Abstract
The fungal peptidyl alkaloids of the tryptoquialanine and fumiquinazoline families are nonribosomally assembled by annulation of the indole side chain of fumiquinazoloine F (FQF) with an alaninyl or aminoisobutyryl unit by monomodular NRPS enzymes containing adenylation, thiolation and condensation (A-T-C) domains. The enzyme pair Af12060 and Af12050 from Aspergillus fumigatus thereby convert FQF to FQA while the homologous enzyme pair TqaH and TqaB from Penicillium aethiopicum make the 2′-epi diastereomer of FQA, differing only in the stereochemistry of one of the C-N bonds formed in the annulation with L-Ala. To evaluate the basis for this stereochemical control we have mixed and matched the flavoprotein oxygenases Af12060 and TqaH with the A-T-C modular enzymes Af12050 and TqaB to show that the NRPS enzymes control the stereochemical outcome. The terminal 50 kDa condensation domains of Af12050 and TqaB are solely responsible for the stereochemical control as shown both by making chimeric (e.g. A-T-C* and A*-T*-C ) forms of these monomodular NRPS enzymes and also by expression, purification and assay of the excised C domains. The Af12050 and TqaB condensation domains are thus a paired set of diastereospecific annulation catalysts that act on the fumiquinazoline F scaffold.
Nonribosomal peptide synthetase (NRPS) assembly lines are prevalent in the biosynthesis of secondary metabolites among a wide variety of bacterial and fungal species. Fungi have been postulated and shown to produce a variety of compounds of intriguing biological activity and structure by NRPS based mechanisms. The peptaibols, a subgroup of peptaibiotics, have been isolated from a variety of soil-dwelling fungi and shown to exhibit antibacterial, antifungal and anti-cancer properties (1-3). Clavicipitaceae family fungi, notably Claviceps purpurea produce a group of multi-cyclic peptides appended to a core structure of D-lysergic acid, several of which (or synthetic derivatives thereof) are in use as agents to treat a number of disorders of the vascular and central nervous systems (4, 5). A group of prenylated indole alkaloids derived from the diketopiperazine brevianamide F (cyclo-L-Trp-L-Pro) including the tryptostatins, which are detected in Aspergillus fumigatus extracts, have been used as promising candidates for the development of anti-cancer drugs (6, 7). Members of the tryptostatin family are also known tremorgenic mycotoxins, which act on the central nervous system of vertebrate animals that feed on food-stocks contaminated with tremorgen-producing molds and can result in tremors, seizures and convulsions (8, 9). Another tremorgenic mycotoxin tryptoquialanine from Penicillium aethiopicum, has recently been shown to be produced by a fungal NRPS-based system (10). Insight into the biosynthesis of tryptoquialanine revealed pathway intermediates of remarkable similarity to the fumiquinazolines, a group of cytotoxic metabolites isolated from the opportunistic human pathogen A. fumigatus (11, 12). These examples highlight the importance of NRPS based mechanisms for the production of an arsenal of secondary metabolites by fungi, thus making their study of potentially vast intellectual and medicinal value.
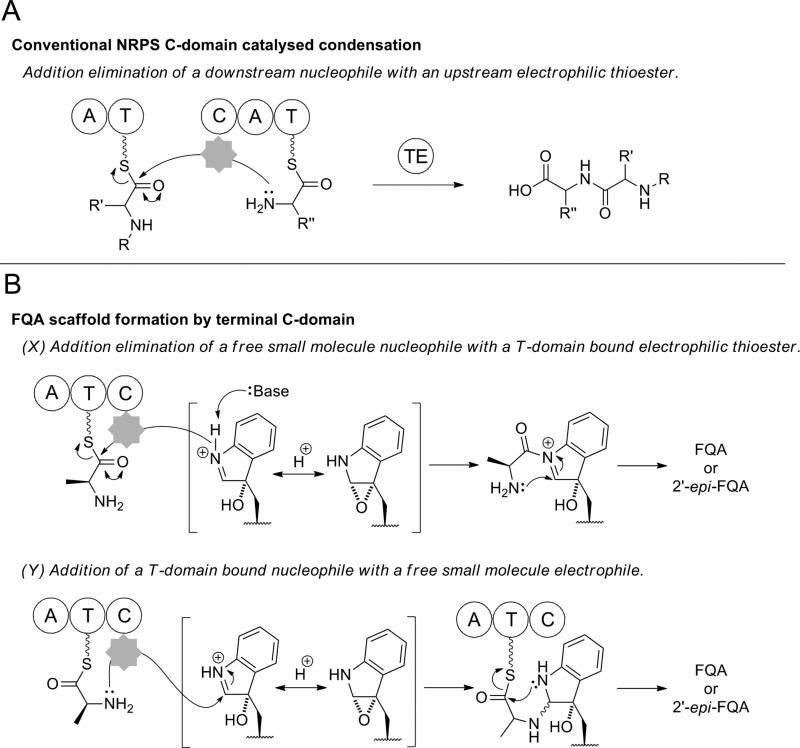
NRPSs are often found as multi-modular megasynthases, where a single massive protein comprised of multiple catalytic domains is involved in the biosynthesis of large peptidyl scaffolds. Minimally a single NRPS module consists of adenylation (A), carrier protein/thiolation (T) and condensation (C) domains. A-domain catalyzed activation of an amino acid unit as the corresponding acyl-AMP mediates the attachment to the 4′-phosphopantetheine (ppt) prosthetic arm of the T-domain to form an acyl-thioester intermediate. The T domain-tethered acyl thioester is then delivered to the active site of a C-domain, which catalyses amide bond formation (13-15). In bacterial NRPSs, the completed peptide is normally transferred to an active site serine of a terminal thioesterase (TE) domain, which mediates chain release by hydrolysis or macrocyclization. A noticeably distinct feature of most fungal NRPSs is the replacement of the TE domain with a terminal condensation, reductive, or thiolation domain (16, 17).
Such terminal C-domains of fungal NRPSs may catalyse the chain release of the peptide via cyclization by mediating the attack of intramolecular or intermolecular nucleophiles. Terminal C-domain mediated cyclizations have been proposed to be key termination steps in the biosynthesis of several fungal NRPS products. The terminal C-domain of the bimodular NRPS (ftmA), which has been shown to be sufficient for the heterologous production of the tryprostatin precursor brevianamide F, appears likely to control the concomitant cyclization and cleavage of the cyclo-L-Trp-L-Pro product (18). Terminal C-domains of fungal NRPSs have also been postulated to control the macrocyclization of large rings during the biosynthesis of cyclosporine in Tolypocladium niveum (19, 20) and ferricrocin, a siderophore essential for Aspergillus nidulans viability (21). Furthermore, the carboxy terminal C-domain of the trimodular NRPS TqaA is postulated to be required for the production of fumiquinazoline F, the tricyclic imidazoindolone precursor of tryptoquialanine (22). However, although terminal C-domains are cited as controlling the cyclization of NRPS based intermediates, there is as yet no experimental evidence to illustrate their proposed catalytic activity.
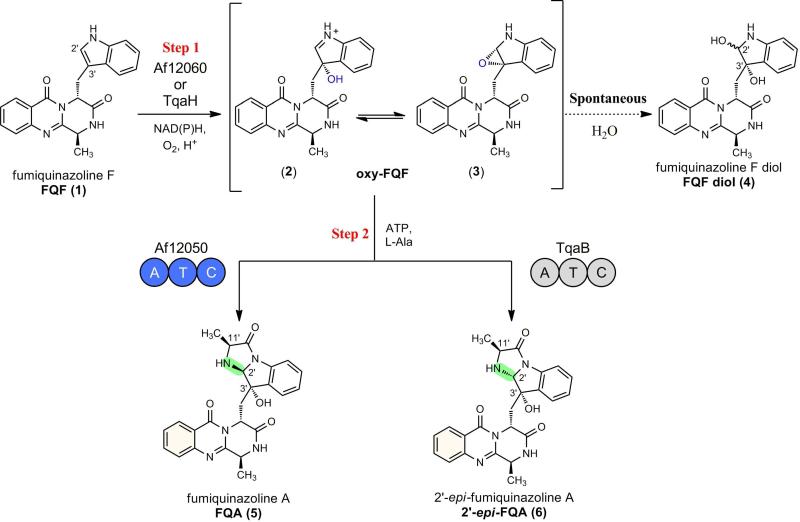
We have recently reported that a monomodular NRPS (of domain structure A-T-C) from the fumiquinazoline pathway in A. fumigatus catalyzes the formation of fumiquinazoline A (FQA) from an activated oxidation product of fumiquinazoline F (FQF) (23). Comparison of FQA biosynthesis to the recently described tryptoquialanine (TQA) pathway of P. aethiopicum reveals closely related enzymatic logic for the generation of early pathway intermediates (10). Both pathways are proposed to use a trimodular NRPS to condense and cyclize Anthranilate (Ant), Trp, and Ala to form FQF, and an FAD-dependent monooxygenase and monomodular NRPS for the oxidative-acylation of FQF to form a tricyclic imidazoindolone framework. In FQA biosynthesis, oxidation across the C2′ and C3′ of the pendant indole of FQF by the monooxygenase Af12060 is followed by alkylation with a unit derived from L-alanine catalyzed by the monomodular NRPS Af12050. In TQA biosynthesis, the monooxygenase TqaH (56/76% identity/similarity to Af12060, Figure S1) and monomodular NRPS TqaB (53/75% identity/similarity to Af12050, Figure S2) are postulated to catalyze an analogous process but yield a product with the opposite stereochemistry at C2′ and a gem-dimethyl at C11′ (the dimethyl group results from the preferential coupling of 2-AIB by TqaB). A small amount of the C2′ epimer of FQA (2′-epi-FQA) has also been observed in P. aethiopicum extracts, arising from TqaB-mediated coupling of L-Ala (10) (Figure 1).
