Abstract
Background and Aims
Lady's slipper orchids (Paphiopedilum) are of high value in floriculture, and interspecific hybridization has long been used for breeding improved cultivars; however, information regarding the genome affinities of species and chromosome pairing behaviour of the hybrids remains almost unknown. The present work analyses the meiotic behaviour of interspecific hybrids by genomic in situ hybridization and cytologically evaluates the genomic relationships among parental species.
Methods
Eight interspecific F1 hybrids of Paphiopedilum species in various subgenera or sections were investigated in this study. The chromosome behaviour in meiosis of these interspecific hybrids was analysed and subjected to genomic in situ hybridization and fluorescent in situ hybridization.
Key Results
Genomic in situ hybridization was demonstrated as an efficient method to differentiate between Paphiopedilum genomes and to visualize the chromosome pairing affinities in interspecific F1 hybrids, clarifying the phylogenetic distances among these species. Comparatively regular chromosome pairing observed in the hybrids of P. delenatii × P. bellatulum, P. delenatii × P. rothschildianum and P. rothschildianum × P. bellatulum suggested high genomic affinities and close relationships between parents of each hybrid. In contrast, irregular chromosome associations, such as univalents, trivalents and quadrivalents occurred frequently in the hybrids derived from distant parents with divergent karyotypes, such as P. delenatii × P. callosum, P. delenatii × P. glaucophyllum, P. rothschildianum × P. micranthum and P. rothschildianum × P. moquetteanum. The existence of multivalents and autosyndesis demonstrated by genomic in situ hybridization in this study indicates that some micro-rearrangements and other structural alterations may also play a part in differentiating Paphiopedilum species at chromosomal level, demonstrated as different chromosome pairing affinities in interspecific hybrids.
Conclusions
The results indicate that genome homology and the interaction of genetic factors, but not chromosome number nor karyotype similarity, determine the chromosome pairing behaviour in Paphiopedilum hybrids.
Keywords: Paphiopedilum, homeologous pairing, autosyndesis, genomic in situ hybridization, pollen formation
INTRODUCTION
The genus Paphiopedilum (known as the lady's slipper orchid) is one of the most popular orchid genera because of its distinctive flower morphology. There are about 75 species occurring in the tropical Asia (ranging from India, through southern China and south-east Asia, to the Philippines and New Guinea). Based on morphological characteristics, karyomorphology and molecular phylogenetic data, Paphiopedilum species are grouped into three subgenera, including Parvisepalum, Brachypetalum and Paphiopedilum (Cribb, 1998). Karyomorphological analysis in Paphiopedilum revealed that, despite a variation in chromosome number (2n = 26–42), the total number of chromosome arms (nombre fondamental or n.f.; Mathey, 1949) in a karyotype is almost conserved as n.f. = 52 (Karasawa and Saito, 1982). The changes in chromosome number and karyotype symmetry were considered as a consequence of Robertsonian centric fission (Jones, 1998). The species with a chromosome complement of 2n = 26 is suggested to be the basic number of this genus. Chromosome numbers vary from 2n = 28–42 and 2n = 30–37 in the sections Barbata and Cochlopetalum of subgenus Paphiopedilum. Moreover, there is an obvious coincidence that those species with more telocentric chromosome often contain more DNA in their genomes (Cox et al., 1998).
During the past century, thousands of interspecific hybridizations have been made to combine various traits, such as the colours, the multiflorous state and heavier flowers, to improve the horticultural values of Paphiopedilum in cultivation. The progeny resulting from intercrossing species between subgenera may be sterile or have low fertility due to irregular chromosome pairing. Although detailed karyotype data are available for Paphiopedilum species, virtually nothing is known about genome affinity and chromosome pairing between parental species of interspecific hybrids. Genome analysis provides valuable information about species relationships and, therefore, plays an important role in plant breeding programmes. The genome affinities between parental species were conventionally appraised according to the chromosome pairing behaviour observed at meiotic MI in F1 hybrids (Singh, 2003). Recently, a molecular cytogenetic technique, genomic in situ hybridization (GISH), has been effectively performed for this purpose, allowing the ancestral genomes to be distinguished, the pairing of homoeologous chromosomes at meiosis to be visualized and even chromosome rearrangements detected (reviewed in Schubert et al., 2001; Maluszynska and Hasterok, 2005). Numerous successful examples have been reported; one of the most famous studies was carried out in the Festuca–Lolium complex (reviewed by Kopecký et al., 2008a). GISH, a method of modified fluorescent in situ hybridization (FISH), uses the total genomic DNA as a probe (Heslop-Harrison et al., 1988; Schwarzacher et al., 1989) and usually with an excess of unlabelled DNA from another species as blocking agent for better differentiation (Anamthawat-Jonsson et al., 1990).
In this study, the efficiency of GISH to differentiate between the parental genomes in meiotic chromosome pairing of the Paphiopedilum F1 hybrids is demonstrated. The results give direct evidence of genome homology between the parental species, and thus provide useful information in the breeding programmes.
MATERIALS AND METHODS
Plant materials
Seven species in various subgenera or sections in the genus Paphiopedilum (Table 1) were taken from the living collection in the greenhouse of the Botanical Garden of the National Museum of Natural Science, Taiwan. Among them, P. delenatii (subgenus Parvisepalum – the primitive group) and P. rothschildianum (section Coryopedilum of subgenus Paphiopedilum) were used as the pod parents. Eight interspecific hybrids of Paphiopedilum (Table 2) were produced by hand pollination. The plants of interspecific F1 hybrids were cultivated in the greenhouse of the National Museum of Natural Science, Taiwan.
