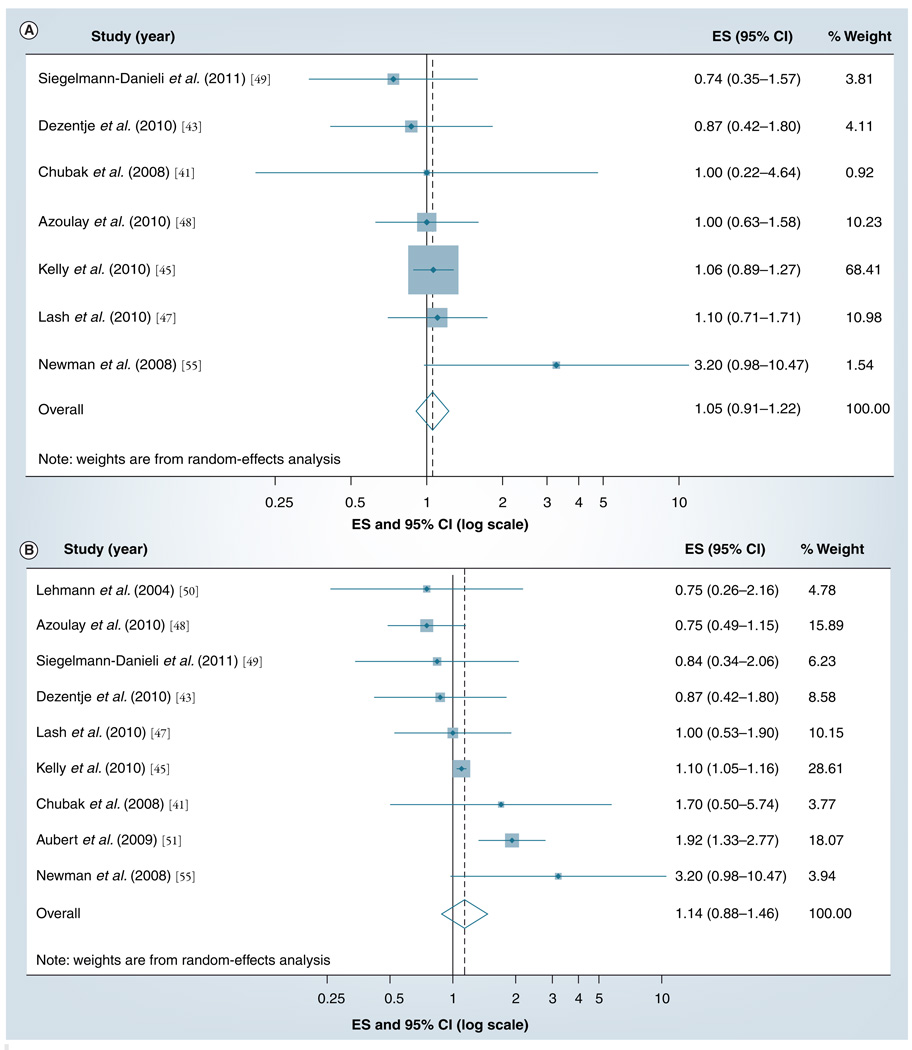
A total of 11 clinical epidemiology studies have investigated the association between taking SSRI medications and breast cancer outcomes among tamoxifen-treated breast cancer patients [41–51] , with fairly heterogeneous results. Three of these studies have overlapping patients and follow-up time [42,46,47], therefore, we included only the most relevant report in the meta-analyses [47]. FIGURE 2A & B show the results of our meta-analyses of the included studies, organized by presupposed strength of CYP2D6 inhibition (Figure 2A shows weak inhibitors, such as citalopram; FIGURE 2B shows strong inhibitors, such as paroxetine). The study with greatest weight was Kelly et al., as indicated by the relative area of its squares on the graphs [45] . The summary random-effects estimate associating breast cancer recurrence with concomitant use of tamoxifen and a weak CYP2D6 inhibitor was 1.05 (95% CI: 0.91–1.22). The summary random-effects estimate associating breast cancer recurrence with concomitant use of tamoxifen and a strong CYP2D6 inhibitor was 1.14 (95% CI: 0.88–1.46). These summary estimates indicate that the overall effect of concurrent use of SSRIs and tamoxifen on breast cancer recurrence is null or small.
Figure 2. The summary effect size and 95% confidence intervals for the association between concurrent use of (A) a weak CYP2D6 inhibitor drug and (B) a strong CYP2D6 inhibitor drug and breast cancer recurrence/survival.
Summary ES and 95% confidence intervals were estimated using a fixed-effects meta-analytical model. All statistical tests were two-sided. The size of each square is an illustrative representation of the study weight. The horizontal lines represent the CIs. The diamond represents the summary ES and 95% CIs.
CI: Confidence interval; ES: Effect size.
Tamoxifen effectiveness in the presence of a potential gene–drug interaction: the example of genetic variants inhibiting CYP2D6 function
As noted previously, and depicted in FIGURE 1, the metabolic pathway of tamoxifen involves a large set of enzymes. The enzymes that participate in 4-hydroxylation are most likely to influence the concentration profile of tamoxifen’s metabolites. Because CYP2D6 is almost entirely responsible for 4-hydroxylation of the most abundant metabolite (N-desmethyltamoxifen), variants that reduce or eliminate CYP2D6 function may have the greatest effect on the profile of metabolite concentrations. Reduced plasma concentrations of 4-hydroxylated metabolites have been reported in women who carry two reduced-function alleles, and intermediate concentrations have been reported among women who carry one reduced-function allele [14,15,17].
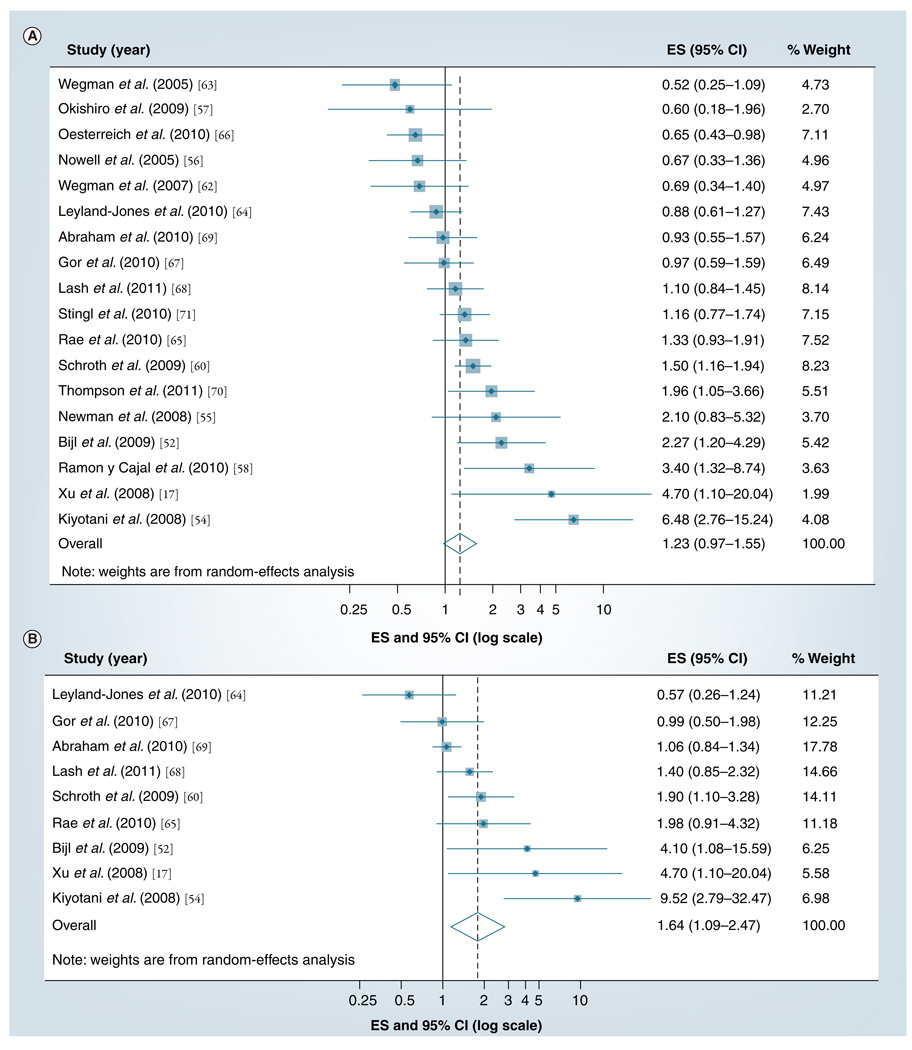
To date, 24 studies have investigated the association between CYP2D6 inhibition by genetic variants and the effectiveness of adjuvant tamoxifen treatment [17,44,52–73] . Two of these studies [53,72] are updates of earlier reports [44,54,59], with overlapping patients and follow-up time, so we only included the updated reports in the meta-analyses. We were unable to extract estimates of association from two studies [61,73] , both of which characterized their results as null. As previously reviewed by our group [8] , the results of these studies are widely heterogeneous (p <0.001, for test of homogeneity) and almost evenly distributed either side of the null. The precision of many of the studies was very poor, as portrayed by the width of the confidence intervals in FIGURE 3A & B. Taken together, the heterogeneity of results and symmetric dispersion about the null should caution against any strong conclusion regarding a causal association.