Figure 1.
Stereochemical divergence of biosynthetic intermediates mediated by the action of a monomodular NRPS, Af12050 or TqaB, and illustration of spontaneous formation of diol shunt metabolite from the oxidized intermediate 2 or 3.
The basis for the difference in stereochemistry at C2′ is likely to be dictated by FQF oxidation (by Af12060 or TqaH) or amino acid coupling (by Af12050 or TqaB). Because the stereochemistry of the C3′ hydroxyl group of FQA is identical to that found in 2′-epi-FQA, we hypothesize that Af12060 and TqaH catalyze the same stereospecific C3′ hydroxylation of FQF. Consequently, we believe that the monomodular NRPS Af12050 or TqaB, and specifically the carboxy terminal C-domain, directs a specific stereochemical outcome at C2′ during amino acid coupling to the oxidized FQF scaffold (Figure 1). Although the consequences of the structural differences between FQA and 11′-dimethyl-2′-epi-FQA with regard to biological activity and effect on downstream enzymatic events are currently unknown, this differentiation provides an interesting example of divergence of secondary metabolite populations and divergent evolution of the corresponding biosynthetic enzymes.
The similarities between FQA from A. fumigatus and 2′-epi-FQA from P. aethiopicum, both in terms of structure and anticipated biosynthetic routes provide an opportunity to study the role of the terminal C-domain of the monomodular NRPSs in catalyzing regio- and stereospecific nucleophilic addition. In this work we reconstitute the activity of both the intact proteins and excised C-domains from Af12050 and TqaB of the fumiquinazoline and tryptoquialanine pathways, respectively, in order to explore the function of the terminal C-domains in the enantioselective annulation of amino acids to indole in fungal alkaloid biosythesis. Our biochemical and bioinformatic analysis presents the first clear evidence of the catalytic activity and biological context of C-terminal condensation domains of fungal monomodular NRPSs.
EXPERIMENTAL PROCEDURES
General Materials and Methods
N-acetylcysteamine, diisopropylethylamine, anhydrous DCM, potassium carbonate, sodium bicarbonate, potassium hydrogen sulphate, coenzyme A trilithium salt, THF, and L-[U-14C]Ala (128 mCi/mmol) were all purchased from Sigma-Aldrich and used as supplied. PyBOP was purchased from ChemPep Inc. N-BOC-L-Ala-OH, and pET expression vectors were purchased from EMD Chemicals. Oligonucleotide primers were purchased from Integrated DNA Technologies (Coralville, IA). PCR reactions were carried out using Phusion High-Fidelity PCR MasterMix (New England Biolabs). Plasmid DNA was propagated in E. coli XL1 Blue (Stratagene), and prepared using the QIAprep Spin Mini Kit (Qiagen). DNA sequencing to confirm the correct construction of expression vectors was performed by Genewiz (South Plainfield, NJ). Protein concentration was determined spectrophotometrically using theoretical extinction coefficients obtained from the online ProtParam tool (24). An Agilent Technologies 6520 Accurate-Mass QTOF instrument was used for high-resolution LC-MS analysis, and a Beckman Coulter System Gold instrument equipped with diode-array detection for reverse-phase HPLC. Liquid scintillation counting was performed with a Beckman Coulter LS 6500 instrument. NMR data were collected on Varian 600 and 400 MHz spectrometers using the residual solvent peak from incomplete deuteration as internal standard (CDCl3, δ 7.26; D2O, δ 4.75; MeCN, δ 1.94).
Synthesis of L-Ala coenzyme A thioester
Potassium carbonate (14 mg, 0.102 mmol, 4.0 equiv) was added to a stirred solution of N-BOC-L-Ala (5 mg, 0.026 mmol, 1.0 equiv) and PyBOP (27 mg, 0.051 mmol, 2.0 equiv) in THF/H2O (50:50, 2 ml) at room temperature. To the resulting clear and colourless solution was added Coenzyme A trilithium salt (20 mg, 0.026 mmol, 1.0 equiv) and the reaction was stirred until completion as judged by LC-MS analysis (circa 2 hours). THF was then removed in vacuo and an equal volume of TFA (approx 1 ml) was added to the aqueous residue at 0 °C. The mixture was then stirred until deprotection was complete (circa 1 hour) to give the named product in 58% yield over 2 steps. Solvent was then removed in vacuo and the residue purified by preparative HPLC on a Beckmann Coulter Gold system equipped with a reverse phase C18 column (Phenomenex Luna, 250 × 21.2 mm, 10 micron) with detection at 234 nm. Solvent system A (water plus 0.1% TFA) and B (acetonitrile plus 0.1% TFA) held at 1% B for 1 minute and then run over a linear gradient of 1-20% over 20 minutes, followed by a gradient of 20-95% B over 2 minutes before a holding at 95% B for 7 minutes, the column was then equilibrated back to initial conditions by returning to 1% B and holding for 6 minutes. The peak with retention time of 15 minutes was collected.
1H-NMR (D2O, 600 MHz) δ: 0.82 (s, 3H), 0.93 (s, 3H), 1.58 (d, J=6.46 Hz, 3H), 2.45 (m, 2H), 3.15 (m, 2H), 3.40 (m, 2H), 3.45 (m, 2H), 3.60 (dd, J=9.39, 4.11 Hz, 1H), 3.83 (dd, J=9.39, 4.11 Hz, 1H), 4.03 (s, 1H), 4.26 (m, 2H), 4.35 (q, J=6.46 Hz, 1H), 4.59 (m, 1H), 4.86 (m, 2 H), 6.21 (m, 1 H), 8.43 (s, 1 H), 8.67 (s, 1 H).
HRMS: m/z calculated for C24H41N8O17P3S: 861.1415 [M + Na]+. Found 861.1417
Synthesis of L-Ala SNAC
Diisopropylethylamine (0.74 ml, 4.24 mmol, 4.0 equiv) was added to a stirred solution of N-BOC-L-Ala (0.20 g, 1.06 mmol, 1.0 equiv) and PyBOP (1.10 g, 2.12 mmol, 2.0 equiv) in DCM (5 ml) at room temperature. To the resulting clear and colorless solution was added N-acetylcysteamine (0.12 ml, 1.16 mmol, 1.1 equiv) and the reaction was monitored by TLC and stirred until completion (circa 1 hours). The solvent was then removed in vacuo and the residue was dissolved in ethyl acetate (25 ml). The organic layer was then washed successively with 5% KHSO4 (3 × 10 ml), 5% NaHCO3 (3 × 10 ml) and brine (10 ml). Extract was then dried over magnesium sulfate and evaporated to dryness in vacuo to leave a residue of colorless oil and white solid which was purified by flash column chromatography on silica gel (50 % - 100 % ethyl acetate/hexane) to give N-BOC-L-Ala SNAC as a hygroscopic white solid (Figure S9A). N-BOC-L-Ala SNAC was dissolved in DCM (1 ml), cooled to 0 °C and TFA (1 ml) was added dropwise. The resulting mixture was stirred at 0 °C for 1 hour. Solvent was then removed in vacuo and the resulting residue was washed with sequentially with DCM, diethyl ether and ethyl acetate/hexane (1:1) to give L-Ala SNAC as colorless sticky oil in 65% yield over 2 steps. A small portion of this material was further purified by preparative HPLC on a Beckmann Coulter Gold system equipped with a reverse phase C18 column (Phenomenex Luna, 250 × 21.2 mm, 10 micron) with detection at 234 nm. Solvent system A (water plus 0.1% TFA) and B (acetonitrile plus 0.1% TFA) held at 1% B for 1 minute and then run over a linear gradient of 1-20% over 20 minutes, followed by a gradient of 20-95% B over 2 minutes before a holding at 95% B for 7 minutes, the column was then equilibrated back to initial conditions by returning to 1% B and holding for 6 minutes. The peak with retention time of 13 minutes was collected.
1H-NMR (D2O, 400 MHz) δ: 1.58 (d, J=7.43 Hz, 3H), 1.94 (s, 3H), 3.11 (m, 1H), 3.21 (m, 1H), 3.40 (td, J=6.26, 2.74 Hz, 2H), 4.36 (m, 1H).