Table 1.
The Paphiopedilum species used for interspecific crosses
| Species | Subgenus* | Section* | 2n | Karyomorphology† |
|
|---|---|---|---|---|---|
| V‡ | I§ | ||||
| P. delenatii | Parvisepalum | – | 26 | 26 | – |
| P. micranthum | Parvisepalum | – | 26 | 26 | – |
| P. bellatulum | Brachypetalum | – | 26 | 26 | – |
| P. rothschildianum | Paphiopedilum | Coryopedilum | 26 | 26 | – |
| P. callosum | Paphiopedilum | Barbata | 32 | 20 | 12 |
| P. glaucophyllum | Paphiopedilum | Cochlopetalum | 36 | 14 | 22 |
| P. moquetteanum | Paphiopedilum | Cochlopetalum | 34 | 16 | 18 |
* The taxonomic system follows Cribb (1998).
† Data adapted from Karasawa and Saito (1982).
‡ Metacentric chromosome.
§ Telocentric chromosome.
Table 2.
The number (percentage) of sporad types in eight Paphiopedilum interspecific hybrids
| Hybrid | Total sporads analysed | Dyad (%) | Dyad* (%) | Triad (%) | Triad* (%) | Tetrad (%) | Tetrad* (%) |
|---|---|---|---|---|---|---|---|
| P. delenatii × P. micranthum | 216 | 4 (1·8) | – | 9 (4·2) | – | 181 (83·8) | 22 (10·2) |
| P. delenatii × P. bellatulum | 254 | 8 (3·1) | 6 (2·4) | 14 (5·5) | 5 (2·0) | 194 (76·4) | 27 (10·6) |
| P. delenatii × P. rothschildianum | 223 | 7 (3·1) | – | 16 (7·2) | 5 (2·2) | 158 (70·9) | 37 (16·6) |
| P. delenatii × P. callosum | 204 | – | – | 12 (5·9) | 7 (3·4) | 135 (66·2) | 50 (24·5) |
| P. delenatii × P. glaucophyllum | 243 | 7 (2·9) | 5 (2·1) | 10 (4·1) | 12 (4·9) | 167 (68·7) | 42 (17·3) |
| P. rothschildianum × P. micranthum | 236 | – | – | 21 (8·9) | 10 (4·2) | 142 (60·2) | 63 (26·7) |
| P. rothschildianum × P. bellatulum | 228 | – | – | 18 (7·9) | – | 176 (77·2) | 34 (14·9) |
| P. rothschildianum × P. moquetteanum | 232 | 5 (2·2) | – | 12 (5·1) | 6 (2·6) | 164 (70·7) | 45 (19·4) |
* Sporads with micronucleus.
Pollen viability
The pollinia at anthesis were collected and stained with 1 % aceto-carmine (1 % carmine in 45 % acetic acid) as previously described (MacFarlane et al., 1989): pollen with positive staining was taken as indicating viability. At least three individuals were analysed.
Chromosome preparation
To analyse chromosome pairing in these F1 hybrids, anthers with pollen mother cells (PMCs) at meiotic MI were collected and fixed in fresh prepared Farmer's fluid (three parts of ethanol to one part of glacial acetic acid). PMCs were macerated with an enzyme mixture containing 6 % cellulose (Onoauka R-10, Yakult Honsha, Japan) and 6 % pectinase (Sigma Chemical Co., St Louis, MO.) in 75 mm KCl, pH 4·0 at 37 °C for 30 min then squashed in 1 % aceto-carmine according to the method of Kao et al. (2006). Slides with good quality MI spreads were stored at –80 °C for GISH and FISH analysis.
GISH and 45rDNA FISH
Genomic DNA from young leaves of accessions was extracted using a DNeasy Plant Mini Kit (Qiagen, Oslo, Norway). Genomic DNA of the pod parents (P. delenatii or P. rothschildianum) was labelled with biotin-16-dUTP by nick translation (Roche Diagnostics GmbH, Penzberg, Germany) and used as the probe. For use as unlabelled blocking, genomic DNA of each pollen parent was fragmented up to 100–300 bp in size by boiling in 0·4 m NaOH for 40 min according to the previously described protocol (Cao et al., 2000). The position of the 45S rRNA gene locus was detected by FISH and mapped as the chromosomal landmark to target the possible homeologous pairing between parental species. Purified DNA of plasmid pTA71 containing a repetitive unit of 45S rRNA gene (rDNA, approx. 9 kb) of Triticum aestivum (Gerlach and Bedbrook, 1979) was labelled with digoxigenin-11-dUTP by nick translation (Roche Diagnostics GmbH) as the FISH probe. GISH and FISH were performed together according to the protocols of Chung et al. (2008) and Schwarzacher and Heslop-Harrison (2000) with minor modifications. Immediately after the slide was taken out of the –80 °C deep freezer, the coverslip on the slide was removed with a razor and the slide was dehydrated in a series of 70 %, 90 % and 100 % ethanol, then dried on a hot plate (60 °C) for 30 min. The chromosome preparations on the slide were denatured in 70 % formamide (in 2× SSC) at 70 °C for 1 min and dehydrated in an ethanol series before the hybridization mixture was applied. The hybridization mixture, 20 µL per slide, contained 20 ng of biotin-labelled genomic DNA, 20 ng of digoxigenin-labelled 45S rDNA, 30–50 % formamide, 10 % dextran sulfate, 2× SSC, 10 mg of salmon sperm DNA and blocking DNA in suitable concentration. The hybridization mixture was denatured at 90 °C for 10 min and left on ice for 5 min before being applied to a slide with denatured chromosome spreads. Hybridization was conducted in a humid box at 37 °C overnight. Stringent washes were done with 2× SSC at room temperature for 5 min, at 45 °C for 10 min, then at room temperature for 5 min. The digoxigenin-labelled probes were detected by anti-digoxigenin-rhodamine (Roche Diagnostics GmbH). The biotin-labelled probes were detected by fluorescein isothiocyanate-conjugated avidin (Vector Laboratories, Burlingame, CA, USA). Chromosomes were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI) in Vectashield (Vector Laboratories). Images were acquired and processed as previously described in Kao et al. (2006) and Chung et al. (2008). For each accession, at least five chromosome complements with good labelling signals were photographed and analysed.