Figure 3. The summary effect size and 95% confidence intervals for the association between (A) inheritance of any and (B) inheritance of two nonfunctional variants of CYP2D6*4 or CYP2D6*10 and breast cancer recurrence.
Summary ES and 95% CIs were estimated using random-effects meta-analytical models. All statistical tests were two-sided. The size of each square is an illustrative representation of the study weight. The horizontal lines represent the CIs. The diamond represents the summary ES and 95% CIs.
CI: Confidence interval; ES: Effect size.
Our quantitative meta-analysis of these studies first compared the risk of breast cancer recurrence or mortality with the inheritance of at least one nonfunctional CYP2D6*4 or CYP2D6*10 allele. The results are presented in FIGURE 3A. The effect estimates reported in the studies range from 0.52 to 6.48, with eight reporting associations below the null and ten reporting associations above the null, and 11 out of the 18 studies including the null in their 95% confidence intervals. The summary random-effects estimate associating breast cancer recurrence with variant CYP2D6 genotype was 1.23 (95% CI: 0.97–1.55). The studies with the greatest weight, as indicated by the relative areas of the squares in the graph, are those with associations nearest to the null. An important exception is the updated analysis [72] of the Goetz [44] and Schroth [59] cohorts. It appears, however, that the association may have been null in the newly accumulated person-time included in the second analysis [74].
Our next quantitative meta-analysis compared the risk of breast cancer recurrence or mortality with the inheritance of two nonfunctional CYP2D6*4 or CYP2D6*10 alleles. The results are presented in FIGURE 3B. The effect estimates reported in the studies range from 0.57 to 9.52, with two reporting associations below the null and seven reporting associations above the null, and five out of the nine studies including the null in their 95% confidence intervals. The summary random-effects estimate associating breast cancer recurrence with variant CYP2D6 genotype was 1.64 (95% CI: 1.09–2.47). This result suggests a non-null association between CYP2D6 inhibition among carriers of two reduced-function alleles and recurrence risk. We note, however, that the three studies with the strongest associations have important design flaws [8]. The two studies with the strongest associations were studies of CYP2D6*10 [17,53], which is a reduced-function allele, but does not eliminate CYP2D6 function. If CYP2D6 inhibition increases the risk of recurrence among tamoxifen-treated patients with two reduced-function alleles, then one would expect the strongest associations to be observed among carriers of two CYP2D6*4 alleles, because that variant eliminates CYP2D6 function.
Discussion
Review of the evidence regarding CYP2D6 inhibition
In both the overview of trial results comparing approximately 5 years of tamoxifen against placebo [1], and in the Arimidex, Tamoxifen, Alone or in Combination trial results comparing approximately 5 years of aromatase inhibitor against tamoxifen [75,76], the 5-year risk of breast cancer recurrence among tamoxifen-treated patients (RT) was approximately 15%. This comparability suggests that the trial populations are approximately exchangeable [77]. If we assume that the recurrence risk in any subgroup of tamoxifen-treated patients (Ri) cannot be greater than the recurrence risk in placebo-treated patients (RP) or less than the recurrence risk in aromatase-treated patients (RAI), then the comparison of any two subgroups (R1/R2) cannot be less than the ratio (RAI/RP; the effect of AI vs placebo) or greater than the ratio (RP/RAI; the effect of placebo vs AI). From their respective analyses at 5 years of follow-up, RAI/RT = 0.79 [75] and RT/RP = 0.59 [1]. We can therefore write the equation as seen in Box 1.
Box 1. Estimated limits on the ratio of recurrence risks between any two tamoxifen-treated subgroups, assuming only a predictive effect of the marker used to create the subgroups.
Any ratio of risks, rates or hazards in tamoxifen-treated subgroups outside these limits suggests an implausible point estimate or requires that the biomarker used to categorize the subgroups has both predictive and prognostic value. To date, no one has postulated a direct effect of CYP2D6 inhibition on the risk of breast cancer recurrence. The only effect, if any, is thought to be mediated through modulation of the profile of metabolite concentrations. CYP2D6 inhibition is therefore hypothesized to predict response to tamoxifen therapy, but not to have any prognostic value in itself. Valid estimates of the relative risk of recurrence in the tamoxifen-treated subgroups created by categorization of CYP2D6 inhibition should be expected, therefore, to fall into the range 0.47–2.15. Of the studies included in our meta-analyses, six out of seven studies (depicted in FIGURE 2A), eight out of nine studies (depicted in FIGURE 2B), 12 out of 17 studies (depicted in FIGURE 3A) and six out of nine studies (depicted in FIGURE 3B) yielded point estimates that fell into the expected range. Confidence intervals of all studies overlapped the range. While some of the incongruity between the results and the expected strength of estimated associations may be explained by chance, the heterogeneity of results and deviation from expectation merits further consideration.
An initial consideration is the potential for the potency of CYP2D6 inhibition to vary from study to study. For example, differences in physicians’ preferences for prescribing specific antidepressants may vary geographically. Citalopram and escitalopram were the most frequent SSRI prescriptions in the Danish study [47], paroxetine and fluoxetine predominated in a North American study [41], and moderate/strong and weak CYP2D6 inhibitors were prescribed in approximately equal proportion in a UK study [48]. Citalopram inhibits CYP2D6 less than most other SSRIs, but despite the variability in prescribing patterns, all three studies yielded near-null results. Similarly, CYP2D6*4 knocks out CYP2D6 function and is the predominant functional variant in Caucasians, whereas CYP2D6*10 reduces CYP2D6 function and is the predominant functional variant in Asians [78]. Studies in both Caucasian and Asian populations have yielded both protective (e.g., [63] and [57]) and causal (e.g., [72] and [53]) associations between CYP2D6 inhibition and breast cancer recurrence. While variation in the population distribution of the potency of inhibition of CYP2D6 variants may contribute to variation in study results, this explanation would only hold if there is a true non-null association. Therefore, the pattern of clinical epidemiology results does not follow the pattern expected if population-dependent variability in potency was important.
As a second consideration, the quality of exposure data used to characterize CYP2D6 inhibition has varied widely. With regard to the studies of drug–drug interactions, sources of medication data include retrospective data from medical record review and prospective data from prescription claims databases. In this context, ‘retrospective’ indicates that data on SSRI prescription use were retrieved after follow-up data on recurrence were recorded, whereas ‘prospective’ indicates that the data on SSRI prescription use were recorded without the knowledge of the subject’s recurrence status. This methodological distinction has important validity implications regarding the potential for differential misclassification bias of the SSRI exposure [79]. Nonetheless, studies of both designs have yielded null (e.g., [50] and [48]) and causal (e.g., [44] and [45]) estimates of the association between SSRI inhibition of CYP2D6 function and breast cancer recurrence.
