Abstract
The first full genome sequences were established in the mid 1990s. Shortly thereafter, genome-scale metabolic network reconstructions appeared. Since that time, we have witnessed an exponential growth in their number and uses. Here I discuss, from a personal point of view, four topics: 1) the placement of metabolic systems biology in the context of broader scientific developments, 2) its foundational concepts, 3) some of its current uses, and 4) some of the expected future developments in the field.
1. Metabolic Systems Biology in the Grand Scheme of Things
Ever since Gregor Mendel discovered discrete quanta of information passed from one generation to the next that determined form and function of an organism, the genotype-phenotype relationship has been of fundamental importance in the life sciences. For monogeneic traits, the genotype-phenotype relationship can be readily understood. However, most phenotypic traits involve multiple gene products. This makes the genotype-phenotype relationship a challenge to reconstruct and understand, given the complex interactions that can form among the gene products.
With the publication of the first full genome sequences in the mid-1990s [1] it became possible, in principle, to identify all the gene products involved in complex biological processes in a single organism. The well-studied biochemistry of metabolic transformations made it possible to reconstruct, on a genome-scale, metabolic networks for a target organism in a biochemically detailed fashion [2,3]. Such metabolic network reconstructions can be converted into a mathematical format yielding mechanistic genotype-phenotype relationships for microbial metabolism [4]. The mathematical format of the underlying biochemical, genetic, and genomic (BiGG) knowledge allows the formulation of genome-scale models (GEMs). GEMs enable the computation of phenotypic traits based on the genetic composition of the target organism [4,5].
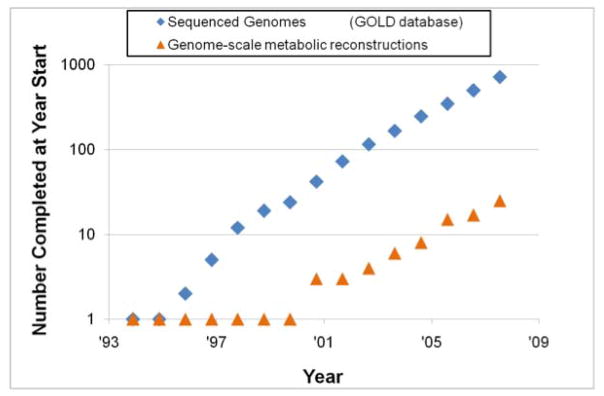
Since the establishment of the first metabolic genome-scale reconstruction in 1999 and in silico models thereof, many more have followed (Figure 1); perhaps most notably for human metabolism in 2007. The scope and content of network reconstructions continues to grow, for instance to include the entire transcription/translation apparatus of a cell [6] and the structural information about the metabolic enzymes [7].
Figure 1.
Growth of genome sequences and genome-scale metabolic reconstructions. The number of network reconstruction has grown exponentially, at a similar pace as genome sequences have appeared [Prepared by Adam Feist and Ines Thiele].
2. Foundations of Metabolic Systems Biology
2.1 The basic paradigm
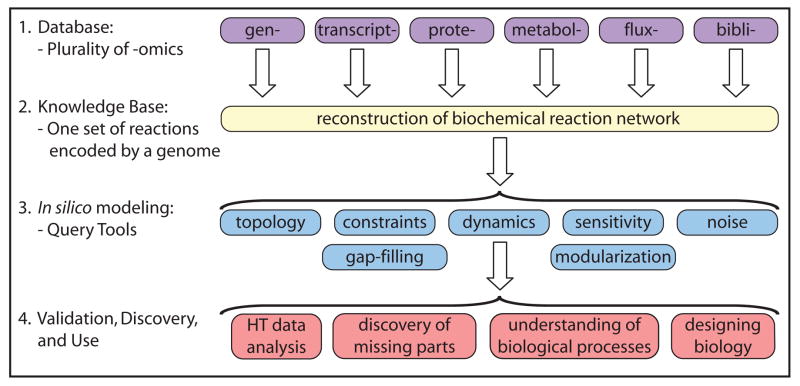
We can now enumerate various cellular components, describe their interactions chemically, formulate a mathematical description of the totality of such interactions, identify the constraints that the resulting network operates under, and apply optimality principles to evaluate likely physiological functions in a given environment. These capabilities provide a consistent framework on which a mechanistic basis for the microbial metabolic genotype-phenotype relationship can be formulated. The underlying process is based on an emerging paradigm to relate the genotype to the phenotype through reconstruction and in silico model building (Figure 2) is comprised of four steps:
Figure 2.