Phylogenetic analysis
To exhibit the relationships of species used in this study, a phylogenetic tree was constructed, based on the nucleotide sequences of 5·8S ribosomal RNA gene and the internal transcribed spacers (ITS1 and ITS2) obtained from the GenBank database. Sequence alignment and phylogenetic analysis were performed with neighbor-joining estimation using GrowTree in Wisconsin Package (SeqWeb 3·1·2)
RESULTS
Sporad types and pollen viability in the hybrids
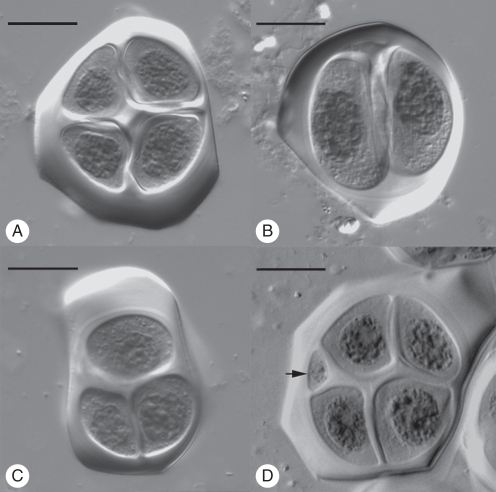
At the end of meiosis, each PMC normally forms a tetrad with four microspores (Fig. 1A). A considerable number of irregular tetrads were observed in these eight interspecific hybrids (Fig. 1B–D). In these eight hybrids, varying amounts between 60·2 % and 83·8 % of normal tetrads were formed. The tetrads with micronuclei were frequently observed in all eight hybrids, ranging from 10·2 % to 26·7 % (Table 2). The amount of stainable pollen, including those which were obviously small, ranged from 47·8 % to 74·1 % in these eight hybrids (Table 3). Most of the unstained pollen was derived from those irregular sporads. A large amount of small pollen which derived from the micronuclei was usually unstained.
Fig. 1.
Sporad types observed in the Paphiopedilum interspecific F1 hybrids using a differential interference contrast microscope: (A) a normal tetrad is enclosed in a thick callose wall; (B) a dyad is enclosed in a thick callose wall; (C) a triad is enclosed in a thick callose wall; (D) an irregular tetrad with a micronucleus (arrow). Scale bars = 10 µm
Table 3.
The number (percentage) of stainable pollen and pollen types in eight Paphiopedilum interspecific hybrids
| No. (%) of stainable pollen counted |
||||
|---|---|---|---|---|
| Hybrid | Total pollen analysed | Stained pollen* | Large pollen† | Small pollen‡ |
| P. delenatii × P. micranthum | 618 | 458 (74·1) | 432 (69·9) | 26 (4·2) |
| P. delenatii × P. bellatulum | 623 | 427 (68·5) | 379 (60·8) | 48 (7·7) |
| P. delenatii × P. rothschildianum | 620 | 376 (60·6) | 308 (49·6) | 68 (11·0) |
| P. delenatii × P. callosum | 608 | 291 (47·8) | 216 (35·5) | 75 (12·3) |
| P. delenatii × P. glaucophyllum | 622 | 349 (56·1) | 291 (46·8) | 58 (9·3) |
| P. rothschildianum × P. micranthum | 618 | 362 (58·6) | 301 (48·7) | 61 (9·9) |
| P. rothschildianum × P. bellatulum | 632 | 453 (71·7) | 419 (66·3) | 34 (5·4) |
| P. rothschildianum × P. moquetteanum | 624 | 416 (66·6) | 364 (58·3) | 52 (8·3) |
* Total number of pollen grains positively stained by aceto-carmine.
† The number of pollen grains positively stained ranged from 12 µm to 15 µm in size.
‡ The number of pollen that were positively stained and were smaller than 8 µm.
Chromosome pairing in the hybrids
The chromosome pairing behaviour at the meiotic MI stage in eight interspecific hybrids was analysed by GISH in this study (Figs 2 and 3). With the exception of P. delenatii × P. micranthum (intra-subgeneric hybrid), the parental genome in the constitution of each of these hybrids could clearly be distinguished by GISH and its configuration at the meiotic MI stage was also revealed (Table 4).
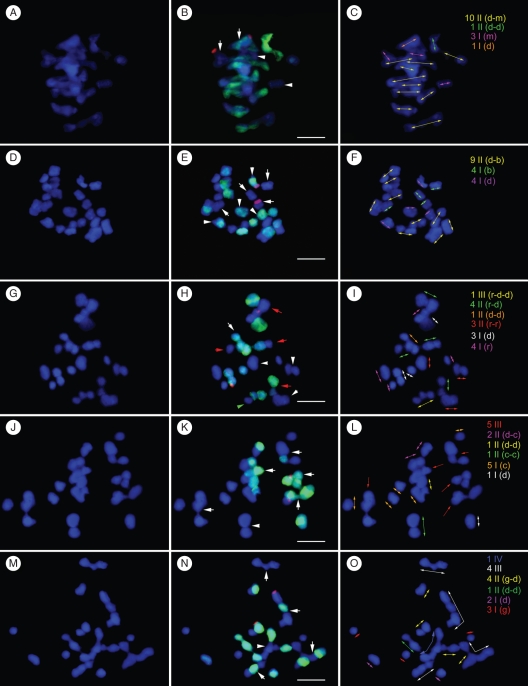
Fig. 2.