HRMS: m/z calculated for C7H14N2O2S: 191.0849 [M + H]+. Found 191.0853
Cloning of TqaH, standalone C- and T-domains, and chimeric constructs
TqaH cDNA was amplified from P. aethiopicum gDNA using reverse transcription (RT)-PCR and cloned into the NdeI and EcoRI restriction sites of the pET28a vector for protein expression with an N-terminal His6-tag (483 residues [53.2 kDa]).
Standalone C- and T-domain constructs were cloned from plasmid DNA (encoding Af12050 (23) and TqaB (10)) into pET30 Xa-LIC and pET46 Ek-LIC vectors, respectively. Domain boundaries were determined by homology detection and structure prediction using the online server HHpred (25), and primers (Table S1) were designed to PCR amplify a nucleotide region corresponding to amino acids: 684-end, Af12050 C-domain (464 residues [53.4 kDa], N-terminal His6-S-tag); 673-end, TqaB C-domain (483 residues [53.4 kDa], N-terminal His6-S-tag); 591-678, Af12050 T-domain (103 residues [11.2 kDa], N-terminal His6-tag); 581-668, TqaB T-domain (103 residues [11.2 kDa], N-terminal His6-tag). The boundaries for the standalone T- and C-domain constructs are annotated on the Af12050/TqaB sequence alignment (Figure S2)
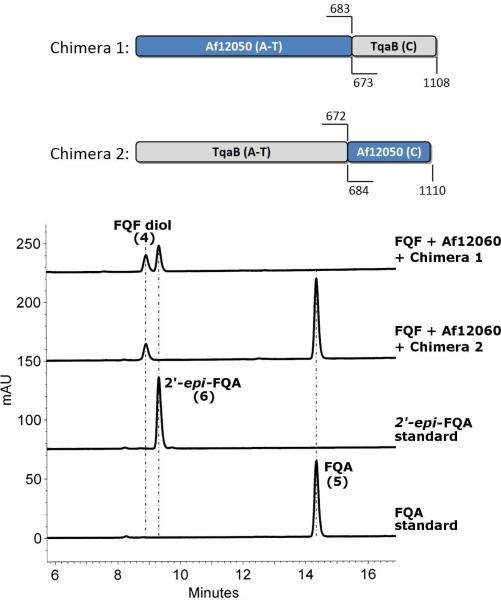
The domain boundaries determined above for the C-domains of Af12050 and TqaB were used for the generation of chimeric constructs by splice overlap extension PCR (see Table S1 for primers). Chimera 1 consists of the A- and T-domains of Af12050 (amino acids 1-683) and the C-domain of TqaB (673-end), while Chimera 2 consists of the A- and T-domains of TqaB (1-672) and the C-domain of Af12050 (684-end) (Figure 3A). The chimeric constructs were cloned into pET52b 3C-LIC to generate proteins containing an N-terminal Strep-tag II and a C-terminal His6-tag (Chimera 1, 1161 residues [127.9 kDa]; Chimera 2, 1159 residues [128.8 kDa]).
Figure 3.
(A) Cartoon representation of the structure of chimeras 1 and 2, numbers indicate the amino acid numbers from the native protein included in each chimera. (B) HPLC assay indicating the chimera mediated formation of FQA or 2′-epi-FQA with detection at 254 nm.
Protein overproduction and purification
Expression constructs for Af12050, TqaB, and Af12060 have been described previously (10, 23). Af12060, TqaH, C-domains and T-domains were overproduced in E. coli BL21-Gold(DE3) cells (Stratagene), while Af12050, TqaB and the chimeras were overproduced in E. coli BAP1 (26) for the production of phosphopantetheinylated (holo) proteins. The production and purification of proteins was performed in a similar manner: 2-4 liters of cells were grown at 37 °C in LB plus the appropriate antibiotic (50 μg/mL kanamycin or 100 μg/mL carbenicillin) to an OD600 between 0.4-0.8, and the temperature lowered to 16 °C prior to induction with 0.2 mM IPTG. Cells were harvested 18-24 hours post-induction by centrifugation, suspended in lysis buffer (25 mM Tris-HCl [pH 7.5], 300 mM NaCl, 20% glycerol, 0.1% Tween 20, 1x protease inhibitor cocktail [SigmaFast, EDTA-free]) and lysed using a EmulsiFlex-C5 homogenizer (Avestin). Insoluble material was removed by centrifugation (35,000g) and soluble protein applied to 1-2 mL of Ni-NTA agarose (Qiagen). Ni-affinity purification was performed by batch binding protein for 30 min at 4 °C, Ni-resin was washed with 2 × 25 mL buffer A (50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 10% glycerol, 0.1 mM EDTA) containing 20 mM imidazole, and protein eluted with buffer A containing 250 mM imidazole (2 × 5 mL). The elutions were pooled and concentrated using centrifugal filtration devices (Amicon), flash-frozen in liquid N2 and stored at -80°C (except for TqaB, which was kept at 4 °C due to instability during freeze/thaw).
Assay for phosphopantetheinylation of the Af12050 and TqaB T-domain constructs with [1-14C]acetyl-CoA
Reactions (50 μL) contained 25 μM [1-14C]acetyl-CoA (0.075 μCi), 250 nM B. subtilis Sfp (27), and 10 μM T-domain in buffer (50 mM sodium phosphate (pH 7.5), 5 mM MgCl2, 100 mM NaCl, 1 mM TCEP, and 5% glycerol). Reactions were initiated by addition of Sfp and incubated at 25 °C for 1, 2.5, 5, or 10 min prior to quenching with 0.5 mL 10% TCA (containing 50 μg BSA). Protein precipitate was pelleted by centrifugation, washed twice with 10% TCA, and dissolved in 80% formic acid for liquid scintillation counting (Figure S3).
Single time-point HPLC-based assays for monooxygenase and NRPS activity
Reaction mixtures were set up containing the required components as listed below. Reactions were initiated by the addition of monooxygenase (Af12060 or TqaH) and incubated at room temperature overnight (14-16 hrs). Reactions were then quenched by the addition of an equal volume acetonitrile, the precipitant removed by centrifugation, and 20 μL samples of the resulting supernatant were injected for HPLC analysis on a Beckmann Coulter Gold system equipped with an Alltima C18 reverse-phase column (150 × 4.6 mm, 5 micron). Solvent system A (water plus 0.1% TFA) and B (acetonitrile plus 0.1% TFA) held at 25% B for 1 minute and then run over a linear gradient of 25-55% B over 20 minutes, followed by a gradient of 55-95% B over 1 minute before a holding at 95% B for 2.5 minutes, the column was then equilibrated back to initial conditions by returning to 25% B and holding for 5 minutes.
Monooxygenase activity assays (Af12060 and TqaH). Reactions (100 μL) contained: 2.5 μM Af12060 or TqaH, 2 mM NADH, and 250 μM FQF in NaPi buffer [50 mM sodium phosphate (pH 7.5), 100 mM NaCl and 5% glycerol]. The reaction containing Af12060 was scaled up to 15 mL to allow for isolation and characterization of FQF diol, the same reaction components and concentrations were used as the small scale with the exception of FQF, which was used at a concentration of 500 μM. FQF diol was purified from the resulting mixture by the following procedure. Enzyme was removed from the incubation by centrifugal filtration (30 kDa MWCO). The filtrate was then purified by preparative HPLC on a Beckmann Coulter Gold system equipped with a reverse phase C18 column (Phenomenex Luna, 250 × 21.2 mm, 10 micron) with detection at 230 nm. Solvent system A (no additive) and B (no additive) held at 30% B for 1 minute and then run over a linear gradient of 30-60% over 20 minutes, followed by a gradient of 60-95% B over 2 minutes before a holding at 95% B for 7 minutes, the column was then equilibrated back to initial conditions by returning to 30% B and holding for 5 minutes. The peak with retention time of 14.5 minutes was collected. This peak was shown to consist of a mixture of diol and multiple dimerization products and further purified by solid phase extraction over a Sep-Pak® C18 cartridge (Waters) according to the following protocol using D2O and d6-acetonitrile. The column was wet with 10 column volumes (CV) of d6-acetonitrile (approximately 6 ml) then flushed with 10 CV of D2O. Sample was loaded to column in 500 μL of D2O and washed with 4 CV of D2O (2.4 ml). Bound products were then eluted from the column and collected as 600 μL fractions using 30% d6-acetonitrile/D2O (4 × 600 μL), 50% d6-acetonitrile/ D2O (4 × 600 μL) and 90% d6-acetonitrile/ D2O (4 × 600 μL). HPLC analysis of the resulting fractions was used to determine the presence of pure FQF diol and the sample was analyzed directly by NMR spectroscopy. For FQF diol spectroscopic characterization and assignments see Table S2. HRMS: m/z calculated for C21H20N4O4: 375.1452 [M-(H2O) + H]+. Found 375.1458
Monomodular NRPS activity assays (Af12050, TqaB, Chimera 1, and Chimera 2). Reactions (100 μL) contained: 2.5 μM Af12060, 5 μM holo-NRPS, 1 mM ATP, 2 mM MgCl2, 1 mM L-Ala, 2 mM NADH, and 250 μM FQF in NaPi buffer. The reaction containing TqaB was scaled up to 30 mL to allow for isolation and characterization of 2′-epi-FQA, the same reaction components and concentrations were used as the small scale with the exception of FQF, which was used at a concentration of 500 μM. 2′-epi-FQA was purified from the resulting mixture by the following procedure. An equal volume of acetonitrile was added to the incubation and the precipitated protein was pelleted by centrifugation. The supernatant was then decanted, the acetonitrile was removed in vacuo and the residual water was removed by lyophilization. The resulting residue was taken up in 25% acetonitrile and water and purified by preparative HPLC on a Beckmann Coulter Gold system equipped with a reverse phase C18 column (Phenomenex Luna, 250 × 21.2 mm, 10 micron) with detection at 254 nm. Solvent system A (water plus 0.1% TFA) and B (acetonitrile plus 0.1% TFA) held at 25% B for 1 minute and then run over a linear gradient of 25-53% over 30 minutes, followed by a gradient of 53-95% B over 1 minute before a holding at 95% B for 5 minutes, the column was then equilibrated back to initial conditions by returning to 25% B and holding for 8 minutes. The peak with retention time of 25 minutes was collected. For 2′-epi-FQA spectroscopic characterization and assignments see Table S3. HRMS: m/z calculated for C24H23N5O4: 446.1823 [M + H]+. Found 446.1824
Standalone C-domain activity assays (Af12050 and TqaB)
The ability of the C-domain constructs to catalyze coupling of alanine to oxy-FQF was tested using L-Ala-SNAC as a free-standing substrate, and also using a tethered L-alanyl-S-T-domain intermediate as substrate (prepared via coupling of L-Ala-CoA to holo-Af12050 T-domain). Reactions using L-Ala-SNAC as substrate contained: 1 μM Af12060, 10 μM C-domain, 1 mM L-Ala-SNAC, 2 mM NADH, and 250 μM FQF and in NaPi buffer. Reactions assaying L-alanyl-S-T-domain as substrate contained: 1 μM Af12060, 10 μM C-domain, 50 μM Af12050 T-domain, 1 μM Sfp (27), 1 mM L-Ala-CoA, 2 mM MgCl2, 2 mM NADH, and 250 μM FQF in NaPi buffer. Prior to initiating the reaction with monooxygenase, the reaction containing L-Ala-CoA was incubated for 10 minutes to allow for the Sfp-catalyzed loading of alanine onto the T-domain. Following 120 min incubation at room temperature, the reactions (100 μL) were quenched with an equal volume of acetonitrile and 20 μL samples analyzed by analytical HPLC as described above.