The four-step paradigm for metabolic systems biology. Adapted from [4]. Prepared by Adam Feist.
Generation of ‘omics’ and collection of literature data on the target organism;
Network reconstruction and the formulation of a BiGG knowledge base;
Conversion of the reconstruction into a mathematical format and the implementation of in silico query tools; and
Enablement of a variety of basic and applied uses.
This fundamental paradigm allows for the first time the genome-scale computation of phenotypic functions of an organism. The establishment of a mechanistic formulation of the most fundamental relationship in biology - the genotype-phenotype relationship - for a limited number of phenotypic functions, differs from that developed for the basic physical laws about a century ago. This mechanistic description must account for both proximal and distal causation. Proximal (or proximate) causation occurs against a fixed genetic background i.e., an individual organism), while distal (or ultimate) causation results from (genomic) changes that occur from generation to generation, i.e., evolution.
2.2 Some basic principles
This basic paradigm has been implemented for a number of organisms and a variety of biological results have been obtained [8,9]. As a result of this successful reduction to practice, one is tempted to try to determine and state the underlying reasons for this success. Below, I attempt to start this process.
Axiom #1: All cellular functions are based on chemistry. A simple but consequential statement, as it implies the fundamental events in a cell can be described by chemical equations. These equations, in turn, come with chemical information and physico-chemical principles.
Axiom #2: Annotated genome sequences along with experimental data enable the reconstruction of genome-scale metabolic networks. The reconstruction process is a grand-scale systematic assembly of information in a quality controlled/quality assured (QC/QA) setting [10] that leads to a BiGG knowledge base, which is a collection of established Biochemical, Genetic, and Genomic data represented by a network reconstruction. The reconstruction process has been reviewed elsewhere [11,12], and a growing number of reconstructions are available (Figure 1).
Axiom #3: Cells function in a context-specific manner. When a cell is placed in a particular environment, it expresses a subset of its genes in response to environmental cues. The abundance of cellular components can be profiled using ‘omic’ methods (ie. transcriptomics, proteomics, metabolomics). Such omic data can be mapped onto a network reconstruction to tailor it to the particular condition being considered.
Axiom #4: Cells operate under a series of constraints. Factors constraining cellular functions fall into four principal categories [4]: physico-chemical (see Axiom #5), topological (molecular crowding effects and steric hindrance), environmental (Axiom #3), and regulatory (basically self-imposed constraints, or restraints). These constraints cannot be violated allowing the estimation of all functional (i.e., physiological) states that a genome-scale reconstruction can achieve. Mathematically, such statements are translated into fundamental subspaces associated with the stoichiometric matrix (S), whose properties can be characterized [4]. In this so-called S matrix, where S stands for stoichiometric, the rows correspond to the network metabolites and the columns to the network reactions. The coefficients of the substrates and products of each reaction are entered in the corresponding cell of the matrix.
Axiom #5: Mass (and energy) is conserved. This statement is one of the basic physical laws. Since all proper chemical equations can be described by stoichiometric coefficients, and since a set of chemical equations can be described by the stoichiometric matrix, S, this means that all steady states (normally close to the homeostatic states of interest) of a network can be described by a simple linear equation, S•v=0, where v is a vector of fluxes through chemical reactions [4]. Thus, the computation of functional states of a network is enabled based on the known underlying chemistry.
Axiom #6: Cells evolve under a selection pressure in a given environment. This Darwinian statement has implicit optimality principles built into it. Consequently, if e know the selection pressure, we can state a so-called objective function and determine optimal states given a network reconstruction and governing constraints.
Each one of these statements by themselves is almost trivial and accepted in various scientific disciplines as being fundamental. Taken together, though, they combine to form the conceptual basis for constraint-based reconstruction and analysis (COBRA), and enable the development of the mechanistic genotype phenotype relationship for metabolism. The recent emergence of genome-scale reconstructions (axiom #2) has proven key to this formulation.