FISH and GISH analyses revealed the meiotic chromosome pairing of Paphiopedilum interspecific F1 hybrids. Meiotic chromosomes were counterstained with DAPI in blue (A, D, G, J, M). Merged images of GISH/FISH results (B, E, H, K, N) show the chromosomes from P. delenatii (in green) and the locations of 45S rDNA loci (in red) of each hybrid. Chromosomes in same/similar configurations were grouped and are separately indicated by arrows in different colours (C, F, I, L, O). (A–C) Chromosome configuration (4I + 11II) at meiotic MI of the hybrid P. delenatii × P. micranthum (2n = 2x = 26). In (B) two univalents (arrows) with an 45S rDNA cluster (red), which originated from each parental genome, are shown. The two univalents without green signals indicated by arrowheads were from P. micranthum. (D–F) Chromosome configuration (8I + 9II) at meiotic late MI of the hybrid P. delenatii × P. bellatulum (2n = 2x = 26). In (E) arrowheads indicate the univalents from P. delenatii and arrows indicate the univalents from P. bellatulum. (G–I) Chromosome configuration (7I + 8II + 1III) at MI of meiosis in the hybrid P. delenatii × P. rothschildianum (2n = 2x = 26). Among the eight bivalents shown in (H), one was the autosyndetic pair of P. delenatii (arrow), three were autosyndetic pairs of P. rothschildianum (arrowheads) and four were homeologous pairs between parents (red arrows). One trivalent is indicated with a green arrow. (J–L) Chromosome configuration (6I + 4II + 5III) at MI of meiosis in the hybrid P. delenatii × P. callosum (2n = 2x = 29). In (K) arrows indicate the trivalents and the arrowhead indicates one P. callosum autosyndetic pair. (M–O) Chromosome configuration (5I + 5II + 4III + 1IV) at MI of meiosis in the hybrid P. delenatii × P. glaucophyllum (2n = 2x = 31). In (N) arrows indicate the trivalents and an arrowhead indicates the quadrivalent. Scale bars = 10 µm
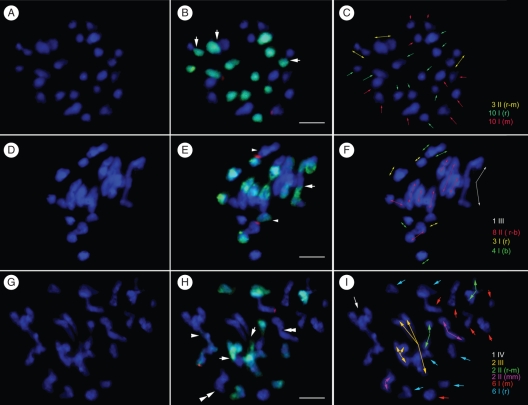
Fig. 3.
FISH and GISH analyses revealed the meiotic chromosome pairing of Paphiopedilum interspecific F1 hybrids. Meiotic chromosomes were counterstained with DAPI in blue (A, D, G). Merged images (B, E, H) showed the chromosomes from P. rothschildianum (in green) and the locations of 45S rDNA loci (in red) of each hybrid. Chromosomes in same/similar configurations were grouped, which were separately indicated by arrows in different colours (C, F, I). (A–C) Chromosome configuration (20I + 3II) at MI of meiosis in the hybrid P. rothschildianum × P. micranthum (2n = 2x = 26). In (B) arrows indicate the bivalents. (D–F) Chromosome configuration (7I + 8II + 1III) at MI of meiosis in the hybrid P. rothschildianum × P. bellatulum (2n = 2x = 26). In (E) arrows indicate the trivalents and chromosomes with 45S rDNA from both parents were separated as univalents (arrowheads). (G–I) Chromosome configuration (12I + 4 II + 2III + 1IV) at MI of meiosis in the hybrid P. rothschildianum × P. moquetteanum (2n = 2x = 30). In (H) arrows indicate the trivalents, an arrowhead indicates the quadrivalent and the double arrowheads indicate two P. moquetteanum autosyndetic pairs. Scale ars = 10 µm.
Table 4.
Chromosome configurations of eight Paphiopedilum interspecific hybrids at metaphase I of meiosis
| Mean (range) chromosome configuration per cell |
||||||
|---|---|---|---|---|---|---|
| Hybrid | 2n | Total PMCs analysed | I | II | III | IV |
| P. delenatii × P. micranthum | 26 | 20 | 3·1 (0–4) | 11·6 (11–13) | – | – |
| P. delenatii × P. bellatulum | 26 | 40 | 6·9 (2–8) | 10·7 (9–12) | – | – |
| P. delenatii × P. rothschildianum | 26 | 48 | 5·6 (4–8) | 9·7 (7–11) | 0·5 (0–2) | – |
| P. delenatii × P. callosum | 29 | 20 | 6·1 (3–9) | 3·5 (2–5) | 5·3 (4–6) | – |
| P. delenatii × P. glaucophyllum | 31 | 35 | 5·7 (4–8) | 5·4 (5–7) | 3·3 (3–4) | 1·1 (0–2) |
| P. rothschildianum × P. micranthum | 26 | 24 | 19·6 (18–22) | 3·2 (2–4) | – | – |
| P. rothschildianum × P. bellatulum | 26 | 36 | 3·8 (3–5) | 8·4 (7–10) | 1·8 (1–3) | – |
| P. rothschildianum × P. moquetteanum | 30 | 28 | 8·8 (6–12) | 6·4 (4–9) | 2·1 (1–3) | 0·7 (0–1) |
In the intra-subgeneric hybrid P. delenatii × P. micranthum (2n = 26), a high genome homology between two species was demonstrated in chromosome pairing with an average of 3·1 univalents (I) and 11·6 bivalents (II) per PMC (Table 4). In GISH experiments, the parental chromosomes could not be clearly distinguished, even when an addition of 50× unlabelled P. micranthum blocking DNA over the probe was added to the hybridization mixture (Fig. 2A–C).