Timecourse HPLC-based assays for 2′-epi-FQA formation
A 250 μL initial reaction was set up and 50 μL aliquots were quenched with an equal volume of acetonitrile at 15, 30, 60, and 120 minutes after addition of Af12060. To follow the non-enzymatic rate of 2′-epi-FQA formation we combined: 1 μM Af12060, 2 mM NADH, 250 μM FQF, and 1 mM L-Ala or 1 mM L-Ala-SNAC or 1 mM L-Ala-CoA in NaPi buffer. The enzymatic rate of 2′-epi-FQA formation was determined by the addition of 10 μM TqaB C-domain. For full-length TqaB we combined: 2.5 μM Af12060, 10 μM holo-TqaB, 1 mM ATP, 2 mM MgCl2, 1 mM L-Ala, 2 mM NADH, and 250 μM FQF in NaPi buffer. Quenched and clarified samples were run on analytical HPLC as described above. For each timepoint, the peak corresponding to 2′-epi-FQA was integrated and converted to a concentration using a standard curve made by injecting 20 μL samples of known concentration (determined using an extinction coefficient calculated for 2′-epi-FQA at 256 nm of 10532 M-1 cm-1). Plotting the concentration of 2′-epi-FQA vs. time allowed for the calculation of initial rate data (Figure S4).
RESULTS
Examination of the effect of monomodular NRPS and monooxygenase
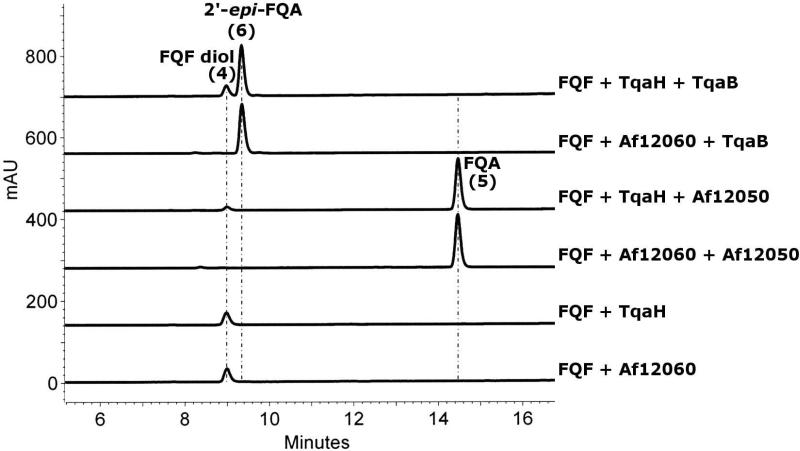
To gain initial insight into the enzymatic requirements for directing the stereochemical course of dual N-C bond formation for alanine coupling to oxy-FQF, we mixed-and-matched combinations of NPRS (Af12050 and TqaB) and monooxygenase (Af12060 and TqaH) and analyzed their respective products. The production and characterization of Af12050 and Af12060 has been previously described (23); for comparative analysis we overproduced TqaH and TqaB as the corresponding His6 tagged proteins from E. coli and purified them via Ni-NTA affinity chromatography. Interestingly, though TqaH and Af12060 share 76% similarity at the amino acid level, their expression profiles, purity, and flavin content vary significantly (Figures S1 and S5). Incubation of FQF with TqaH generated a peak at a retention time of 9 min that is identical to that seen upon incubation of FQF with Af12060 (Figure 2). Isolation and characterization of this compound was hindered by rapid decomposition to a complex mixture of apparent dimerization products upon concentration and lyopholization (post preparative HPLC). However, removal of the dimerization products by SPE (Sep-Pak® C18) allowed us to identify this product by 1D and 2D NMR experiments as a diol shunt product (FQF diol, Figure 1). The FQF diol arises from the quenching of the predicted unstable oxy-FQF epoxide/hydroxy imine product of Af12060 (or TqaH) with water. Co-incubation of TqaH with the monomodular NRPS from A. fumigatus, Af12050, resulted in the production of a compound matching the retention time and UV pattern (Figure S6) of FQA as previously characterized from an in vitro reconstitution of the natural Af12060 + Af12050 system (Figure 2). By comparison when the monomodular NRPS TqaB is co-incubated with either monooxygenase (Af12060 or TqaH) a new peak is produced with a retention time of 9.4 minutes (Figure 2). This compound was isolated in sufficient quantity for investigation by NMR spectroscopy and structurally characterized as the C2′ epimer of FQA (2′-epi-FQA). These experiments clearly illustrate that it is the activity of the monomodular NRPS that determines the stereochemical outcome of this reaction and not that of the monoxygenase.
Figure 2.
HPLC analysis detected at 254 nm of reactions containing 2.5 μM monoxygenase (Af12060 or TqaH), 2 mM NADH, and 250 μM FQF in NaPi buffer, in addition to 5 μM holo-NRPS (Af12050 or TqaB), 1 mM ATP, 2 mM MgCl2 and 1 mM L-Ala when both monoxygenase and monomodular NRPS are included.
Construction of C-domain swapped chimeric proteins
With the knowledge that the monomodular NRPSs Af12050 and TqaB (and not the monooxygenases) direct the stereochemical outcome at C2′ of FQA and epi-FQA, respectively, we then wanted to test the hypothesis that the divergent NRPS-based activities could be specifically attributed to the carboxy terminal C-domains of Af12050 and TqaB. In order to investigate this hypothesis we constructed two hybrid (or chimeric) proteins. The C-domains of TqaB and Af12050 were swapped, resulting in one protein containing the A and T domains from Af12050 and the C-domain from TqaB (Chimera 1) and another protein with the A and T domains from TqaB and the C-domain from Af12050 (Chimera 2) (Figure 3A). For cloning purposes, the linker region between T- and C-domains was defined by a combination of sequence alignment and secondary/tertiary structure prediction using HHpred (25). In particular, modeling of tertiary structure using the bidomain TycC T5-C6 structure as template (PDB ID 2jgp, (28)) indicated an unstructured linker of 30 residues for both Af12050 (amino acids 669-698, TASV. . .RKSQ) and TqaB (amino acids 659-687, VKQP. . .AKCQ). To allow for flexibility with regard to the predicted vs. actual structure of the corresponding T- and C-domains, a C-domain swap point of Q684 of Af12050 and Q673 of TqaB was chosen which lies near the middle of the predicted unstructured linker region. Hybrid constructs were then generated using splice overlap extension PCR.