With regard to the studies of CYP2D6–tamoxifen interaction, sources of genotyped DNA have included blood samples collected at diagnosis, archived tumor specimens and blood samples collected well after diagnosis (e.g., up to 11 years postdiagnosis [54]). When follow-up time begins at diagnosis, although blood samples were only collected well after diagnosis [53,54,58], a study is susceptible to immortal person-time bias. Another important consequence of the source of the DNA is the potential to comprehensively genotype the CYP2D6 gene. DNA extracted from whole blood is of higher quality, allowing the use of more sophisticated and comprehensive genotyping, such as use of the AmpliChip® (a commercially available CYP2D6 comprehensive genotyping tool [80]). DNA extracted from archived tumor or adjacent normal tissue is of lower quality, currently precluding the use of the AmpliChip. Comprehensive genotyping of the CYP2D6 gene is important because CYP2D6*4 is not the only variant that eliminates CYP2D6 function. Other alleles associated with no enzymatic activity include CYP2D6*3 through *8, *11 through *16, *18 through *20, *38, *40, *42 and *44. Similarly, CYP2D6*10 is not the only variant that reduces CYP2D6 function without eliminating it. Other alleles associated with reduced enzyme function include CYP2D6*9, *10, *17, *29, *36, *37 and *41 [39].
The studies included in this article have varied widely in how comprehensively they genotyped CYP2D6 (TABLE 1). A total of six studies genotyped only CYP2D6*4 or only CYP2D6*10; while the others genotyped at least one other functional variant. In general, studies that more comprehensively genotyped the CYP2D6 gene reported higher relative risks of breast cancer recurrence or breast cancer-specific mortality associated with CYP2D6 inhibition. This pattern would be expected if failure to comprehensively genotype CYP2D6 resulted in substantial nondifferential misclassification of the CYP2D6 functional phenotype, a phenomenon reported with empirical evidence in two studies [70,72]. In the Schroth study, approximately a third of the breast cancer patients were misclassified with regard to presumed CYP2D6 function when it was based on only the *4 mutation, compared with when it was based on comprehensive genotyping using the AmpliChip [72]. The relative risk associating breast cancer recurrence with poor CYP2D6 function increased from a nearly null association (1.3; 95% CI: 0.5–3.7) when based on only the *4 mutation to a strong positive association (2.9; 95% CI: 1.4–6.1) when based on more than 30 mutations assayed by the AmpliChip.
Table 1.
List of studies associating CYP2D6 inhibition by genotype with breast cancer recurrence.
| Study (year) | RR | *2 | *3 | *4 | *5 | *6 | *7 | *9 | *10 | *14 | *17 | *18 | *20 | *21 | *35 | *36 | *41 | UM | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wegman et al. (2005) | 0.52 | X | [63] | ||||||||||||||||
| Okishiro et al. (2009) | 0.60 | X | [57] | ||||||||||||||||
| Nowell et al. (2005) | 0.67 | X | X | X | [56] | ||||||||||||||
| Wegman et al. (2007) | 0.69 | X | [62] | ||||||||||||||||
| Leyland-Jones et
al. (2010 ) |
0.88 | X | X | X | X | X | X | X | [64] | ||||||||||
| Gor et al. (2010) | 0.97 | X | [67] | ||||||||||||||||
| Abraham et al. (2010) | 1.01 | X | X | X | X | X | X | X | [69] | ||||||||||
| Lash et al. (2011) | 1.1 0 | X | [68] | ||||||||||||||||
| Stingl et al. (2010) | 1.16 | X | [71] | ||||||||||||||||
| Rae et al. (2010) | 1.33 | X | X | X | X | X | X | X | [65] | ||||||||||
| Goetz et al. (2007) | 1.91 | X | X | Y | Y | [44,60] | |||||||||||||
| Thompson et
al. (2011) |
1.96 | X | X | X | X | X | X | X | X | X | X | X | [70] | ||||||
| Newman et al. (2008) | 2 .1 0 | X | X | X | X | [55] | |||||||||||||
| Schroth et al. (2007) | 2.24 | Y | Y | X | X | Y | Y | Y | X | Y | X | [59, 60] | |||||||
| Bijl et al. (2009) | 2.27 | X | [52] | ||||||||||||||||
| Ramon y Cajal et
al. (2010) |
3.40 | X | X | X | X | X | X | X | X | X | X | X | X | [58] | |||||
| Xu et al. (2008) | 4.70 | X | [17] | ||||||||||||||||
| Kiyotani et al.
(2008, 2010) |
6.65 | X | X | X | X | X | X | Y | X | Y | [53, 54] |
Studies are ranked from lowest RR point estimate to highest, and plotted against the CYP2D6 alleles that were genotyped.
RR: Relative risk; UM: Ultrametabolizer; X: Genotyped; Y: Genotyped in an update of the same or an overlapping cohort.
The large cohort study of Abraham et al. [69], which genotyped the most prevalent CYP2D6 functional alleles and yielded a null result, somewhat counters the results of these two studies. In addition, two studies nested within major adjuvant treatment trials and with broad – but not comprehensive – genotyping of CYP2D6 were presented at the 2010 San Antonio Breast Cancer Symposium (TX, USA). A total of seven CYP2D6 alleles were genotyped in the Arimidex, Tamoxifen, Alone or in Combination trial on 588 patients who received tamoxifen only and 615 patients who received anastrozole [65]. The Breast International Group (BIG 1–98) trial genotyped eight CYP2D6 alleles in almost 5000 postmenopausal hormone-responsive breast cancer patients randomized to either letrozole or tamoxifen [64]. Both trials reported a near-null association between reduced CYP2D6 function and breast cancer recurrence.
In our large population-based case-control study of polymorphisms in the CYP2D6 gene and breast cancer recurrence [68], we implemented a quantitative bias analysis to account for the lack of comprehensive genotyping data (only CYP2D6*4 was genotyped). In this analysis, we assumed that cases of recurrence were more likely to carry alleles with reduced-function than were controls. All of the parameters of the bias model were informed by published external data sources. Consistent with the Abraham study [69] and the studies presented at the 2010 San Antonio Breast Cancer Symposium [64,65], our bias analysis suggests that comprehensive genotyping of CYP2D6 would have had little effect on the near-null results.
A third consideration is the potential for tamoxifen adherence to vary across studies and within categories of CYP2D6 inhibition, which may partially explain the heterogeneity of reported associations. Approximately half of tamoxifen treated patients do not complete the intended duration of their tamoxifen therapy [81]. Failure to complete the intended course is related to recurrence risk [82], especially in conjunction with CYP2D6 genotype [70], and in fact, may be caused by CYP2D6 genotype [83]. If lack of adherence is caused by genotype and in turn causes recurrence, then adherence would be a causal intermediate between CYP2D6 genotype and recurrence. Results adjusted for adherence would be more biased than without adjustment, usually towards the null [84]. Although adjustment for a causal intermediate is a well-known error in epidemiologic research [85], and we have made the argument earlier with specific regard to the association between CYP2D6 inhibition and recurrence [8], reviews continue to erroneously raise failure to control for adherence as a problem in the body of literature on the topic [86].
Evidence for other aspects of tamoxifen metabolism to modify its effectiveness
The polymorphic variants of CYP2D6 have been the most studied enzymes in tamoxifen’s metabolic pathway, probably due to 4-hydroxy-N-desmethyl-tamoxifen binding to the ER with 100-fold higher affinity than tamoxifen and being present in the serum at higher concentrations than 4-hydroxytamoxifen. Nonetheless, as previously noted [8,26], with standard-dose regimens, tamoxifen and its metabolites are present at such abundant concentrations that they overwhelm the receptor. Thus, even among poor metabolizers (CYP2D6*4/*4 and women simultaneously taking paroxetine), tamoxifen and its metabolites should still exert their anti-tumorigenic effects. Future perspectives on this topic might focus on the complete metabolic pathway, which might allow the identifcation of gene–gene interactions that sufficiently affect the profile of tamoxifen metabolites to have a clinical impact. In the only such comprehensive evaluation of the tamoxifen metabolic pathway to date [87], which was conducted in the prevention setting, CYP2D6 variants were not strongly related to breast cancer occurrence. However, when the entire pathway was considered, CYP2D6 was identified as a node that interacted with variant alleles in the other tamoxifen-metabolizing genes. In the following section we briefly review candidate genes and the evidence suggesting that their variants might affect the profile of tamoxifen metabolites.