Chromosome pairing in the inter-subgeneric hybrid P. delenatii × P. bellatulum (2n = 26), with an average of 6·9 univalents and 10·7 bivalents per PMC, was relatively lower than that in the intra-subgeneric hybrid P. delenatii × P. micranthum (2n = 26) (Table 4). The 13 chromosomes derived from P. delenatii could be distinguished as they displayed strong green hybridization signals when the 50× unlabelled P. bellatulum blocking DNA over the probe was added (Fig. 2D–F).
In the hybrid P. delenatii × P. rothschildianum (2n = 26), chromosome pairing ranged from seven to eleven bivalents with an average of 9·7 per PMC. A mean of 5·6 univalents and 0·5 trivalents (III) per PMC was scored (Table 4 and Fig. 2G–I). The addition of 30× unlabelled P. rothschildianum blocking DNA over the probe to the hybridization mixture was sufficient to distinguish the parental chromosomes. Autosyndesis could be observed in this hybrid (Fig. 2H) with at least two P. delenatii autosyndetic pairs and one P. rothschildianum autosyndetic pair detected.
In the hybrid P. delenatii × P. callosum (2n = 29), chromosome pairing was poor, ranging from two to five bivalents with an average of 3·5 per PMC (Fig. 2J–L). A relatively high frequency of univalents (an average of 6·1 per PMC) and trivalents (an average of 5·3 per PMC) was observed in PMC of this hybrid (Table 4). Only 10× unlabelled P. callosum blocking DNA over the probe to the hybridization mixture was sufficient to distinguish the parental chromosomes. In this hybrid, autosyndesis frequently occurred and was usually involved in the formation of trivalents (Fig. 2K).
In the hybrid P. delenatii × P. glaucophyllum (2n = 31), the formation of bivalents between parental chromosomes was relatively low, ranging from five to seven with an average of 5·4 bivalents per PMC (Fig. 2M–O). The univalents, trivalents (III) and quadrivalents (IV) occurred frequently instead. The mean chromosome configuration of this hybrid was 5·7I + 5·4II + 3·3III + 1·1IV per PMC (Table 4). The addition of 10× unlabelled P. glaucophyllum blocking DNA over the probe to the hybridization mixture was enough to distinguish the parental chromosomes. This hybrid also displayed a highly frequent autosyndesis in the formation of trivalents (Fig. 2N).
In the hybrid P. rothschildianum × P. micranthum (2n = 26), meiotic pairing was characterized by a large number of univalents with an average of 19·6 per PMC. No trivalents or quadrivalents were observed (Table 4 and Fig. 3A–C). The 13 chromosomes derived from P. rothschildianum were identified displaying strong green hybridization signals when the 30× unlabelled P. micranthum blocking DNA over the probe was added (Fig. 3B). Figure 3B showed only three homoeologous bivalents pairing at MI stage. Autosyndesis was not observed in this hybrid.
GISH results readily revealed several bivalents at the MI stage as heterogeneous pairing in the hybrid P. rothschildianum × P. bellatulum (2n = 26) (Fig. 3D–F). A number of univalents and trivalents were observed. The mean chromosome configuration of this hybrid was 3·8I + 8·4II + 1·8III per PMC (Table 4). The addition of 30× unlabelled P. bellatulum blocking DNA over the probe to the hybridization mixture was enough to differentiate the parental chromosomes.
In the hybrid P. rothschildianum × P. moquetteanum (2n = 30), meiotic pairing ranged from four to nine bivalents with an average of 6·4 per PMC (Fig. 3G–I). GISH results showed that bivalents were mainly formed by heterogeneous pairing between P. rothschildianum and P. moquetteanum, with only a few bivalents derived from autosyndesis. Univalents, trivalents and quadrivalents were also observed. The mean chromosome configuration of this hybrid was 8·8I + 6·4II + 2·1III + 0·7IV per PMC (Table 4). The addition of 30× unlabelled P. moquetteanum blocking DNA over the probe to the hybridization mixture was enough to differentiate the parental chromosomes.
Homeologous pairing tracing by 45S rDNA-FISH
Each Paphiopedilum species used in this study has one 45S rDNA locus (Lee and Chung, 2008). In every hybrid investigated here, rDNA sites were detected at the distal ends of two separated chromosomes by FISH. However, in combination with the labelling signals of GISH, the chromosomes with 45S rDNA sites (nucleolar chromosomes) from each parental species rarely paired as heterogeneous bivalents at the MI stage in these eight F1 hybrids (Figs 2 and 3).
DISCUSSION
Sporad types and pollen viability in the hybrids
In Table 2, the percentages of abnormal sporad types varied in the eight interspecific hybrids, which may be attributed to their frequency of irregular meiotic pairing. The low percentages of stainable pollen in the hybrids (Table 3) may be due to the subsequent degeneration of the sporads without balanced genomes as previously reported in Phalaenopsis (Aoyama et al., 1994) and in lily (Lim et al., 2001).