The enzymatic activity of Chimera 1 was demonstrated in vitro by incubation with FQF, monoxygenase (Af12060), and the required cofactors and shown to produce two peaks, one peak of retention time, HRMS and characteristic UV pattern matching that of the FQF diol and the other matching that of 2′-epi-FQA. Incubation of Chimera 2 produced two peaks, one corresponding to FQF diol and the other matching the retention time and UV pattern of FQA (Figure 3). The ability of the chimeric constructs to activate and load a variety of amino acids was investigated by reconstituting their activity in vitro in the presence of L-Ala, D-Ala or 2-aminoisobutyric acid (2-AIB). Chimera 1 efficiently activates, loads and condenses L-Ala with the FQF-derived oxidized species to give 2′-epi-FQA. 11′-epi-2′-epi-FQA is also produced by Chimera 1 when using D-Ala but at a level much below that seen for 2′-epi-FQA. A very small amount of a compound with mass and UV pattern consistent with 11′-dimethyl-2′-epi-FQA is produced by Chimera 1 when using 2-aminoisobutyric acid in place of alanine (Figure S7). Similar substrate specificity investigations with Chimera 2 show the efficient activation, loading and condensation of L-Ala, D-Ala and 2-aminoisobutyric acid to give FQA, 11′-epi-FQA, and 11′-dimethyl-FQA all in substantial quantity (Figure S8). The activities of the chimeric constructs demonstrate that it is the C-terminal C-domain of the monomodular NRPSs (Af12050 and TqaB) that account for and control their observed stereochemical control.
Demonstration of activity of stand-alone C-domains from Af12050 and TqaB
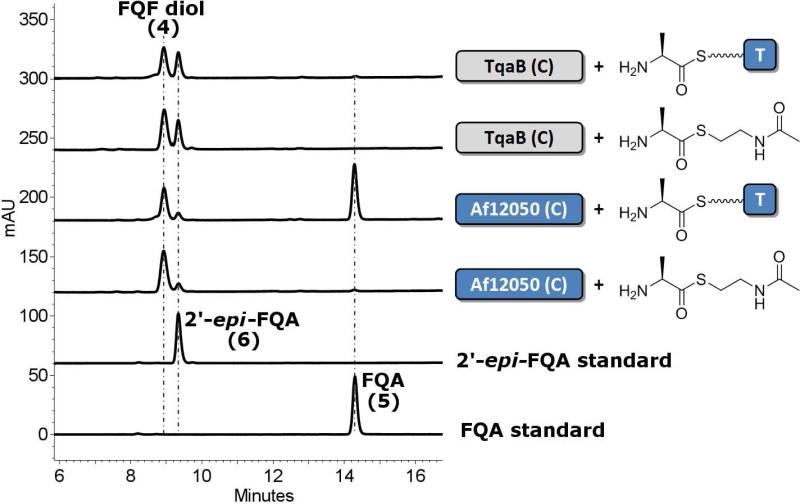
To specifically probe the catalytic activity of the respective C-domains from Af12050 and TqaB we over-expressed the C-domain regions of TqaB and Af12050 as standalone His6-tagged proteins from E. coli (Af12050 C-domain, 53.4 kDa, residue 684-end; TqaB C-domain 53.4 kDa, residue 673-end). Concurrently, we cloned and expressed the T-domains from both TqaB and Af12050. The structure prediction described above for the generation of the chimeric constructs was used as the basis for cloning these standalone proteins. The ability of the excised T-domains to be phosphopantetheinylated was tested by incubating apo-protein with Sfp (a promiscuous phosphopantetheinyl transferase) and 14C-acetyl-CoA. Interestingly, although both proteins were expressed from E. coli in soluble form, only the T-domain from Af12050 was observed to be labeled by 14C-acetyl-CoA (26% after 10 min compared to 1.3% for TqaB T-domain, Figure S3). MALDI-MS analysis of TqaB T-domain revealed a mixture of both apo and holo forms in approximately a 2:1 ratio. Therefore, it is not clear why the apo TqaB T-domain present did not react with acetyl-CoA in vitro. With these protein constructs in hand we synthesized an L-Ala Coenzyme A thioester (L-Ala CoA) via standard peptide coupling procedures, and generated an L-alanyl-S-T-domain substrate by incubating L-Ala CoA with the Af12050 T-domain and Sfp. Addition of TqaB C-domain to L-alanyl-S-T-domain resulted in the conversion of FQF to two new peaks corresponding to FQF-diol and to 2′-epi-FQA (as confirmed by comparison with authentic standards). Incubating Af12050 C-domain with L-alanyl-S-T-domain resulted in the production of three new peaks; two of large magnitude indicated the presence of large quantities of FQA and the FQF-diol, and one smaller peak which showed the formation of some 2′-epi-FQA (Figure 4).
Figure 4.
HPLC analysis of incubations of stand alone C-domains from TqaB or Af12050 with L-Ala SNAC (shown as chemical structure) and L-alanyl-S-T-domain (shown as T-domain tethered cartoon).
N-acetylcysteamine derived L-Ala thioester (L-Ala SNAC) was also synthesized via PyBOP mediated peptide coupling. Comparison of the products observed after the incubation of TqaB C-domain with L-Ala SNAC and FQF with authentic standards allowed them to be confirmed as FQF-diol and 2′-epi-FQA, in agreement with the analogous standalone T- and C-domain experiment. In contrast, when Af12050 C-domain was incubated with FQF and L-Ala SNAC the result observed did not match that seen with the equivalent T-domain tethered substrate as only FQF diol was produced in significant quantity, while a small peak indicated the presence of a small amount of 2′-epi-FQA (Figure 4).
DISCUSSION
Fuminquinazoline A (FQA) produced by A. fumigatus contains an interesting pendant indole moiety derived from a tryptophan unit that has been modified by the stereospecific addition of L-alanine to form a new 5-membered ring resulting in a tricyclic 5-5-6 imidazoindolone framework. FQA has been shown to be the direct precursor to two further previously identified metabolites fuminquinazoline C (FQC) and fuminquinazoline D (FQD) (29). Recently, characterization of intermediates from the tryptoquialanine pathway in P. aethiopicum has revealed a close similarity in the early biosynthetic steps to intermediates of the FQA pathway in A. fumigatus. Intermediates on pathway to tryptoquialanine which share a remarkable resemblance to FQA have been observed in mutants of P. aethiopicum and appear to be substrates for downstream enzymes which mediate their transformation to tryptoquialanine. Notably, 2′-epi-FQA identified in P. aethiopicum only differs from FQA in the stereochemistry at C2′ of the imidazoindolone moiety. The stereochemical difference between FQA and 2′-epi-FQA may influence the downstream fate of these two molecules to ultimately give FQC/FQD and tryptoquialanine, respectively. FQA formation from oxy-FQF in A. fumigatus is controlled by the action of a monomodular NRPS from the fumiquinazoline pathway (Af12050). Where as the biosynthesis of 2′-epi-FQA in P. aethiopicum has been proposed to be controlled by the action of TqaB, based on its close homology to Af12050. The opposite stereochemistry at C2′ of the metabolites observed in A. fumigatus and P. aethiopicum poses the question of how the cyclization of an amino acid derived unit to generate the observed 5-membered ring is controlled: specifically how the stereochemistry of addition of the amine nucleophile to the C2′ of the indole is differentially directed in FQA and 2′-epi-FQA biosynthesis.
Enzymatic assays of both TqaH and Af12060 revealed the product of each to be identical, whereas the activity of the A-T-C tridomain-containing Af12050 and TqaB resulted in two different products, FQA and 2′-epi-FQA, respectively. These data confirm the crucial role that the monomodular NRPS plays in directing the stereospecific formation of the C2′ carbon-nitrogen bond.
To determine whether the C-domain of the NRPS was in fact responsible for the facial specific addition as we proposed required the study of C-domain activity in a non-native environment. Characterization of the chimeric constructs showed that the enantiospecificity of FQA scaffold formation is consistent with that of the parent protein from which the C-domain of each chimeric construct was taken. The A-domain of Af12050 has previously been shown to preferentially activate L-Ala over D-Ala, which is in line with the observed amino acid coupling to oxy-FQF to form FQA in A. fumigatus (23). On the other hand, the A-domain of TqaB exhibits much broader substrate specificity, activating not only 2-aminoisobutyric acid as observed in tryptoquialanine but also both L-Ala and D-Ala (10). The activity of both the Af12050 and TqaB A-domains was maintained in Chimera 1 and Chimera 2 (Figures S7 and S8), illustrating the persistence of A-domain substrate specificity and also the ability of the terminal C-domains to accept and couple non-cognate amino acids for the generation of novel ‘unnatural’ natural products: 11′-dimethyl-FQA and 11′-epi-FQA (Chimera 2), and 11′-epi-2′-epi-FQA (Chimera 1).
To our knowledge, this is the first reported example of C-domain swapping yielding functional multidomain NRPS assembly lines. Previously, the N-terminal C-domain of the enterobactin synthetase EntF was excised and activity reconstituted in vitro (30), adenylation domain swapping has been performed (31, 32), and whole modules and domains of the tyrocidine synthetase have been fused to generated hybrid enzymes (33). In our case we recognize that the swapped domains are the carboxy terminal domains of monomodular proteins that are highly homologous, which facilitates the process of generating functionally intact hybrid proteins.