Cytochrome P450 2C19 plays a role in metabolizing tamoxifen into 4-hydroxytamoxifen and N-desmethyltamoxifen (FIGURE 1) [88,89]. Three polymorphic variants of CYP2C19 have been identified (CYP2C19*2, *3 and *17). In contrast to CYP2D6*4, which eliminates enzymatic activity, the *17 variant of CYP2C19 confers increased gene transcription and subsequently increased enzymatic activity [90]. The result could be an increase in the rate of tamoxifen activation in women carrying the allele. Gjerde et al. reported that increased CYP2C19 activity was associated with increased serum concentrations of 4-hydroxytamoxifen, and that CYP2D6 inhibition affects the ratio of 4-hydroxytamoxifen to tamoxifen, but not among individuals with the CYP2C19*17 variant [88]. In addition to reporting higher rates of recurrence among women with genetic inhibition of CYP2D6, Schroth et al. found that carriers of the C Y P2C19*17 variant had a lower risk of breast cancer recurrence, with the strongest effect seen in homozygotes [59]. Regarding the other CYP2C19 variants, a study of Japanese breast cancer patients reported that CYP2C19*2 and CYP2C19*3 were not predictive indicators of response to tamoxifen treatment [57]. By contrast, a Dutch group reported that the CYP2C19*2 variant predicted better survival in tamoxifen-treated breast cancer patients [91].
In the absence of functional CYP2D6, CYP2C9 takes the lead in catalyzing the formation of 4-hydroxy-N-desmethyl-tamoxifen, via initial 4-hydoxylation of tamoxifen [21,24] followed by demethylation catalyzed by CYP3A4 and CYP3A5 (FIGURE 1). Genetic variants of CYP2C9 (CYP2C9*2 and CYP2C9*3) reduce its catalytic ability and lower the production of 4-hydroxytamoxifen [24]. Thus a combination of lower CYP2D6 activity and reduced-function CYP2C9 may reduce an individual’s ability to metabolize tamoxifen to 4-hydroxy-N-desmethyl-tamoxifen. However, combined inheritance of these nonfunctional or reduced-function alleles is likely to occur at a very low frequency.
While most research has focused on the metabolic enzymes involved in 4-hydroxylation of tamoxifen, which confers 100-fold greater binding affinity with the ER, demethylation is also an important step in the ultimate production of 4-hydroxy-N-desmethyl-tamoxifen. CYP3A4 and CYP3A5 primarily catalyze this metabolic step (FIGURE 1) [10]. CYP3A4*1B is a polymorphism in the untranslated region upstream of the DNA coding sequence, and thus affects mRNA expression levels. Although it is expected to have little functional consequence [92], carriers of this allele were at increased risk for recurrence in the only study that has evaluated it [67]. CYP3A5*3, which results in a truncated protein, affects the profile of tamoxifen metabolites [88], and has been related to improved disease-free survival in one study [62], marginally poorer survival in a second study [67] and had a null association in a third [93].
The secondary tamoxifen metabolites are sulfonated in a reaction catalyzed by sulfotransferases (especially SULT1A1) [94] or glucoronidated in a reaction catalyzed by UDP-glucoronosyltransferases (especially UGT1A8, UGT1A10, UGT2B7 and UGT2B15 ) [16]. Sulfonation and glucoronidation facilitate excretion by increasing the metabolite’s water solubility and reduce the metabolite’s activity because addition of the charged sulfonate or glucoronyl moiety prevents binding to the ER (FIGURE 1). Sulfonated tamoxifen metabolites are also desulfonated to their active forms by SULT1A1 and SULT1E1 (estrogen sulfotransferase; see FIGURE 1) [95].
Sulfotransferase 1A1 is polymorphic, with a wild-type allele (SULT1A1*1) and a variant allele (SULT1A1*2) [96,97]. The sulfotransferase produced by the variant allele has twofold reduced activity [98] and reduced thermostability [99], which results in a lower enzyme concentration. Women with reduced SULT1A1 activity are expected to have higher concentrations of the secondary tamoxifen metabolites, because they cannot deactivate them as rapidly as women with the fully functional gene product. The profile of tamoxifen metabolite concentrations consistent with this expectation has been observed in vivo [14,15]. In three studies of breast cancer patients treated with tamoxifen, women with the SULT1A1*2 variant had a higher hazard of recurrence than women with the wild-type allele in one study [56], a lower risk of recurrence in a second study [63] and approximately the same risk of recurrence in the third study [62].
Among the UGT enzymes that play a role in the detoxification of tamoxifen, UGT1A10 has a polymorphic variant (UGT1A10139Lys) [16], UGT2B7 has a variant allele (UGT2B7*2) with a prevalence of approximately 50%, and both variant alleles confer reduced enzymatic activity [16,100]. UGT2B15 has a variant allele (UGT2B15*2 ) with at least a twofold higher rate of catalytic activity [101] . UGT1A8 has two polymorphic variants (UGT1A8*2 and UGT1A8*3) [16] – the UGT1A8*2 variant has activity similar to the wild type, but the UGT1A8*3 allele has significantly reduced activity compared with the wild type [16]. UGT2B7 is thought to be the most active hepatic UGT enzyme with respect to tamoxifen metabolism [13]. Despite the high prevalence of some of the mutations (e.g., UGT2B7*2) and the compelling evidence for the role of UGTs in tamoxifen metabolism [13], the impact of inter-individual variation of these enzymes on tamoxifen metabolism and breast cancer recurrence has only been investigated in two published studies [56,62] and one conference abstract [66]. Women with the increased-function UGT2B15*2 variant are expected to have lower concentrations of the secondary tamoxifen metabolites because they deactivate them more rapidly than women with the wild-type allele. Consistent with this expectation, two of the studies of breast cancer patients treated with tamoxifen reported that women with the variant allele had a higher rate of recurrence than women with the normal function allele, although the difference was imprecisely measured. By contrast, the conference abstract reported better survival among women with high UGT2B7 activity (the genotype associated with increased elimination of endoxifen, so expected to have poorer survival). More detailed examination of these findings awaits publication of the full report.
Taken together, these data illustrate the importance of considering the entire metabolic pathway for tamoxifen. Given the complexity of the metabolic pathway, it seems unlikely that the key to tamoxifen resistance lies within allelic variation of a single gene, even if that gene (CYP2D6) encodes a major enzyme in its metabolic pathway. Rather, the combined effect of variation in all enzymes involved in its metabolic pathway is likely to determine treatment effectiveness, if the metabolic profile has any impact at all. As demonstrated by the recent work of Dunn et al. [87], the combined effect of several changes in the function of metabolic enzymes within the entire tamoxifen metabolic pathway may be the key to evaluating its effectiveness.
Expert commentary
Cytochrome P450 enzymes metabolize tamoxifen to more active intermediate metabolite forms [10], each with its own binding affinity to the ER. All of the enzymes in this pathway are polymorphic, and the changes in function related to genotypes contribute to interindividual differences in tamoxifen metabolite concentrations in the serum [14–16]. Most clinical epidemiology studies examining the associations between gene variants and breast cancer outcomes have focused on CYP2D6.
Epidemiologic studies associating reduced CYP2D6 function with risk of breast cancer recurrence have reported widely heterogeneous results, and no adequate explanation has been proffered for this variability. We have previously addressed criticisms based on small sample size, survivor and other selection biases, potential for uncontrolled confounding by prognostic markers, and information bias arising from retrospective or absent information on use of CYP2D6-inhibiting medications or from noncentralized testing of ER expression [8]. In this article, we have also considered and rejected as complete explanations the variable potency of CYP2D6 inhibition, the variable quality of exposure information on CYP2D6 inhibition and failure to control for adherence to tamoxifen over the full duration of its intended course.
While the inconsistent pattern of associations remains, we note that the most recent large and high-quality studies have consistently reported near-null associations [48,64,65,68,69]. In addition, summary estimates of the association have consistently been near-null [8,102,103], and remain near-null in our quantitative meta-analyses.
The compelling molecular and pharmacologic hypotheses have prompted some to advocate the implementation of CYP2D6 genotyping of breast cancer patients who are candidates for tamoxifen therapy in routine clinical practice. However, as our article and meta-analyses indicate, the results of clinical epidemiologic scientific studies present no sound foundation for this recommendation. Overall, there is little variation in the effectiveness of tamoxifen among individuals with varying degrees of CYP2D6 inhibition induced either by receipt of inhibiting medications or by functional polymorphisms in the CYP2D6 gene. It is possible that the variations in the profile of metabolite concentrations associated with CYP2D6 inhibition are of no consequence with regard to breast cancer recurrence and survival. It is also possible that the focus on CYP2D6 inhibition has masked the complexity of the true underlying biology, which requires a more complete assessment of the function of many genes whose products metabolize tamoxifen.