2.3 Practicing COBRA
The COBRA approach [4,5] has been widely used to analyze network reconstructions. It uses stoichiometric information about biochemical transformations taking place in a target organism to construct the model. While a metabolic reconstruction is unique to the target organism, one can derive many different condition-specific models from a single reconstruction. This conversion of a metabolic reconstruction of an organism into models requires the imposition of physicochemical and environmental constraints to define systems boundaries. The conversion also includes the transformation of the reaction list into a computable, mathematical matrix format (e.g., using the COBRA toolbox [13]).
Formulation of BiGG knowledge-bases
The four-step procedure to reconstruct genome-scale metabolic networks has been detailed elsewhere [11,12,14]. Briefly, all metabolic functions encoded by an organism’s genome are systematically retrieved, curated, and translated into a list of biochemical reactions that comprise the network. The association between the biochemical reactions and the catalyzing gene products is achieved using Boolean logic through the gene-protein-reaction (GPR) associations [12]. Further metabolic functions supported by experimental data can be included without gene association. Extensive QA/QC procedures ensure that the BiGG knowledge base is self-consistent, comprehensive, and exhibits similar physiological properties as the target organism [10]. A BiGG knowledge-base effectively represents a 2- dimensional annotation of a genome [15] and represents the implementation of axioms #1 and #2.
From a knowledge-base to a GEM
BiGG knowledge-bases can be converted into GEMs (genome-scale models) by implementing axioms #3, #4, and #5. While BiGG accounts for genome-scale information, a GEM can represent the capacities of a cell in a particular environmental and genetic state. Subsequently, there are different possible GEMs that can be derived from a given organism’s reconstruction. In this conversion, the BiGG knowledge-base is represented in a mathematical format, S.
System boundaries are defined around the entire reaction network. Exchange reactions are added to all transportable extracellular metabolites and can be constrained in simulations to represent different environmental conditions. Demand reactions are added, including the biomass reaction, that details all precursors and their fractional contributions to a cell’s macromolecular composition, as well as any maintenance energy requirements [16].
Computational tools
A broad spectrum of methods has been developed under this umbrella [4,5,14], collectively called COBRA methods. Constraint-based Metabolic models can be imported into Matlab in SBML format. COBRA methods can then be applied [13]. With a growing number of metabolic reconstructions available (see http://systemsbiology.ucsd.edu/In_Silico_Organisms/Other_Organisms) and the accessibility of COBRA tools, the number of practitioners in this field is growing.
In a recent review [8], the uses of the E. coli GEM were classified into five categories: 1) metabolic engineering (i.e., genome-scale synthetic biology), 2) gap-filling (i.e., systematic generation of hypothesis), 3) phenotypic screens (data analysis), 4) determining network properties (in silico systems biology) and 5) evolutionary studies (i.e., COBRA methods can analyze both proximal and distal causation). Similarly, a review of applications to other organisms, [9], classifies them as: 1) contextualization of high-throughput data, 2) guidance of metabolic engineering, 3) directing hypothesis-driven discovery, 4) interrogation of multispecies relationships, and 5) network property discovery. Thus, the basic and applied uses of reconstructions and associated GEMs are growing.
3. Biological Science in the Era of Systems Biology
Genome-scale reconstructions enable biological science to proceed in fundamentally new ways. Here I discuss four new possibilities.
3.1 Integration of high-throughput data
Omics data can be analyzed using a reconstruction as a scaffold. As stated above, a reconstruction is a BiGG knowledge-base, and if ‘omics’ data are mapped onto the reconstruction, it enables the analysis of the ‘omics’ data against the curated knowledge about the target organism as a context.
Several examples of this use of reconstructions have been demonstrated. Tissue-specific expression profiling data has been mapped against the reconstruction of the global human metabolic map [17] to yield draft reconstructions of tissue-specific metabolic networks in humans. Proteomic data from the human cardiac mitochondria has been used to form tissue-specific organelle models [18] and used for the analysis of SNPs through the use of cosets [19]. The use of reconstructions to analyze expression-profiling data from E. coli in many studies has been reviewed recently [20]. In a similar fashion, metabolomic and fluxomic data can be analyzed, e.g., see [21].
Thus, curated genome-scale reconstructions provide a new way to analyze ‘omics’ data. This ability is likely to help with revealing the information content in an ‘omics’ data set, as purely statistical approaches have proven to be somewhat limited.