Genomic differentiation by GISH
The components and organization of repetitive sequences on chromosomes determine the efficiency of GISH to differentiate between the parental genomes (Anamthawat-Jonsson and Reader, 1995). Therefore, an excess of unlabelled DNA from the parental genome other than the probe-derived genome is often needed as blocking agent for better differentiation (Anamthawat-Jonsson et al., 1990). In this study, the chromosomal composition of the meiotic configurations in interspecific hybrids of Paphiopedilum could be successfully identified by GISH with a suitable concentration of blocking DNA in the hybridization mixture.
The phylogenetic distance between parental genomes (Fig. 4) reflects the ratio of blocking DNA to probe needed in the hybridization mixture of GISH. The failure to discriminate parental chromosomes in the hybrid P. delenatii × P. micranthum (Fig. 2B) may be due to the close distance between both parental genomes of this intra-subgeneric hybrid shown in Fig. 4. In the inter-subgeneric hybrids P. delenatii × P. rothschildianum, P. rothschildianum × P. micranthum, P. rothschildianum × P. bellatulum and P. rothschildianum × P. moquetteanum, parental chromosomes were discriminated successfully using a 1 : 30 ratio of the probe and blocking DNA in the hybridization mixture. An addition of 10× unlabelled P. callosum or P. glaucophyllum DNA was sufficient to discriminate the P. delenatii chromosomes from P. callosum or P. glaucophyllum chromosomes in their hybrids. These results demonstrate that P. delenatii, P. callosum and P. glaucophyllum are remotely related species, which agreed with the conclusions regarding the phylogenetic relationships according to their rDNA ITS sequences (Fig. 4; Cox et al., 1997). An addition of 50× unlabelled P. bellatulum DNA in the hybridization mixture as blocking was needed to distinguish P. delenatii chromosomes in the inter-subgeneric hybrid of P. delenatii × P. bellatulum by GISH. Paphiopedilum bellatulum is closer to P. delenatii than the other species investigated (Fig. 4.). The subgenus Parvisepalum (P. delenatii) and the subgenus Brachypetalum (P. bellatulum) are closely related sister groups (Cox et al., 1997) and possess comparable karyotypes: 26 meta- or sub-metacentric chromosomes (Karasawa, 1986). These characteristics contribute to the heterogeneous pairing between P. delenatii and P. bellatulum (Fig. 2D–F).
Fig. 4.
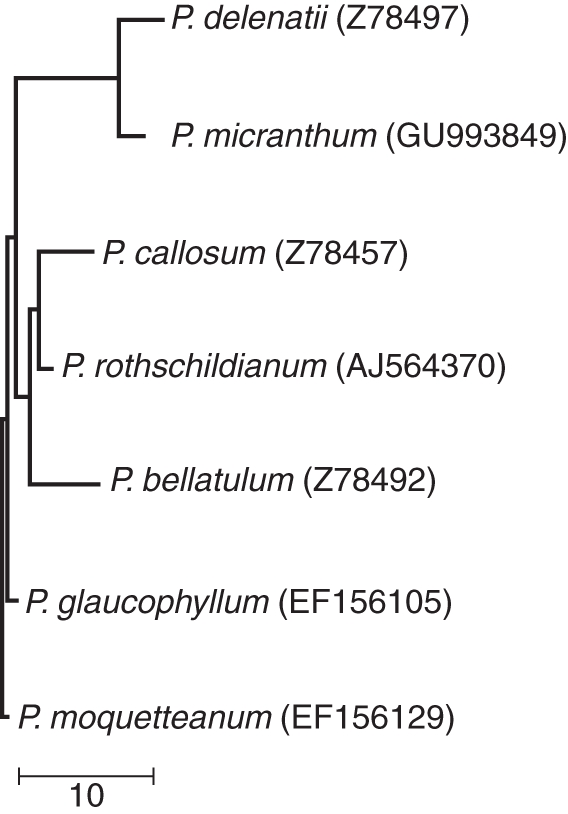
Phylogenetic tree of Paphiopedilum species based on nucleotide sequences of 5·8S rDNA, ITS1 and ITS2 obtained from the GenBank database. The accession number of the sequence used to represent each species is indicated in parenthesis. Scale bar = substitutions per 100 bases.
Karyotype variation and chromosome pairing
High irregularities in meiotic pairing were observed in the hybrids of P. delenatii × P. callosum and P. delenatii × P. glaucophyllum (Table 4), reflecting the distant relationships between their parental species (Fig. 4; Cox et al., 1997). In addition, the divergent karyotypes of these parental species may also cause high irregularity in meiosis. Species in sections Barbata and Cochlopetalum depart from the constant karyotypic pattern, ranging from 2n = 28 to 42 (n.f. = 52) in section Barbata, and 2n = 32 to 36 (n.f. = 50) in section Cochlopetalum (Cox et al., 1998). The increase in chromosome numbers in Paphiopedilum species has been proposed to be the result of a centric fission mechanism (Duncan and MacLeod, 1949, 1950; Jones, 1998). The low frequency of bivalents found in these hybrids, suggesting that structural alterations in addition to centric fission have occurred in P. callosum and P. glaucophyllum, thus hindered their chromosomes from homoeologous pairing with P. delenatii.