Five-year view
Although breast cancer prevention remains a considerable public health challenge, effective screening and ever-advancing therapies continue to improve the prognosis of breast cancer patients. More than 30 years ago, anti-estrogen therapy was understood to be most effective against tumors that expressed the ER. This result presaged the era of personalized medicine, and personalization of breast cancer therapies will probably continue to provide a model for individualized treatment regimens. With regard to the identification of breast cancer patients who are, or are not, good candidates for tamoxifen therapy by virtue of their capacity to metabolize the drugs, research will probably develop along one of three paths.
First, it is possible that metabolic capacity does affect tamoxifen’s anti-estrogenic potency, but that the focus on CYP2D6 inhibition has told only a part of this story. Future research will have to investigate the complete metabolic pathway and multiple variant functional alleles in multiple metabolic enzymes. The complexity of modeling the complete pathway, and the requisite study size to evaluate combinations of genotypes that number in the tens of thousands, currently preclude such studies. However, in vitro models could be generated to incorporate combination profiles of these genetically variable metabolic enzymes. Such in vitro models could provide Bayesian priors, that is, a system to categorize breast cancer patients whose genotypes have similar metabolic profiles. These categorizations would reduce the requisite study size, and might allow a more complete evaluation of the association between metabolic capacity and tamoxifen effectiveness. The research model is very similar to the one already applied to CYP2D6 inhibition, in which observed variations in metabolic profile associated with CYP2D6 genotype or CYP2D6-inhibiting medications provided a prior basis for evaluating the association between these characteristics and breast cancer outcomes in tamoxifen-treated breast cancer patients. The challenge in the coming years will be to extend that model to multiple metabolic enzymes, each with functional variants that could affect the metabolic capacity to different extents.
Second, it is possible that CYP2D6 inhibition, or other modes of suboptimal tamoxifen metabolism, only conveys an increased risk of breast cancer in a subset of patients. Premenopausal women are a logical candidate subset because tamoxifen remains a guideline therapy only in these women. Furthermore, their higher endogenous concentration of estrogen suggests that impeded competition of tamoxifen metabolites for binding to the ER would be most relevant. No clinical epidemiology study has been restricted to premenopausal women or presented a subanalysis limited to premenopausal women. Wu et al. recently proposed that low concentrations of endoxifen may increase recurrence risk in tamoxifen-treated women but only among those whose tumors do not express ER-β [104]. This second subgroup, now demarcated by two biomarkers (genetic inhibition of CYP2D6 and lack of ERβ expression), will require a very large study population in order to precisely estimate the three-way interaction between drug (tamoxifen), gene (CYP2D6 genotype) and protein (ER-β expression).
Finally, it is possible that research focused on CYP2D6 inhibition has already told the complete story. These studies now appear now to be converging on a null or small association. Regardless of metabolic capacity, tamoxifen and its metabolites may be present at the site of action in sufficient concentrations to adequately antagonize growth stimulation by the ER. If that idea turns out to be correct, then it is likely that genotype-guided tamoxifen therapy will not come to fruition.
Key issues.
Tamoxifen is metabolized by several polymorphic enzymes from its administered form to more active metabolites. Genetic variation in these polymorphic enzymes contributes to interindividual differences in tamoxifen metabolite concentrations in the serum.
The CYP2D6 enzyme is one of several enzymes catalyzing the formation of tamoxifen’s metabolites. CYP2D6 has several polymorphic variants, which confer a fully functional, reduced-function or nonfunctional enzyme.
The therapeutic dose of tamoxifen is set such that it and its metabolites overwhelm estrogen in competing for the estrogen receptor, depriving the breast tumor cell of estrogen-induced growth stimulation. A few-fold reduction in the concentration of one active metabolite, owing to genetic variation or drug-induced inhibition of CYP2D6, is unlikely to affect the ability of tamoxifen to block estrogen-induced growth stimulation.
There is little evidence to suggest variation in the effectiveness of tamoxifen among individuals with varying degrees of CYP2D6 inhibition induced either by the receipt of inhibiting medications or by functional polymorphisms in the CYP2D6 gene.
Given that several enzymes play a role in the tamoxifen metabolic pathway, comprehensive genotyping of the CYP2D6 gene and of genes encoding other enzymes involved in the entire tamoxifen metabolic pathway may be key to evaluating the genotype-guided effectiveness of tamoxifen.
To date, the clinical epidemiology studies investigating the outcome of tamoxifen treatment in breast cancer patients have mainly focused on only one of these polymorphic enzymes, namely, cytochrome P450 2D6.
Acknowledgments
This work was supported by grants from the US National Cancer Institute at the NIH (grant number R01 CA118708), the Danish Cancer Society (grant number DP06117) and the Danish Medical Research Council (grant number DOK 1158859).The Department of Clinical Epidemiology at Aarhus University Hospital, Denmark, is involved in studies with funding from various companies as research grants to, and administered by, Aarhus University. These include grants from the Lundbeck Foundation and H Lundbeck A/S (a manufacturer of citalopram and escitalopram). These grants have no direct relation to the present study and supported none of the work reported herein.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
- 1.Early Breast Cancer Trialists Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2. Burstein HJ, Prestrud AA, Seidenfeld J, et al. American Society of Clinical Oncology clinical practice guideline: update on adjuvant endocrine therapy for women with hormone receptor-positive breast cancer. J. Clin. Oncol. 2010;28(23):3784–3796. doi: 10.1200/JCO.2009.26.3756. • Guidelines for clinical practice recommend tamoxifen therapy for premenopausal women with tumors that express the estrogen receptor, and some postmenopausal women. Cytochrome P450 2D6 genotyping is not recommended.
- 3.Goldhirsch A, Ingle JN, Gelber RD, et al. Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann. Oncol. 2009;20(8):1319–1329. doi: 10.1093/annonc/mdp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pirmohamed M. Acceptance of biomarker-based tests for application in clinical practice: criteria and obstacles. Clin. Pharmacol. Ther. 2010;88(6):862–866. doi: 10.1038/clpt.2010.245. [DOI] [PubMed] [Google Scholar]
- 5.Wolf DM, Gottardis MM, Jordan VC. Tamoxifen-resistant growth. In: Jordan VC, editor. Long-Term Tamoxifen Treatment for Breast Cancer. WI, USA: University of Wisconsin Press; 1994. pp. 181–198. [Google Scholar]
- 6.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer. 2009;9(9):631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 7.Henriksen KL, Sonne-Hansen K, Kirkegaard T, et al. Development of new predictive markers for endocrine therapy and resistance in breast cancer. Acta Oncol. 2008;47(4):795–801. doi: 10.1080/02841860802026993. [DOI] [PubMed] [Google Scholar]
- 8. Lash TL, Lien EA, Sorensen HT, et al. Genotype-guided tamoxifen therapy: time to pause for reflection? Lancet Oncol. 2009;10(8):825–833. doi: 10.1016/S1470-2045(09)70030-0. • While gene–drug and drug–drug interactions affect the concentration profile of tamoxifen’s metabolites, these changes may have little clinical impact on tamoxifen’s effectiveness. The clinical epidemiology to date is widely heterogeneous, without adequate explanation, and with an average association near the null.
- 9.Briest S, Stearns V. Tamoxifen metabolism and its effect on endocrine treatment of breast cancer. Clin. Adv. Hematol. Oncol. 2009;7(3):185–192. [PubMed] [Google Scholar]
- 10.Desta Z, Ward BA, Soukhova NV, et al. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J. Pharmacol. Exp. Ther. 2004;310(3):1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 11.Lim YC, Desta Z, Flockhart DA, et al. Endoxifen (4-hydroxy-N-desmethyltamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxytamoxifen. Cancer Chemother. Pharmacol. 2005;55(5):471–478. doi: 10.1007/s00280-004-0926-7. [DOI] [PubMed] [Google Scholar]
- 12.Coezy E, Borgna JL, Rochefort H. Tamoxifen and metabolites in MCF7 cells: correlation between binding to estrogen receptor and inhibition of cell growth. Cancer Res. 1982;42(1):317–323. [PubMed] [Google Scholar]
- 13.Lazarus P, Blevins-Primeau AS, Zheng Y, et al. Potential role of UGT pharmacogenetics in cancer treatment and prevention: focus on tamoxifen. Ann. NY Acad. Sci. 2009;1155:99–111. doi: 10.1111/j.1749-6632.2009.04114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gjerde J, Hauglid M, Breilid H, et al. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann. Oncol. 2008;19(1):56–61. doi: 10.1093/annonc/mdm434. • Polymorphisms in genes whose enzymes metabolize tamoxifen affect the concentration profile of tamoxifen’s metabolites.