3.2 Gap-filling
BiGG knowledge bases are not complete; they have ‘gaps’ in them. These gaps come in at least two fundamental varieties. First, there can be a missing reaction or a path between two metabolites in the reconstruction, and secondly, a metabolite can be detected that has no connections to the network – representing an ‘island’ on the metabolic map. The former can be filled with a gap-filling process [22]. Missing links have been discovered this way [23] and the methods developed to date for gap-filling have been described [24]; the latter calls for the discovery of a new pathway. Computational tools for suggesting missing pathways have been recently described [25,26].
These methods may be viewed as representing interesting computational and algorithmic challenges. However, these developments are more profound and fundamental. They represent the computational generation of hypotheses using genome-scale BiGG knowledge-bases. The reconstruction is used as a context for analyzing data and determining a candidate explanation for discrepancies between experimental data and computational predictions. In fact, mixed-integer linear programming (MILP) algorithms represent the generation of the ‘most parsimonious’ hypothesis as they can be used to grade the complexity of the candidate explanations. In the initial study of this kind, the simplest explanation was always found to be the correct one [22].
3.3 Understanding complex biological phenomena
Phenotypic functions rely on the coordinated and simultaneous action of multiple gene products. This makes complex biological processes hard to comprehend. In addition, with changes over generations, such comprehension may be even more challenging. COBRA tools enable the computation of both proximal and distal causation at a genome-scale, and thus possesses the potential to provide a framework for a deep understanding of complex biological phenomena.
This expectation is perhaps being realized through work on bacterial adaptation. The ability of GEMs to predict the outcome of bacterial adaptation to new nutritional environments [27], even in the face of gene deletions [28], opens up new and fascinating avenues to study fundamental biological phenomena. The network level predictions are made using GEMs and phenotypic functions. Fortunately, the third generation sequencing methods enable the full resequencing of microbial genomes following adaptation [29,30]. With available allelic replacement methods the causality of the mutations can be assessed [30,31].
The convergence of these developments, COBRA, GEMs, cheap re-sequencing, and allelic replacement, allows the study of evolution in a laboratory setting. This fascinating prospect is likely to help us understand the plasticity and functions of bacterial genomes better than before. For instance, the RNA polymerase has been shown to be a highly mutable enzyme [31] and we are now beginning to determine the objective functions that wild type strains seem to adhere to [32]. Taken together, this means that teleology could be studied in an experimental setting.
Thus, metabolic systems biology is enabling a totally new scientific pursuit. Although the application of COBRA methods has been to bacterial adaptation, one might expect similar applications to pathogenic processes. For instance, carcinogenesis has been conceptualized as the alleviation of a series of constraints [33] and metabolism is now seen as an integral part of this process [34]. Thus, the analysis of the adjustment of metabolic processes to enable this pathologic state is likely to be possible using COBRA methods.
3.4. Metabolic engineering
Microbial metabolism has been and is being modified to achieve practical ends through willful genetic manipulation of a wild type organism to generate a production strain. This field is called metabolic engineering; with a foundational paper by JE Bailey appearing in 1991 [35]. A recent review describes three phases in the history of metabolic engineering [36]: first is the use of random mutagenesis and screening; second, the use of targeted genetic manipulations to achieve ‘local’ results in the function of a network; and third, the use of GEMs to perform ‘global,’ or genome-scale analysis of genetic manipulations. Algorithms have been developed to perform the computation of genetic changes to achieve such global results [37–39].
The engineering dictum, ‘there is nothing more practical than reliable theory,” is at the foundation of engineering design and practice. Good computational models accelerate design processes, and minimize prototyping, testing, and experimentation. The fact that the third phase in metabolic engineering has been ushered in through the use of GEMs signifies that we are now beginning to contemplate and practice synthetic biology at the genome-scale. Many commercial enterprises that are developing sustainable technologies benefit from this capability.
4. Beyond Metabolism in Microbes
With the successes of metabolic systems biology in microbes and the statement of some of its underlying axioms, it is natural to wonder what will happen next in this field. The foray into human metabolism seems like an obvious extension with the clear challenges of greater organismic complexity and function. i.e., what are the objectives of various metabolic functions in man?