Meiotic pairing in the hybrid of P. delenatii × P. rothschildianum is higher than that in the hybrid P. rothschildianum × P. micranthum (Table 4). Though both P. micranthum and P. delenatii belong to subgenus Parvisepalum and demonstrate a high homology between their genomes (Fig. 2A–C and Table 4), they showed different meiotic chromosome configurations and GISH performance when crossed with P. rothschildianum (Figs. 2G–I and 3A–C and Table 4). Breeding records also show that it is easier to get progenies from the hybrid of P. delenatii × P. rothschildianum than from the hybrid of P. rothschildianum × P. micranthum when backcrossing to the parent. The success in genome differentiation by GISH in the hybrid P. delenatii × P. rothschildianum (Fig. 2G–I) also reflects that the genomic components and organizations of these two parental species must be substantially different. In spite of their disparity in genomic components and organizations, homoeologous pairing frequency was high in their hybrids. These results suggest that chromosome pairing in Paphiopedilum hybrids is attributed to not only the genome homology but also to the interaction of genetic factors. One possible explanation is that the genetic factors required for chromosome pairing in P. micranthum and P. rothschildianum are incompatible. Kopecký et al. (2008b) analysed meiotic behaviour of the Lolium–Festuca hybrids by GISH and suggested that, although the repetitive DNA sequences might diverge markedly during evolution of the two genera, the sequences involved in chromosome pairing were conserved enough to facilitate regular pairing partner recognition and crossing-over.
Quadrivalents were found in the hybrids P. delenatii × P. glaucophyllum (Fig. 2M–O) and P. rothschildianum × P. moquetteanum (Fig. 3G–I). It is worthy to note that the total number of chromosome arms of section Cochlopetalum (P. moquetteanum and P. glaucophyllum) would account for an n.f. of 50 rather than 52. It is proposed that the ‘ancestral’ karyotype has lost either a single metacentric or two telocentric chromosomes prior to divergence of extant species in this section (Cox et al., 1998). The lack of a single metacentric or two telocentric chromosomes in section Cochlopetalum genomes may hamper the normal chromosome association with the other species with an n.f. of 52 during meiosis.
Chromosome pairing of the hybrid progenies
Two kinds of homeologous pairing were displayed in these Paphiopedilum hybrids by GISH. Most of the pairings involved chromosomes from different parental genomes (allosyndetic pairing) and some involved chromosomes within the same genome (autosyndesis). Depending upon the time of evolutionary divergence, related species may have highly differentiated homoeologous genomes. Therefore, the homoeologous chromosome pairings may be attributable to accumulated changes in the chromosomal structure through evolution (Armstrong and Keller, 1982) or to genetic regulations on homoeologous pairing (Prakash, 1974), such as the Ph1 locus in wheat (Triticum aestivum) (Griffiths et al., 2006). The homoeologous chromosome pairings indicate the relative affinities between the parental genomes of the hybrids. Using classical chromosome staining methods, it would not be possible to identify the origin of chromosomes involved in each meiotic configuration (i.e. univalent, bivalent or other multivalents). GISH reveals the chromosomal composition of the meiotic configurations and, thus, determines the actual meiotic affinities of the respective genomic components between parental species (allosyndesis) and may indicate the existence of duplicated genomes in these species (autosyndesis).
In this study, autosyndesis was observed in the hybrids P. delenatii × P. rothschildianum, P. delenatii × P. callosum, P. delenatii × P. glaucophyllum (Fig. 2H, K, M) and P. rothschildianum × P. moquetteanum (Fig. 3H). Autosyndetic pairing suggested the existence of genome duplication in these Paphiopedilum species. Trivalents or quadrivalents observed in these Paphiopedilum hybrids (Table 4) suggested that translocations have been involved in the differentiation of Paphiopedilum species.
The number and distribution of 45S rDNA loci may vary among closely related species (see references in Chung et al., 2008). According to a previous examination by FISH, each parental species used in this study has one 45S rDNA locus (Lee and Chung, 2008). Two rDNA loci in a separated univalent were detected in each of Paphiopedilum interspecific hybrids investigated here, suggesting that chromosomes with 45S rDNA loci in these Paphiopedilum species were less homologous.
In conclusion, GISH analysis is an efficient method to visualize cytologically the genomic affinities and homeologous differentiations in Paphiopedilum species. Therefore, GISH may help in studying the phylogenetic relationships among Paphiopedilum species and in tracing the origin and the introgression of parental genomes of interspecific hybrids. The present results indicate that chromosome pairing in Paphiopedilum hybrids is subject to genome homology and the interaction of genetic factors, while chromosome number and karyotype similarity are less involved. In addition to centric fission, which is known as the major mechanism involved in karyotype evolution in the genus Paphiopedilum, the present results indicated that other structural rearrangements may also play roles in differentiating Paphiopedilum species at chromosomal level.
ACKNOWLEDGEMENTS
This work was supported by Academia Sinica, Taiwan, ROC to Mei-Chu Chung [94S-1503] and National Science Council, Taiwan, ROC [NSC 99–2313-B-178-001] and National Museum of Natural Science, Taiwan, ROC to Yung-I Lee.