- 15. Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J. Natl Cancer Inst. 2005;97(1):30–39. doi: 10.1093/jnci/dji005. • Selective serotonin reuptake inhibitor medications and polymorphisms in genes whose enzymes metabolize tamoxifen affect the concentration profile of tamoxifen’s metabolites.
- 16.Blevins-Primeau AS, Sun D, Chen G, et al. Functional significance of UDP-glucuronosyltransferase variants in the metabolism of active tamoxifen metabolites. Cancer Res. 2009;69(5):1892–1900. doi: 10.1158/0008-5472.CAN-08-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Sun Y, Yao L, et al. Association between CYP2D6 *10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann. Oncol. 2008;19(8):1423–1429. doi: 10.1093/annonc/mdn155. [DOI] [PubMed] [Google Scholar]
- 18.Jordan VC, Collins MM, Rowsby L, et al. A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J. Endocrinol. 1977;75(2):305–316. doi: 10.1677/joe.0.0750305. [DOI] [PubMed] [Google Scholar]
- 19.Lien EA, Solheim E, Kvinnsland S, et al. Identification of 4-hydroxy-N-desmethyltamoxifen as a metabolite of tamoxifen in human bile. Cancer Res. 1988;48(8):2304–2308. [PubMed] [Google Scholar]
- 20.Adam HK, Douglas EJ, Kemp JV. The metabolism of tamoxifen in human. Biochem. Pharmacol. 1979;28(1):145–147. doi: 10.1016/0006-2952(79)90283-1. [DOI] [PubMed] [Google Scholar]
- 21.Crewe HK, Ellis SW, Lennard MS, et al. Variable contribution of cytochromes P450 2D6, 2C9 and 3A4 to the 4-hydroxylation of tamoxifen by human liver microsomes. Biochem. Pharmacol. 1997;53(2):171–178. doi: 10.1016/s0006-2952(96)00650-8. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Borges S, Jason RD, et al. A penalized mixture model approach in genotype/ phenotype association analysis for quantitative phenotypes. Cancer Inform. 2010;9 doi: 10.4137/cin.s3493. 993-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stearns V, Johnson MD, Rae JM, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J. Natl Cancer Inst. 2003;95(23):1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 24.Coller JK, Krebsfaenger N, Klein K, et al. The influence of CYP2B6, CYP2C9 and CYP2D6 genotypes on the formation of the potent antioestrogen Z-4-hydroxytamoxifen in human liver. Br. J. Clin. Pharmacol. 2002;54(2):157–167. doi: 10.1046/j.1365-2125.2002.01614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallicchio L, Tkaczuk K, Lord G, et al. Medication use, tamoxifen (TAM), and TAM metabolite concentrations in women with breast cancer. Cancer Lett. 2004;211(1):57–67. doi: 10.1016/j.canlet.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Jordan VC. Metabolites of tamoxifen in animals and man: identification, pharmacology, and significance. Breast Cancer Res. Treat. 1982;2(2):123–138. doi: 10.1007/BF01806449. [DOI] [PubMed] [Google Scholar]
- 27.Biglia N, Torta R, Roagna R, et al. Evaluation of low-dose venlafaxine hydrochloride for the therapy of hot flushes in breast cancer survivors. Maturitas. 2005;52(1):78–85. doi: 10.1016/j.maturitas.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Henry NL, Rae JM, Li L, et al. Association between CYP2D6 genotype and tamoxifen-induced hot flashes in a prospective cohort. Breast Cancer Res. Treat. 2009;117(3):571–575. doi: 10.1007/s10549-009-0309-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demissie S, Silliman RA, Lash TL. Adjuvant tamoxifen: predictors of use, side effects, and discontinuation in older women. J. Clin. Oncol. 2001;19(2):322–328. doi: 10.1200/JCO.2001.19.2.322. [DOI] [PubMed] [Google Scholar]
- 30.Baum M, Budzar AU, Cuzick J, et al. Anastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early breast cancer: first results of the ATAC randomised trial. Lancet. 2002;359(9324):2131–2139. doi: 10.1016/s0140-6736(02)09088-8. [DOI] [PubMed] [Google Scholar]
- 31.Fisher B, Costantino JP, Wickerham DL, et al. Tamoxifen for the prevention of breast cancer: current status of the National Surgical Adjuvant Breast and Bowel Project P-1 study. J. Natl Cancer Inst. 2005;97(22):1652–1662. doi: 10.1093/jnci/dji372. [DOI] [PubMed] [Google Scholar]
- 32.Deitcher SR, Gomes MP. The risk of venous thromboembolic disease associated with adjuvant hormone therapy for breast carcinoma: a systematic review. Cancer. 2004;101(3):439–449. doi: 10.1002/cncr.20347. [DOI] [PubMed] [Google Scholar]
- 33.Chien AJ, Goss PE. Aromatase inhibitors and bone health in women with breast cancer. J. Clin. Oncol. 2006;24(33):5305–5312. doi: 10.1200/JCO.2006.07.5382. [DOI] [PubMed] [Google Scholar]
- 34.Swerdlow AJ, Jones ME. Tamoxifen treatment for breast cancer and risk of endometrial cancer: a case-control study. J. Natl Cancer Inst. 2005;97(5):375–384. doi: 10.1093/jnci/dji057. [DOI] [PubMed] [Google Scholar]
- 35.Crewe HK, Lennard MS, Tucker GT, et al. The effect of selective serotonin re-uptake inhibitors on cytochrome P4502D6 (CYP2D6) activity in human liver microsomes. Br. J. Clin. Pharmacol. 1992;34(3):262–265. doi: 10.1111/j.1365-2125.1992.tb04134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeppesen U, Gram LF, Vistisen K, et al. Dose-dependent inhibition of CYP1A2, CYP2C19 and CYP2D6 by citalopram, fluoxetine, fluvoxamine and paroxetine. Eur. J. Clin. Pharmacol. 1996;51(1):73–78. doi: 10.1007/s002280050163. [DOI] [PubMed] [Google Scholar]
- 37.Cronin-Fenton D, Lash TL, Sorensen HT. Selective serotonin reuptake inhibitors and adjuvant tamoxifen therapy: risk of breast cancer recurrence and mortality. Future Oncol. 2010;6(6):877–880. doi: 10.2217/fon.10.65. [DOI] [PubMed] [Google Scholar]
- 38.Sideras K, Ingle JN, Ames MM, et al. Coprescription of tamoxifen and medications that inhibit CYP2D6. J. Clin. Oncol. 2010;28(16):2768–2776. doi: 10.1200/JCO.2009.23.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borges S, Desta Z, Li L, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin. Pharmacol. Ther. 2006;80(1):61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 40.Borges S, Desta Z, Jin Y, et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J. Clin. Pharmacol. 2010;50(4):450–458. doi: 10.1177/0091270009359182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chubak J, Buist DS, Boudreau DM, et al. Breast cancer recurrence risk in relation to antidepressant use after diagnosis. Breast Cancer Res. Treat. 2008;112(1):123–132. doi: 10.1007/s10549-007-9828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahern TP, Pedersen L, Cronin-Fenton DP, et al. No increase in breast cancer recurrence with concurrent use of tamoxifen and some CYP2D6-inhibiting medications. Cancer Epidemiol. Biomarkers Prev. 2009;18(9):2562–2564. doi: 10.1158/1055-9965.EPI-09-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dezentje VO, van Blijderveen NJ, Gelderblom H, et al. Effect of concomitant CYP2D6 inhibitor use and tamoxifen adherence on breast cancer recurrence in early-stage breast cancer. J. Clin. Oncol. 2010;28(14):2423–2429. doi: 10.1200/JCO.2009.25.0894. [DOI] [PubMed] [Google Scholar]