We have already shown that similar bottom-up reconstructions of the transcription-translation machinery can be achieved [6]. Given the chemically detailed representation of this process on a genome-scale, COBRA approaches could be used for the analysis of systems properties. Several chemically detailed reconstructions of signaling systems have appeared [6,40] that could be analyzed using COBRA tools. New genome-scale data types (transcription start sites, tiled arrays for expression profiling, ChIP-chip, and proteomic methods) are now yielding the data that is needed to reconstruct transcriptional regulatory networks at a genome-scale [41]. Other data types, such as phosphoproteomics [42,43] and rapid metabolic regulatory mechanisms [44] will expand the scope of reconstruction of regulatory phenomena.
5. Closing
The emergences of mechanistic genotype-phenotype relationships that can account for dual causality are likely to have a broad impact on the life sciences. A number of metabolic networks have already been reconstructed for target organisms. New biological science is being performed with these reconstructions. The new avenues that have been opened up by COBRA tools and a growing number of GEMs are probably just at the early stages of their exploration.
This piece is written as a personal account of the development of the field of genome-scale systems biology of metabolism. It is not meant to be a comprehensive view of the field but a discussion of the four topics stated in the summary. Although most of the references are to the author’s own work, many of them are reviews citing the major developments in the field.
Acknowledgments
The author is thankful of the many individuals that have entered the COBRA field and contributed significantly to it. Early adopters include Jens Nielsen, Sang Yup Lee, Costas Maranas, Steve Oliver, Bas Teusink, George Church, and Daniel Segre, entering the field in the early 2000s. Since then, many have followed, making this a vibrant and growing field with an increasing list of accomplishments. The author has been supported by the NIGMS, NIAID, NHRI, and the DOE, and is a co-founder of Genomatica.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–498. 507–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Edwards JS, Palsson BO. Systems properties of the Haemophilus influenzae Rd metabolic genotype. Journal of Biological Chemistry. 1999;274:17410–6. doi: 10.1074/jbc.274.25.17410. [DOI] [PubMed] [Google Scholar]
- 3.Edwards JS, Palsson BO. The Escherichia coli MG1655 in silico metabolic genotype: Its definition, characteristics, and capabilities. Proc Natl Acad Sci U S A. 2000;97:5528–5533. doi: 10.1073/pnas.97.10.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palsson BO. Systems biology: properties of reconstructed networks. Cambridge University Press; New York: 2006. [Google Scholar]
- 5.Price ND, Reed JL, Palsson BO. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Microbiol. 2004;2:886–897. doi: 10.1038/nrmicro1023. [DOI] [PubMed] [Google Scholar]
- 6.Thiele I, Jamshidi N, Fleming RM, Palsson BO. genome-scale reconstruction of Escherichia coli’s transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization. PLoS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Thiele I, Palsson BO, Ostermann A, Godzik A. Structural genomics provides an atomic level view of the metabolic network of Thermotoga maritima. Science. 2009 doi: 10.1126/science.1174671. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feist AM, Palsson BO. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotech. 2008;26:659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papin JA, Oberhardt M, Palsson BO. Applications of Genome-Scale Metabolic Reconstructions. Molecular Systems Biology. 2009 doi: 10.1038/msb.2009.77. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nature Protocols. 2009 doi: 10.1038/nprot.2009.203. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feist AM, Herrgard MJ, Theile I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol. 2009;7:129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nat Rev Genet. 2006;7:130–41. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- 13.Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, Herrgard MJ. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox. Nat Protocols. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 14.Durot M, Bourguignon PY, Schachter V. Genome-scale models of bacterial metabolism: reconstruction and applications. FEMS Microbiol Rev. 2009;33:164–190. doi: 10.1111/j.1574-6976.2008.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palsson BO. Two-dimensional annotation of genomes. Nat Biotechnol. 2004;22:1218–9. doi: 10.1038/nbt1004-1218. [DOI] [PubMed] [Google Scholar]