Expression and purification of the excised 53.4 kDa C-domains from Af12050 and TqaB in soluble form allowed us to conclusively show that the C-domain of either enzyme is all that is required for directing the stereospecific formation of an FQA (or 2′-epi-FQA) product. T-domain loading with acetyl-CoA mediated by Sfp (a phosphopantetheinyl transferase from Bacillus subtilis) was only successful with the T-domain of Af12050 (and not that of TqaB). The mixture of both apo and holo forms of TqaB T-domain presumably results from the activity of an E. coli 4′-phosphopantetheinyl transferase (PPTase), which shows some low level activity on the non-native T-domain. However, given that of the loading of Af12050 T-domain preceded smoothly we did not pursue the reason for the inactivity of the TqaB T-domain further. Presumably the inactivity of TqaB T-domain is due to misfolding or other structurally irregularities, as perhaps is suggested by the poor E. coli based overproduction of this protein. Furthermore, we reasoned that given the high sequence similarity between Af12050 and TqaB T-domain regions the T-domain used should have no bearing on the observed result. Aminoacyl-N-acetylcysteamine thioesters (aminoacyl-SNACs) have been shown on several occasions to act as efficient small molecule substrates of NRPS C-domains (34). Therefore in order to devise a small-molecule based experiment complementary to the co-incubation of standalone T and C domains we also synthesized L-Ala SNAC. Purified C-domain from TqaB was observed to specifically generate 2′-epi-FQA from FQF plus either L-Ala loaded T-domain (L-alanyl-S-T-domain) or L-Ala SNAC. By contrast, Af12050 C-domain only gave FQA when using L-alanyl-S-T-domain as substrate; when using the small molecule (T-domain mimic) SNAC derivative only the shunt metabolite FQF-diol was observed in significant quantity. The appearance of a small amount of 2′-epi-FQA in Af12050 C-domain incubations with both L-Ala loaded T-domain and L-Ala SNAC provoked our interest in the mechanism of its formation. Was the Af12050 C-domain losing some degree of stereo-specificity as a result of catalyzing an in trans like condensation? To this end, the level of spontaneous condensation (in the absence of TqaB C-domain) of L-alanyl-S-T-domain and L-Ala SNAC with oxy-FQF was investigated by a time course study. These experiments revealed a background level of spontaneous 2′-epi-FQA formation approximately one third the rate of C-domain catalyzed condensation with both L-alanyl-S-T-domain and L-Ala SNAC (Figure S4). Spontaneous (uncatalyzed) attack of the thioester substrates (L-Ala SNAC and L-alanyl-S-T-domain) by the latent nucleophilic indole –NH after dearomatization of the indole ring by epoxidation/hydroxylation, appears to explain this ‘background’ level of 2′-epi-FQA formation. Such an attack would be favored by the stability of the resulting amide bond and subsequent intramolecular ring closure to give the kinetically favored 5-membered ring (to give 2′-epi-FQA) would be predicted to occur rapidly. Crude quantitative analysis of the levels of spontaneous formation of 2′-epi-FQA, using UV analysis (and standard curve calibration) to quantify 2′-epi-FQA show that the levels of spontaneous formation account for the observed 2′-epi-FQA in incubations of Af12050 C-domain with L-alanyl-S-T-domain and L-Ala SNAC.
The condensation of a T-domain-tethered amino acid to the oxy-FQF scaffold by the action of the terminal condensation domains of Af12050 and TqaB could proceed through two distinct pathways depending on the order of N-C bond formation. Both of these pathways differ significantly from standard C-domain chemistry (Figure 5). Classically a C-domain would catalyze the formation of an amide bond between a T-domain tethered upstream acyl-thioester intermediate with a nucleophile tethered to a downstream T-domain (Figure 5A) (13). The biosynthesis of FQA (or 2′-epi-FQA) from FQF could be envisaged to proceed via one of two routes, both of which presumably proceed by an initial C-domain mediated addition, followed by a spontaneous intramolecular cyclization. Initial thioester cleavage by a C-domain catalyzed addition of the indole derived secondary amine followed by spontaneous ring closure by attack of the L-alanine derived primary amine aligns well with the conventional function of C-domain mediation of directing nucleophilic attack on a T-domain bound thioester (Figure 5B, pathway X). Indeed the observation of a C-domain catalyzing condensation with a free soluble nucleophile has been previously reported, for example in the biosynthesis of pseudomonine where condensation of a T-domain bound intermediate with a free amine monomer has been shown (35), or vibriobactin biosynthesis where a soluble small-molecule derived from norspermidine acts as the incoming nucleophile (36). Alternatively, attack of the L-alanine primary amine may precede a subsequent ring closure by intramolecular spontaneous amide bond formation (Figure 5B, pathway Y). This would require the C-domain to catalyze addition of a thioester bound nucleophile to a free soluble electrophile, via a process currently unprecedented in the chemistry of C-domain catalysis.
Figure 5.
Schematic representation of canonical C-domain catalysis in comparison to the two proposed enzymatic routes to FQA imidazoindolone scaffold formation
Analogs of L-Ala SNAC which disfavored attack at C2′ of oxy-FQF (to form the stereospecific C-N bond) or attack of the oxidized indole -NH (to give the observed amide bond) were both synthesized (Figure S9B) in an attempt to probe the route to the dual N-C bond formation observed during the biosynthesis of the FQA imidazoindolone scaffold. Incubation of TqaB C-domain with N-acetylcysteamine thioesters derived from propionic acid or L-lactic acid, in which nucleophilic attack at C2′ of oxy-FQF is either impossible or severely hindered, both failed to yield any detectable products. L-Ala SNAC analogues in which the thioester is replaced by a thioether or amide bond in order to prevent or hinder amide bond formation to give FQA also failed to give any detectable products, despite the activity of TqaB C-domain with L-Ala SNAC. It is not clear why there were no products observed with either set of SNAC analogs, perhaps the C-domain exhibits a strong specificity for L-Ala such that even close structural homologues are not accepted. At this stage the apparent lack of a viable substrate analog has prevented a detailed mechanistic study of this dual N-C bond formation event.
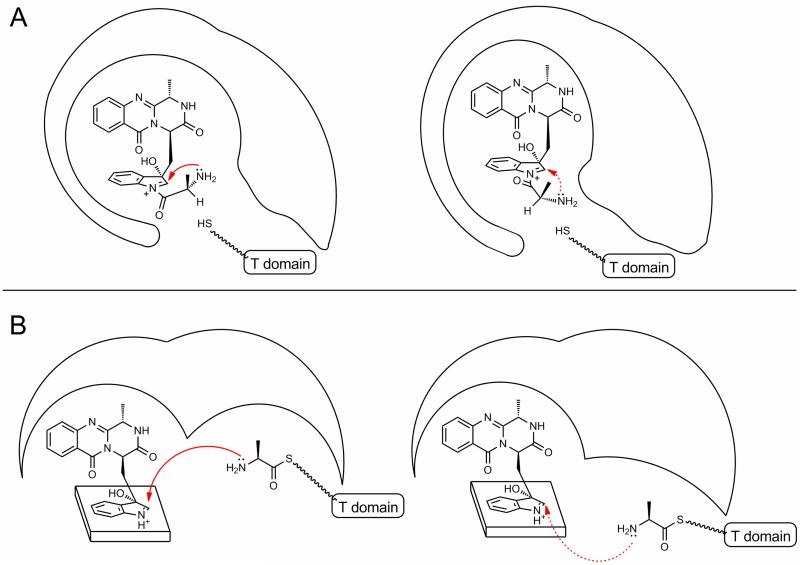
It seems that a minor configurational adjustment to the binding of the L-Ala derived moiety or an adjustment to the position of the FQF derived intermediate within the TqaB C-domain compared to that of Af12050 could direct the attack of the L-Ala amino group to opposite faces of the oxidized-indole species (Figure 6), resulting in the observed stereochemical difference at C2′. Analysis of the C-domains of TqaB and Af12050 by homology modeling suggests the proposed FQF binding pocket (as deduced from the relative position of the acceptor binding site from the point of T-domain attachment) is remarkably similar in both enzymes. Conversely, the presumed donor binding pocket of the incoming upstream T-domain shows some significant differences in residue composition and structure. These observations lead to the supposition that the binding position of FQF in both enzymes is approximately equal and therefore the differential binding of the L-Ala derived moiety directs the stereospecificity of the observed reaction. In order to support this proposal, future investigations that shed light on C-domain structure and FQF/L-Ala binding would be essential.
Figure 6.
Cartoon illustration of proposed routes of C-domain mediated stereospecific C-N bond formation. (A) Stereospecific formation of C-N bond controlled by the required conformation of the amide L-Ala moiety in the C-domain active site. (B) Facial specificity of primary amine addition controlled by positioning of incoming T-domain.