- 44.Goetz MP, Knox SK, Suman VJ, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res. Treat. 2007;101(1):113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 45.Kelly CM, Juurlink DN, Gomes T, et al. Selective serotonin reuptake inhibitors and breast cancer mortality in women receiving tamoxifen: a population based cohort study. BMJ. 2010;340:c693. doi: 10.1136/bmj.c693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lash TL, Pedersen L, Cronin-Fenton D, et al. Tamoxifen’s protection against breast cancer recurrence is not reduced by concurrent use of the SSRI citalopram. Br. J. Cancer. 2008;99(4):616–621. doi: 10.1038/sj.bjc.6604533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lash TL, Cronin-Fenton D, Ahern TP, et al. Breast cancer recurrence risk related to concurrent use of SSRI antidepressants and tamoxifen. Acta Oncol. 2010;49(3):305–312. doi: 10.3109/02841860903575273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azoulay L, Dell’aniello S, Huiart L, et al. Concurrent use of tamoxifen with CYP2D6 inhibitors and the risk of breast cancer recurrence. Breast Cancer Res. Treat. 2010 doi: 10.1007/s10549-010-1162-y. [DOI] [PubMed] [Google Scholar]
- 49.Siegelmann-Danieli N, Kurnik D, Lomnicky Y, et al. Potent CYP2D6 inhibiting drugs do not increase relapse rate in early breast cancer patients treated with adjuvant tamoxifen. Breast Cancer Res. Treat. 2011;125(2):505–510. doi: 10.1007/s10549-010-1008-7. [DOI] [PubMed] [Google Scholar]
- 50.Lehmann D, Nelsen J, Ramanath V, et al. Lack of attenuation in the anti-tumor effect of tamoxifen by chronic CYP isoform inhibition. J. Clin. Pharmacol. 2004;44(8):861–865. doi: 10.1177/0091270004266618. [DOI] [PubMed] [Google Scholar]
- 51.Aubert RE, Stanek EJ, Yao J, et al. Risk of breast cancer recurrence in women initiating tamoxifen with CYP2D6 inhibitors. J. Clin. Oncol. 2009;27 (Abstract 18S) [Google Scholar]
- 52.Bijl MJ, van Schaik RH, Lammers LA, et al. The CYP2D6*4 polymorphism affects breast cancer survival in tamoxifen users. Breast Cancer Res. Treat. 2009;118(1):125–130. doi: 10.1007/s10549-008-0272-2. [DOI] [PubMed] [Google Scholar]
- 53.Kiyotani K, Mushiroda T, Imamura CK, et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J. Clin. Oncol. 2010;28(8):1287–1293. doi: 10.1200/JCO.2009.25.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiyotani K, Mushiroda T, Sasa M, et al. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci. 2008;99(5):995–999. doi: 10.1111/j.1349-7006.2008.00780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newman WG, Hadfield KD, Latif A, et al. Impaired tamoxifen metabolism reduces survival in familial breast cancer patients. Clin. Cancer Res. 2008;14(18):5913–5918. doi: 10.1158/1078-0432.CCR-07-5235. [DOI] [PubMed] [Google Scholar]
- 56.Nowell SA, Ahn J, Rae JM, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res. Treat. 2005;91(3):249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 57.Okishiro M, Taguchi T, Jin KS, et al. Genetic polymorphisms of CYP2D6*10 and CYP2C19*2,*3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifen. Cancer. 2009;115(5):952–961. doi: 10.1002/cncr.24111. [DOI] [PubMed] [Google Scholar]
- 58.Ramon y Cajal T, Altes A, Pare L, et al. Impact of CYP2D6 polymorphisms in tamoxifen adjuvant breast cancer treatment. Breast Cancer Res. Treat. 2010;119(1):33–38. doi: 10.1007/s10549-009-0328-y. [DOI] [PubMed] [Google Scholar]
- 59.Schroth W, Antoniadou L, Fritz P, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J. Clin. Oncol. 2007;25(33):5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 60.Schroth W, Goetz MP, Hamann U, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302(13):1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toyama T, Yamashita H, Sugiura H, et al. No association between CYP2D6*10 genotype and survival of node-negative Japanese breast cancer patients receiving adjuvant tamoxifen treatment. Jpn. J. Clin. Oncol. 2009;39(10):651–656. doi: 10.1093/jjco/hyp076. [DOI] [PubMed] [Google Scholar]
- 62.Wegman P, Elingarami S, Carstensen J, et al. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9(1):R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wegman P, Vainikka L, Stal O, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7(3):R284–R290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leyland-Jones B, Regan MM, Bouzyk M, et al. Presented at: San Antonio Breast Cancer Symposium. San Antonio, TX, USA: 2010. Dec, Outcome according to CYP2D6 genotype among postmenopausal women with endocrine-responsive early invasive breast cancer randomized in the BIG 1–98 trial; pp. 8–12. (Abstract S1–8) [Google Scholar]
- 65.Rae JM, Drury S, Hayes DF, et al. Presented at: San Antonio Breast Cancer Symposium. San Antonio, TX, USA: 2010. Dec, Lack of correlation between gene variants in tamoxifen metabolizing enzymes with primary endpoints in the ATAC trial; pp. 8–12. (Abstract S1–7) [Google Scholar]
- 66.Oesterreich S, Hilsenbeck SH, Skaar T, et al. Presented at: San Antonio Breast Cancer Symposium. San Antonio, TX, USA: 2010. Dec, Correlations between genetic variants in CYP2D6 and UGT2B7 and survival in breast cancer patients treated with or without tamoxifen: results from a large cohort study; pp. 8–12. (Abstract P04-02-01) [Google Scholar]
- 67.Gor PP, Su HI, Gray RJ, et al. Cyclophosphamide-metabolizing enzyme polymorphisms and survival outcomes after adjuvant chemotherapy for node-positive breast cancer: a retrospective cohort study. Breast Cancer Res. 2010;12(3):R26. doi: 10.1186/bcr2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lash TL, Cronin-Fenton D, Ahern TP, et al. CYP2D6 inhibition and breast cancer recurrence in a population-based study in Denmark. J. Natl Cancer Inst. 2011;103(6):489–500. doi: 10.1093/jnci/djr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abraham JE, Maranian MJ, Driver KE, et al. CYP2D6 gene variants: association with breast cancer specific survival in a cohort of breast cancer patients from the United Kingdom treated with adjuvant tamoxifen. Breast Cancer Res. 2010;12(4):R64. doi: 10.1186/bcr2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson AM, Johnson A, Quinlan P, et al. Comprehensive CYP2D6 genotype and adherence affect outcome in breast cancer patients treated with tamoxifen monotherapy. Breast Cancer Res. Treat. 2011;125(1):279–287. doi: 10.1007/s10549-010-1139-x. [DOI] [PubMed] [Google Scholar]
- 71.Stingl JC, Parmar S, Huber-Wechselberger A, et al. Impact of CYP2D6*4 genotype on progression free survival in tamoxifen breast cancer treatment. Curr. Med. Res. Opin. 2010;26(11):2535–2542. doi: 10.1185/03007995.2010.518304. [DOI] [PubMed] [Google Scholar]
- 72. Schroth W, Hamann U, Fasching PA, et al. CYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: expanded polymorphism coverage improves risk stratification. Clin. Cancer Res. 2010;16(17):4468–4477. doi: 10.1158/1078-0432.CCR-10-0478. • Comprehensive genotyping of the CYP2D6 gene increases the strength of association between reduced-function and breast cancer recurrence.