- 16.Feist AM, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3 doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat Biotechnol. 2008;26:1003–10. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- 18.Vo TD, Greenberg HJ, Palsson BO. Reconstruction and functional characterization of the human mitochondrial metabolic network based on proteomic and biochemical data. J Biol Chem. 2004;279:39532–40. doi: 10.1074/jbc.M403782200. [DOI] [PubMed] [Google Scholar]
- 19.Jamshidi N, Palsson BO. Systems biology of SNPs. Mol Syst Biol. 2006;2:38. doi: 10.1038/msb4100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis NE, Cho BK, Knight EM, Palsson BO. Gene expression profiling and the use of genome-scale in silico models of Escherichia coli for analysis: providing context for content. J Bacteriol. 2009;191:3437–44. doi: 10.1128/JB.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamshidi N, Palsson BO. Formulating genome-scale kinetic models in the post-genome era. Mol Sys Biol. 2008;4:171. doi: 10.1038/msb.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed JL, et al. Systems approach to refining genome annotation. Proc Natl Acad Sci U S A. 2006;103:17480–4. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuhrer T, Chen L, Sauer U, Vitkup D. Computational prediction and experimental verification of the gene encoding the NAD+/NADP+-dependent succinate semialdehyde dehydrogenase in Escherichia coli. J Bacteriol. 2007;189:8073–8078. doi: 10.1128/JB.01027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breitling R, Vitkup D, Barrett MP. New surveyor tools for charting microbial metabolic maps. Nat Rev Microbiol. 2008;6:156–61. doi: 10.1038/nrmicro1797. [DOI] [PubMed] [Google Scholar]
- 25.Hatzimanikatis V, Li C, Ionita JA, Henry CS, Jankowski MD, Broadbelt LJ. Exploring the diversity of complex metabolic networks. Bioinformatics. 2005;21:1603–9. doi: 10.1093/bioinformatics/bti213. [DOI] [PubMed] [Google Scholar]
- 26.Kumar VS, Maranas CD. GrowMatch: An Automated Method for Reconciling In Silico/InVivo Growth Predictions PLoS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ibarra RU, Edwards JS, Palsson BO. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–9. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- 28.Fong SS, Palsson BO. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat Genet. 2004;36:1056–58. doi: 10.1038/ng1432. [DOI] [PubMed] [Google Scholar]
- 29.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–32. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 30.Herring CD, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 31.Conrad T, Palsson BO. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009 doi: 10.1186/gb-2009-10-10-r118. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer E, Sauer U. Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat Genet. 2005;37:636–40. doi: 10.1038/ng1555. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 34.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey JE. Toward a science of metabolic engineering. Science. 1991;252:1668–75. doi: 10.1126/science.2047876. [DOI] [PubMed] [Google Scholar]
- 36.Park JH, Lee SY. Towards systems metabolic engineering of microorganisms for amino acid production. Curr Opin Biotech. 2008;19:454–460. doi: 10.1016/j.copbio.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Burgard AP, Pharkya P, Maranas CD. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84:647–57. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 38.Patil KR, Rocha I, Forster J, Nielsen J. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinformatics. 2005;6:308. doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pharkya P, Burgard AP, Maranas CD. OptStrain: a computational framework for redesign of microbial production systems. Genome Res. 2004;14:2367–76. doi: 10.1101/gr.2872004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papin JA, Palsson BO. The JAK-STAT Signaling Network in the Human B-Cell: An Extreme Signaling Pathway Analysis. Biophysical Journal. 2004;87:37–46. doi: 10.1529/biophysj.103.029884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho BKKZ, Qiu Y, Knight EM, Park YS, Barrett CL, Gao Y, Palsson BO. Elucidation of Meta-structure of a Bacterial Genome. Nat Biotechnol. 2009 Submitted. [Google Scholar]
- 42.Macek B, Mann M, Olsen JV. Global and site-specific quantitative phosphoproteomics: principles and applications. Annu Rev Pharmacol Toxicol. 2009;49:199–221. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- 43.Soufi B, Jers C, Hansen ME, Petranovic D, Mijakovic I. Insights from site-specific phosphoproteomics in bacteria. Biochim Biophys Acta. 2008;1784:186–92. doi: 10.1016/j.bbapap.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 44.Ralser M, Wamelink MM, Latkolik S, Jansen EE, Lehrach H, Jakobs C. Metabolic reconfiguration precedes transcriptional regulation in the antioxidant response. Nat Biotechnol. 2009;27:604–5. doi: 10.1038/nbt0709-604. [DOI] [PubMed] [Google Scholar]