If formation of FQA proceeds via pathway X (Figure 5B) then presumably the stereochemistry of the spontaneous addition of the L-alanine primary amine could be controlled by the observed enzyme conformation strictly controlling the orientation of the pendant L-alanine immediately prior to and after thioester bond cleavage (Figure 6A). If catalysis proceeds via pathway Y (Figure 5B) then control of the path of the incoming phosphantetheinyl arm to either the top or bottom face of the oxidized indole would seem the logical mode of stereochemical control (Figure 6B). Although intuitively it seems beneficial for the stereospecific addition to occur first such that it is catalyzed by C-domain activity, a reaction of an upstream T-domain bound nucleophile with a free soluble electrophile has not previously been observed to be mediated by a C-domain.
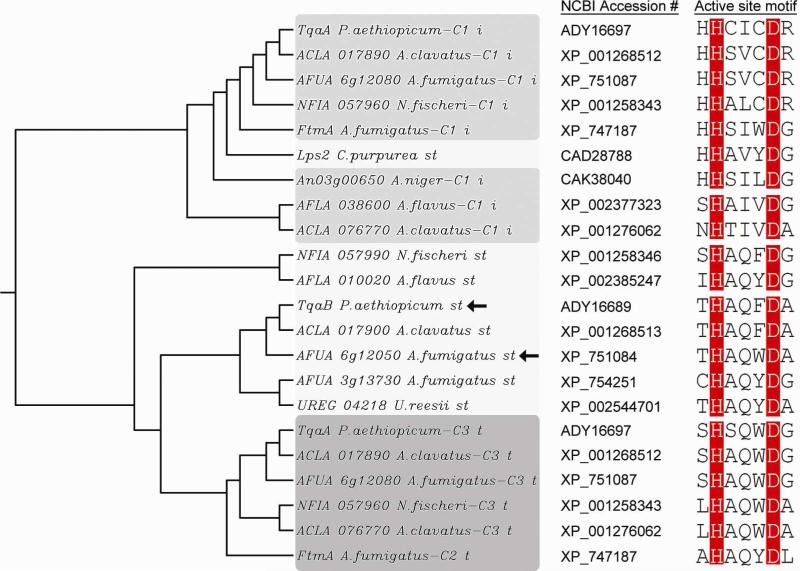
The unique functionality of Af12050 and TqaB to act in concert with multimodular NRPS machinery for amino acid coupling prompted a search of sequenced fungal genomes for similar proteins (monomodular NRPSs with domain organization A-T-C) in order to identify functionally similar C-domains, and to better understand the occurrence and divergence of this terminal domain as part of monomodular NRPSs. While monomodular NRPSs of A-T-R domain organization are widespread among fungal species (due in large part to the involvement of this module in fungal lysine biosynthesis (37, 38)), monomodular systems ending in C-domains are much less common and are restricted almost exclusively to the Eurotiomycetes class, most notably among the genus Aspergillus. In addition to Af12050 and TqaB, 23 monomodular NRPSs were found with domain structure A-T-C. Fifteen of these were identified from Aspergillus (or Neosartorya) genomes, two from Uncinocorpus reesii, one from Podospora anserina and four in total from the Basidiomycota Schizophyllum commune and Puccinia graminis. Of the 23 found, only four are clustered next to multimodular synthetases. ACLA_017900 (part of a putative tryptoquivaline cluster) and NFIA_057990 (part of a putative fiscalin A cluster) are next to trimodular NRPSs that are homologous to Af12080 (39). AFLA_010020 and AO090103000223 (orthologous genes) are found adjacent to hybrid PKS-NRPSs. In these four cases the proximity of the mono- and multimodular NRPSs suggests a sequential action for peptide-based framework construction. For the remaining examples of A-T-C proteins, a biosynthetic role is less clear. Interestingly, in 20 out of 23 cases, the catalytically important second histidine of the canonical C-domain active site motif HHxxxDG remains intact (40), while in all cases the structurally integral aspartic acid is conserved (Figure S10).
A simple phylogenetic analysis of the terminal C-domains of monomodular NRPSs revealed a relatively divergent population when analyzed independently (Figure S10); however, this population generally grouped separate from the internal and terminal C-domains of multimodular NRPSs (Figure 7). As anticipated, the terminal C-domains of monomodular NRPSs are more closely related to terminal C-domains of multimodular systems than to internal C-domains. Overall, our sequence-based search and analysis reveal that Af12050 and TqaB represent a small subset of monomodular NRPSs of domain architecture A-T-C. Future studies investigating enzymatic logic similar to that catalyzed by Af12050 and TqaB will be of value to more fully characterize the intriguing functionality of this subset of terminal C-domains. Additionally, as the number of known Af12050 and TqaB homologs increases it is intriguing to postulate that it may be possible to provide a sequence-based prediction of the stereospecificity of C-N bond formation in analogous couplings of amino acids to oxidized-indolic scaffolds.
Figure 7.
Neighbor-joining phylogenetic tree and active site motif of select fungal C-domains, highlighting the divergent grouping of C-domains of monomodular NRPSs (abbreviated “st” for “single-module terminal”) from those internal (“i”) and C-terminal (“t”) of multimodular synthetases. Arrows highlight the two C-domains biochemically characterized in this work. Sequence alignment and tree construction were performed using Clustal W (41). (For a comprehensive phylogenomic analysis of fungal NRPSs see Bushley and Turgeon (42) and Cramer Jr., et al.(17))
A series of metabolites have been identified from a variety of fungal sources which appear to be derived from amino acid coupling similar to that catalyzed by Af12050 and TqaB (for representative examples see Figure S11A). The population of analogous compounds with relative stereochemistry matching that of FQA (in which the relative orientation of the oxy-indole derived hydroxyl to the C-N bond formed during the amino acid addition is trans) and those with relative stereochemistry matching that of 2′-epi-FQA (in which the relative orientation is trans) appears to be approximately equal. It is interesting to reason that bioinformatic analysis of the currently unidentified gene clusters responsible for the biosynthesis of chaetominine and asperlicin would reveal a monomodular NRPS with a terminal C-domain more similar to that of Af12050 than that of TqaB. Whereas lumpidin and fiscalin biosynthetic gene clusters may reveal a monomodular NRPS with a terminal C-domain more similar to that of TqaB than that of Af12050 (Figure S11B). Such that all the terminal C-domains catalyzing amino acid addition trans to the neighboring hydroxyl are more closely related than those catalyzing the opposite relative stereochemical addition and vice versa.
In conclusion, herein we describe the utilization of homologous enzymes from two different biosynthetic pathways to illustrate the mediation of stereospecific C-N bond formation by a catalytic action of a fungal terminal C-domain. Investigations with full length proteins, C-domain swapped chimeric proteins and the corresponding stand-alone C-domains with the required substrates all prove the C-domain is necessary and sufficient to control the stereochemistry around the observed 5-membered ring. The mechanism of control over the stereospecific addition remains unclear despite efforts to probe the order of steps involved.
Phylogenetic comparison terminal C-domains observed in these types of systems illustrates a clear division from that of canonical C-domains involved in typical amide bond formation. This suggests a clear evolutionary pathway for C-domains of this type devised to increase the complexity of NRPS synthesized peptides by tailoring with carefully moieties of strictly controlled stereochemistry.
Supplementary Material
ACKNOWLEDGMENT
We thank Elizabeth Sattely for providing purified Bacillus subtilis Sfp and Steven Malcolmson for helpful discussion.
Abbreviations
- 2-AIB
2-aminoisobutyric acid
- A-
adenylation
- AMP
adenosine-5′-monophosphate
- Ant
anthranilate (2-aminobenzoate)
- ATP
adenosine-5′-triphospate
- BOC
tert-butyloxycarbonyl
- BSA
bovine serum albumin
- CoA
coenzyme A
- CV
column volumes
- CPM
counts per minute
- DAD
diode-array detector
- DCM
dichloromethane
- DMSO
dimethyl sulfoxide
- EDTA
ethylenediaminetetraacetic acid
- epi
epimer
- FAD
flavin adenine dinucleotide
- FQA
fumiquinazoline A
- FQC
fumiquinazoline C
- FQD
fumiquinazoline D
- FQF
fumiquinazoline F
- gem
geminal
- ESI
electrospray ionization
- HPLC
high-performance liquid chromatography
- HRMS
high resolution mass spectra
- IPTG
isopropyl-β-D-galactopyranoside
- LB
Luria-Bertani medium
- LC-MS
liquid chromatography mass spectrometry
- LIC
Ligation Independent Cloning
- MeCN
acetonitrile
- MWCO
molecular weight cut-off
- NADH
Nicotinamide adenine dinucleotide
- Ni-NTA
nickel nitrilotriacetic acid-agarose
- NMR
nuclear magnetic resonance
- NRPS
nonribosomal peptide synthetase
- PCR
polymerase chain reaction
- PDB ID
Protein Data Bank identifier
- PPT
4′-phosphopantetheine
- PPtase
phosphopantetheinyl transferase
- PyBOP
benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate
- QTOF
quadrupole time-of-flight
- RT-PCR
reverse transcription polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SNAC
N-acetylcysteamine thioester
- SPE
solid-phase extraction
- TCA
trichloroacetic acid
- TFA
trifluoroacetic acid
- THF
tetrahydrofuran
- TQA
tryptoquialanine
- Tris
Tris(hydroxymethyl)aminomethane
Footnotes
This work is supported in part by National Institutes of Health Grant GM20011 (to C.T.W.), F32GM090475 (postdoctoral fellowship to B.D.A.) and 1R01GM092217 (to Y.T.).