- 73.Kim H, Shin HC, Yom CK, et al. Presented at: San Antonio Breast Cancer Symposium. San Antonio, TX, USA: 2010. Dec, Lack of significant association between CYP2D6 polymorphisms and clinical outcomes of adjuvant tamoxifen therapy; pp. 8–12. (Abstract PD05-08) [Google Scholar]
- 74.Lash TL. Association between CYP2D6 polymorphisms and breast cancer outcomes. JAMA. 2010;303(6):516. doi: 10.1001/jama.2010.93. [DOI] [PubMed] [Google Scholar]
- 75.Howell A, Cuzick J, Baum M, et al. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. Lancet. 2005;365(9453):60–62. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 76.Cuzick J, Sestak I, Baum M, et al. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the ATAC trial. Lancet Oncol. 2010;11(12):1135–1141. doi: 10.1016/S1470-2045(10)70257-6. [DOI] [PubMed] [Google Scholar]
- 77.Greenland S, Robins JM. Identifiability, exchangeability, and epidemiological confounding. Int. J. Epidemiol. 1986;15(3):413–419. doi: 10.1093/ije/15.3.413. [DOI] [PubMed] [Google Scholar]
- 78.Myrand S, Sekiguchi K, Man M, et al. Pharmacokinetics/genotype associations for major cytochrome P450 enzymes in native and first- and third-generation Japanese populations: comparison with Korean, Chinese, and Caucasian populations. Clin. Pharmacol. Ther. 2008;84(3):347–361. doi: 10.1038/sj.clpt.6100482. [DOI] [PubMed] [Google Scholar]
- 79.Rothman KJ, Greenland S, Lash TL. Types of epidemiologic studies. In: Rothman KJ, Greenland S, Lash TL, editors. Modern Epidemiology. 3rd Edition. PA, USA: Lippincott Williams & Wilkins; 2008. pp. 95–97. [Google Scholar]
- 80.Rebsamen MC, Desmeules J, Daali Y, et al. The AmpliChip CYP450 test: cytochrome P450 2D6 genotype assessment and phenotype prediction. Pharmacogenomics J. 2009;9(1):34–41. doi: 10.1038/tpj.2008.7. [DOI] [PubMed] [Google Scholar]
- 81.Owusu C, Buist DS, Field TS, et al. Predictors of tamoxifen discontinuation among older women with estrogen receptor-positive breast cancer. J. Clin. Oncol. 2008;26(4):549–555. doi: 10.1200/JCO.2006.10.1022. [DOI] [PubMed] [Google Scholar]
- 82.Geiger AM, Thwin SS, Lash TL, et al. Recurrences and second primary breast cancers in older women with initial early-stage disease. Cancer. 2007;109(5):966–974. doi: 10.1002/cncr.22472. [DOI] [PubMed] [Google Scholar]
- 83.Rae JM, Sikora MJ, Henry NL, et al. Cytochrome P450 2D6 activity predicts discontinuation of tamoxifen therapy in breast cancer patients. Pharmacogenomics J. 2009;9(4):258–264. doi: 10.1038/tpj.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cole SR, Hernan MA. Fallibility in estimating direct effects. Int. J. Epidemiol. 2002;31(1):163–165. doi: 10.1093/ije/31.1.163. [DOI] [PubMed] [Google Scholar]
- 85.Glymour M, Greenland S. Causal diagrams. In: Rothman KJ, Greenland S, Lash TL, editors. Modern Epidemiology. 3rd Edition. PA, USA: Lippincott Williams & Wilkins; 2008. pp. 183–212. [Google Scholar]
- 86.Henry NL, Hayes DF, Rae JM. Oncology. 14. Vol. 23. Williston Park: 2009. CYP2D6 testing for breast cancer patients: is there more to the story? 1236, 1243, 1249. [PubMed] [Google Scholar]
- 87. Dunn BK, Greene MH, Kelley JM, et al. Novel pathway analysis of genomic polymorphism-cancer risk interaction in the breast cancer prevention trial. Int. J. Mol. Epidemiol. Genet. 2010;1(4):332–349. • Analysis of the complete pathway of tamoxifen metabolism provides additional insight into the association between tamoxifen metabolism and breast cancer outcomes.
- 88.Gjerde J, Geisler J, Lundgren S, et al. Associations between tamoxifen, estrogens, and FSH serum levels during steady state tamoxifen treatment of postmenopausal women with breast cancer. BMC Cancer. 2010;10:313. doi: 10.1186/1471-2407-10-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coller JK, Krebsfaenger N, Klein K, et al. Large inter individual variability in the in vitro formation of tamoxifen metabolites related to the development of genotoxicity. Br. J. Clin. Pharmacol. 2004;57(1):105–111. doi: 10.1046/j.1365-2125.2003.01970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li-Wan-Po A, Girard T, Farndon P, et al. Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant CYP2C19*17. Br. J. Clin. Pharmacol. 2010;69(3):222–230. doi: 10.1111/j.1365-2125.2009.03578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ruiter R, Bijl MJ, van Schaik RH, et al. CYP2C19*2 polymorphism is associated with increased survival in breast cancer patients using tamoxifen. Pharmacogenomics. 2010;11(10):1367–1375. doi: 10.2217/pgs.10.112. [DOI] [PubMed] [Google Scholar]
- 92.Keshava C, McCanlies EC, Weston A. CYP3A4 polymorphisms - potential risk factors for breast and prostate cancer: a HuGE review. Am. J. Epidemiol. 2004;160(9):825–841. doi: 10.1093/aje/kwh294. [DOI] [PubMed] [Google Scholar]
- 93.Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J. Clin. Oncol. 2005;23(36):9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 94.Falany CN, Wheeler J, Oh TS, et al. Steroid sulfation by expressed human cytosolic sulfotransferases. J. Steroid Biochem. Mol. Biol. 1994;48(4):369–375. doi: 10.1016/0960-0760(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 95.Glatt H, Boeing H, Engelke CE, et al. Human cytosolic sulphotransferases: genetics, characteristics, toxicological aspects. Mutat. Res. 2001;482(1–2):27–40. doi: 10.1016/s0027-5107(01)00207-x. [DOI] [PubMed] [Google Scholar]
- 96.Tabrett CA, Coughtrie MW. Phenol sulfotransferase 1A1 activity in human liver: kinetic properties, interindividual variation and re-evaluation of the suitability of 4-nitrophenol as a probe substrate. Biochem. Pharmacol. 2003;66(11):2089–2097. doi: 10.1016/s0006-2952(03)00582-3. [DOI] [PubMed] [Google Scholar]
- 97.Coughtrie MW, Gilissen RA, Shek B, et al. Phenol sulphotransferase SULT1A1 polymorphism: molecular diagnosis and allele frequencies in Caucasian and African populations. Biochem. J. 1999;337(Pt 1):45–49. [PMC free article] [PubMed] [Google Scholar]
- 98.Nowell S, Ambrosone CB, Ozawa S, et al. Relationship of phenol sulfotransferase activity (SULT1A1) genotype to sulfotransferase phenotype in platelet cytosol. Pharmacogenetics. 2000;10(9):789–797. doi: 10.1097/00008571-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 99.Raftogianis RB, Wood TC, Otterness DM, et al. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem. Biophys. Res. Commun. 1997;239(1):298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- 100.McCarroll SA, Hadnott TN, Perry GH, et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38(1):86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 101.Levesque E, Beaulieu M, Green MD, et al. Isolation and characterization of UGT2B15(Y85): a UDP-glucuronosyltransferase encoded by a polymorphic gene. Pharmacogenetics. 1997;7(4):317–325. doi: 10.1097/00008571-199708000-00007. [DOI] [PubMed] [Google Scholar]
- 102.Lash TL, Ahern TP, Cronin-Fenton D, et al. Modification of tamoxifen response: what have we learned? J. Clin. Oncol. 2008;26(10):1764–1765. doi: 10.1200/JCO.2007.15.5432. [DOI] [PubMed] [Google Scholar]
- 103.Seruga B, Amir E. Cytochrome P450 2D6 and outcomes of adjuvant tamoxifen therapy: results of a meta-analysis. Breast Cancer Res. Treat. 2010;122(3):609–617. doi: 10.1007/s10549-010-0902-3. [DOI] [PubMed] [Google Scholar]
- 104.Wu X, Subramaniam M, Grygo SB, et al. Estrogen receptor-β sensitizes breast cancer cells to the anti-estrogenic actions of endoxifen. Breast Cancer Res. 2011;13(2):R27. doi: 10.1186/bcr2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 201.NCCN Practice Guidelines in Oncology, Breast Cancer – Version 2.2011. Invasive breast cancer, adjuvant endocrine therapy. National Comprehensive Cancer Network, Inc. 2011 www.nccn.org.
- 202.CYP2D6 allele nomenclature. www.cypalleles.ki.se/cyp2d6.htm.