LITERATURE CITED
- Anamthawat-Jonsson K, Reader SM. Preannealing of total genomic DNA probes for simultaneous genomic in situ hybridization. Genome. 1995;38:814–816. doi: 10.1139/g95-104. [DOI] [PubMed] [Google Scholar]
- Anamthawat-Jonsson K, Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS. Discrimination between closely related Triticeae species using genomic DNA as a probe. Theoretical and Applied Genetics. 1990;79:721–728. doi: 10.1007/BF00224236. [DOI] [PubMed] [Google Scholar]
- Aoyama M, Kojima K, Kobayashi M. Morphology of microspores in Phalaenopsis F1 hybrids. Kinki Chugoku Agricultural Research. 1994;88:49–53. [in Japanese] [Google Scholar]
- Armstrong KC, Keller WA. Chromosome pairing in haploids of Brassica oleracea. Canadian Journal of Genetics and Cytology. 1982;24:735–739. [Google Scholar]
- Cao M, Sleper DA, Dong F, Jiang J. Genomic in situ hybridization (GISH) reveals high chromosome pairing affinity between Lolium perenne and Festuca mairei. Genome. 2000;43:398–403. [PubMed] [Google Scholar]
- Chung MC, Lee YI, Cheng YY, Chou YJ, Lu CF. Chromosomal polymorphism of ribosomal genes in the genus Oryza. Theoretical and Applied Genetics. 2008;116:745–753. doi: 10.1007/s00122-007-0705-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AV, Pridgeon AM, Albert VA, Chase MW. Phylogenetics of the slipper orchids (Cypripedioideae: Orchidaceae): nuclear rDNA ITS sequences. Plant Systematics and Evolution. 1997;208:197–223. [Google Scholar]
- Cox AV, Abdelnour GJ, Bennett MD, Leitch IJ. Genome size and karyotype evolution in the slipper orchids (Cypripedioideae:Orchidaceae) American Journal of Botany. 1998;85:681–687. [PubMed] [Google Scholar]
- Cribb P. The genus Paphiopedilum. 1998 Kew: Royal Botanical Gardens/Borneo: Natural History Publications. [Google Scholar]
- Duncan RE, Macleod RA. The chromosomes of the continental species of Paphiopedilum with solid green leaves. American Orchid Society Bulletin. 1949;18:84–89. [Google Scholar]
- Duncan RE, Macleod RA. The chromosomes of Eremantha tesselata. American Orchid Society Bulletin. 1950;19:137–142. [Google Scholar]
- Gerlach WL, Bedbrook JR. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Research. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Sharp R, Foote TN, et al. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploidy wheat. Nature. 2006;439:749–752. doi: 10.1038/nature04434. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T, Leitch AR, Anamthawat-Jonsson K, Bennett MD. A method of identifying DNA sequences in chromosomes of plants. 1988 European patent application number 8828130·8 (2 December 1988; see http://worldwide.espacenet.com/numberSearch?locale=en_EP. ) [Google Scholar]
- Jones K. Robertsonian fusion and centric fission in karyotype evolution of higher plants. Botanical Review. 1998;64:237–289. [Google Scholar]
- Kao FI, Cheng YY, Chow TY, et al. An integrated map of Oryza sativa L. chromosome 5. Theoretical and Applied Genetics. 2006;112:891–902. doi: 10.1007/s00122-005-0191-0. [DOI] [PubMed] [Google Scholar]
- Karasawa K. Karyomorphological studies on nine taxa of Paphiopedilum. Bulletin of the Hiroshima Botanical Garden. 1986;8:23–42. [Google Scholar]
- Karasawa K, Saito K. A revision of the genus Paphiopedilum (Orchidaceae) Bulletin of the Hiroshima Botanical Garden. 1982;5:1–69. [Google Scholar]
- Kopecký D, Lukaszewski AJ, Doležel J. Cytogenetics of Festulolium (Festuca × Lolium hybrids) Cytogenetic and Genome Research. 2008a;120:370–383. doi: 10.1159/000121086. [DOI] [PubMed] [Google Scholar]
- Kopecký D, Lukaszewski AJ, Dolezel J. Meiotic behaviour of individual chromosomes of Festuca pratensis in tetraploid Lolium multiflorum. Chromosome Research. 2008b;16:987–998. doi: 10.1007/s10577-008-1256-0. [DOI] [PubMed] [Google Scholar]
- Lee YI, Chung MC. Identification of genome relationships among Paphiopedilum species by genomic and fluorescent in situ hybridization. Acta Horticulturae. 2008;766:331–334. [Google Scholar]
- Lim KB, Ramanna MS, de Jong JH, Jacobsen E, van Tuyl JM. Indeterminate meiotic restitution (IMR): a novel type of meiotic nuclear restitution mechanism detected in interspecific lily hybrids by GISH. Theoretical and Applied Genetics. 2001;103:219–230. [Google Scholar]
- MacFarlane SWH, Jones JK, Sebastiampillai AR. Pollen storage of Fragaria and Potentilla. Euphytica. 1989;41:65–69. [Google Scholar]
- Maluszynska J, Hasterok R. Identification of individual chromosomes and parental genomes in Brassica juncea using GISH and FISH. Cytogenetic and Genome Research. 2005;109:310–314. doi: 10.1159/000082414. [DOI] [PubMed] [Google Scholar]
- Mathey T. Les chromosomes des vertebres. Lausanne: F. Rouge; 1949. [Google Scholar]
- Prakash S. Haploid meiosis and origin of Brassica tournefortii Gouan. Euphytica. 1974;23:591–595. [Google Scholar]
- Schubert I, Fransz PF, Fuchs J, de Jong JH. Chromosome painting in plants. Methods in Cell Science. 2001;23:57–69. [PubMed] [Google Scholar]
- Schwarzacher T, Heslop-Harrison JS. Practical in situ hybridization. Oxford: Bio Scientific Publishers; 2000. [Google Scholar]
- Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS. In situ localization of parental genomes in a wide hybrid. Annals of Botany. 1989;64:315–324. [Google Scholar]
- Singh RJ. Plant cytogenetics. 2nd edn. Boca Raton, F: CRC Press; 2003. pp. 277–306. [Google Scholar]