SUPPORTING INFORMATION PARAGRAPH
Figures providing sequence alignments of TqaH/Af12060 (Figure S1) and TqaB/Af12050 (Figure S2), comparison of loading of Af12050 and TqaB standalone T-domains (Figure S3), timecourses of 2′-epi-FQA formation both spontaneous and enzymatic (Figure S4). Depictions of protein purity illustrated by SDS-PAGE and estimation of flavin content of monoxygenases Af12060 and TqaH (Figure S5), and UV absorbance spectra of FQ scaffold compounds (Figure S6). Figures showing substrate specificity of chimera 1 and chimera 2 (Figures S7 and S8), synthesis of L-Ala SNAC and analogs (Figure S9), phylogenetic analysis of fungal monomodular A-T-C NRPSs (Figure S10) and providing additional examples of fungal metabolites with relative stereochemistries equally to FQA or 2′-epi-FQA (Figure S11). Tables showing primers used in this work (Table S1) and NMR spectroscopic characterization of FQF diol (Table S2) and 2′-epi-FQA (Table S3). This supplementary material may be accessed free of charge online at http://pubs.acs.org.
REFERENCES
- 1.Daniel J. F. d. S., Rodrigues Filho E. Peptaibols of Trichoderma. Nat. Prod. Rep. 2007;24:1128–1141. doi: 10.1039/b618086h. [DOI] [PubMed] [Google Scholar]
- 2.Pruksakorn P, et al. Trichoderins, novel aminolipopeptides from a marine spongederived Trichoderma sp., are active against dormant mycobacteria. Bioorg. Med. Chem. Lett. 2010;20:3658–3663. doi: 10.1016/j.bmcl.2010.04.100. [DOI] [PubMed] [Google Scholar]
- 3.Peltola J, et al. Biological effects of Trichoderma harzianum peptaibols on mammalian cells. Appl. Environ. Microbiol. 2004;70:4996–5004. doi: 10.1128/AEM.70.8.4996-5004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBattista C. Antidepressant agents. In: Katzung BG, editor. Basic & Clinical Pharmacology. Eleventh Edition McGraw-Hill Companies; New York: 2009. [Google Scholar]
- 5.Meltzer H. Antipsychotic agents & lithium. In: Katzung BG, editor. Basic & Clinical Pharmacology. Eleventh Edition McGraw-Hill Companies; New York: 2009. [Google Scholar]
- 6.Zhao S, et al. Biological activity of the tryprostatins and their diastereomers on human carcinoma cell lines. J. Med. Chem. 2002;45:1559–1562. doi: 10.1021/jm0155953. [DOI] [PubMed] [Google Scholar]
- 7.Rabindran SK, et al. Fumitremorgin C reverses multidrug resistance in cells transfected with the breast cancer resistance protein. Cancer Res. 2000;60:47–50. [PubMed] [Google Scholar]
- 8.Cole RJ. Fungal tremorgens. J. Food Prot. 1981;44:715–722. doi: 10.4315/0362-028X-44.9.715. [DOI] [PubMed] [Google Scholar]
- 9.Valdes JJ, et al. Aflatrem: a tremorgenic mycotoxin with acute neurotoxic effects. Environ. Health Perspect. 1985;62:459–463. doi: 10.1289/ehp.8562459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao X, et al. Fungal indole alkaloid biosynthesis: genetic and biochemical investigation of the tryptoquialanine pathway in Penicillium aethiopicum. J. Am. Chem. Soc. 2011;133:2729–2741. doi: 10.1021/ja1101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Numata A, et al. Fumiquinazolines, novel metabolites of a fungus isolated from a saltfish. Tetrahedron Lett. 1992;33:1621–1624. [Google Scholar]
- 12.Frisvad JC, et al. Metabolomics of Aspergillus fumigatus. Med. Mycol. 2009;47:S53–S71. doi: 10.1080/13693780802307720. [DOI] [PubMed] [Google Scholar]
- 13.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 14.Schwarzer D, et al. Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 2003;20:275–287. doi: 10.1039/b111145k. [DOI] [PubMed] [Google Scholar]
- 15.Strieker M, et al. Nonribosomal peptide synthetases: structures and dynamics. Curr. Opin. Struct. Biol. 2010;20:234–240. doi: 10.1016/j.sbi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Eisfeld K. Non-ribosomal peptide synthetases of fungi. In: Anke T, Weber D, editors. Physiology and Genetics: Selected Basic and Applied Aspects. Springer-Verlag; Heidelberg: 2009. [Google Scholar]
- 17.Cramer RA, Jr, et al. Phylogenomic analysis of non-ribosomal peptide synthetases in the genus Aspergillus. Gene. 2006;383:24–32. doi: 10.1016/j.gene.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Maiya S, et al. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. ChemBioChem. 2006;7:1062–1069. doi: 10.1002/cbic.200600003. [DOI] [PubMed] [Google Scholar]
- 19.Weber G, et al. The peptide synthetase catalyzing cyclosporine production in Tolypocladium niveum is encoded by a giant 45.8-kilobase open reading frame. Curr. Genet. 1994;26:120–125. doi: 10.1007/BF00313798. [DOI] [PubMed] [Google Scholar]
- 20.Sussmuth R, et al. Fungal cyclooligomer depsipeptides: from classical biochemistry to combinatorial biosynthesis. Nat. Prod. Rep. 2011;28:99–124. doi: 10.1039/c001463j. [DOI] [PubMed] [Google Scholar]
- 21.Eisendle M, et al. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding L-ornithine N 5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC) Mol. Microbiol. 2003;49:359–375. doi: 10.1046/j.1365-2958.2003.03586.x. [DOI] [PubMed] [Google Scholar]
- 22.Gao X, Tang Y. Personal communication. 2011 [Google Scholar]
- 23.Ames BD, et al. Enzymatic processing of fumiquinazoline F: a tandem oxidativeacylation strategy for the generation of multicyclic scaffolds in fungal indole alkaloid biosynthesis. Biochemistry. 2010;49:8564–8576. doi: 10.1021/bi1012029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkins MR, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 25.Soding J, et al. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeifer BA, et al. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 27.Quadri LE, et al. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 28.Samel SA, et al. Structural and functional insights into a peptide bond-forming bidomain from a nonribosomal peptide synthetase. Structure. 2007;15:781–792. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Ames BD, et al. 2010. Unpublished observations.
- 30.Roche ED, Walsh CT. Dissection of the EntF condensation domain boundary and active site residues in nonribosomal peptide synthesis. Biochemistry. 2003;42:1334–1344. doi: 10.1021/bi026867m. [DOI] [PubMed] [Google Scholar]
- 31.Stachelhaus T, et al. Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science. 1995;269:69–72. doi: 10.1126/science.7604280. [DOI] [PubMed] [Google Scholar]
- 32.Fischbach MA, et al. Directed evolution can rapidly improve the activity of chimeric assembly-line enzymes. Proc. Natl. Acad. Sci. USA. 2007;104:11951–11956. doi: 10.1073/pnas.0705348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mootz HD, et al. Construction of hybrid peptide synthetases by module and domain fusions. Proc. Natl. Acad. Sci. USA. 2000;97:5848–5853. doi: 10.1073/pnas.100075897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehmann DE, et al. Aminoacyl-SNACs as small-molecule substrates for the condensation domains of nonribosomal peptide synthetases. Chem. Biol. 2000;7:765–772. doi: 10.1016/s1074-5521(00)00022-3. [DOI] [PubMed] [Google Scholar]
- 35.Sattely ES, Walsh CT. A latent oxazoline electrophile for N−O−C bond formation in pseudomonine biosynthesis. J. Am. Chem. Soc. 2008;130:12282–12284. doi: 10.1021/ja804499r. [DOI] [PubMed] [Google Scholar]
- 36.Keating TA, et al. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry. 2000;39:15513–15521. doi: 10.1021/bi001651a. [DOI] [PubMed] [Google Scholar]
- 37.Ehmann DE, et al. Lysine biosynthesis in Saccharomyces cerevisiae: mechanism of α-aminoadipate reductase (Lys2) involves posttranslational phosphopantetheinylation by Lys5. Biochemistry. 1999;38:6171–6177. doi: 10.1021/bi9829940. [DOI] [PubMed] [Google Scholar]
- 38.Torruella G, et al. The evolutionary history of lysine biosynthesis pathways within eukaryotes. J. Mol. Evol. 2009;69:240–248. doi: 10.1007/s00239-009-9266-x. [DOI] [PubMed] [Google Scholar]
- 39.Ames BD, Walsh CT. Anthranilate-activating modules from fungal nonribosomal peptide assembly lines. Biochemistry. 2010;49:3351–3365. doi: 10.1021/bi100198y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rausch C, et al. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol. Biol. 2007;7:78. doi: 10.1186/1471-2148-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 42.Bushley K, Turgeon BG. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol. Biol. 2010;10:26. doi: 10.1186/1471-2148-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.