Abstract
Epithelial antimicrobial activity may protect the lung against inhaled pathogens. The bactericidal/permeability-increasing protein family has demonstrated antimicrobial activity in vitro. PLUNC (palate, lung and nasal epithelium carcinoma associated) is a 25 kDa secreted protein that shares homology with bactericidal/permeability-increasing proteins and is expressed in nasopharyngeal and respiratory epithelium. The objective of this study was to determine whether PLUNC can limit Pseudomonas aeruginosa infection in mice. Transgenic mice (Scgb1a1-hPLUNC) were generated in which human PLUNC (hPLUNC) was directed to the airway epithelium with the Scgb1a1 promoter. The human PLUNC protein (hPLUNC) was detected in the epithelium throughout the trachea and bronchial airways and in bronchoalveolar lavage fluid (BALF). BALF from transgenic mice exhibited higher antibacterial activity than that from wild type littermates in vitro. Following in vivo P. aeruginosa challenge, Scgb1a1-hPLUNC transgenic mice displayed enhanced bacterial clearance. This was accompanied by a decrease in neutrophil infiltration and cytokine levels. More importantly, the over-expressed hPLUNC in Scgb1a1-hPLUNC transgenic mouse airway significantly enhanced mouse survival against P. aeruginosa induced respiratory infection. These data indicate that PLUNC is a novel antibacterial protein that likely plays a critical role in airway epithelium mediated innate immune response.
Introduction
The ability of the host to avoid infection depends largely on mechanisms of innate immunity. This rapid-response system acts efficiently without prior exposure to a pathogen (1) and is initiated when bacterial products are detected. Lipopolysaccharide (LPS), a cell wall component of gram negative bacteria, is an agonist for innate immune response through activation of the toll-like receptor 4 (TLR4) signaling cascade. Exposure to LPS results in a production of pro-and anti-inflammatory mediators by myeloid lineage and other cell types including epithelial cells. In humans, bactericidal/permeability-increasing protein (BPI) and LPS-binding protein (LBP) can bind LPS and modulate the host response to gram negative bacterial infections. Although BPI and LBP belong to the same protein family (i.e. BPI protein family, their functions are antagonistic: LBP exhibits pro-inflammatory activity while BPI displays anti-inflammatory activity and direct bactericidal action (2–3). Two additional lipid transfer LPS-binding plasma proteins, cholesteryl ester transfer protein (CETP) and phospholipid transfer protein (PLTP) are included in the BPI protein family based on predicted structural similarity (4–5).
Based on predicted structural homology, palate, lung and nasal epithelium associated protein (PLUNC) has been added to the BPI protein family. PLUNC protein is expressed specifically in the nasopharyngeal and respiratory epithelium (6–8). The PLUNC genes are clustered on human chromosome 20 and encode eight different proteins that share some predicted structural similarity (9). PLUNC or Short PLUNC protein 1 (SPLUNC1) is the first identified PLUNC protein that has multiple alternative names including secretory protein in upper respiratory tracts (SPURT), lung-specific protein X (LUNX), and nasopharyngeal carcinoma-related protein (NASG).
All BPI protein family members are defined by a similarly predicted three dimensional structure with a single predicted disulphide bond and a conserved exon structure (7). However, the amino acid homology among members of the BPI protein family is typically around 15%–30% (10). The biological function of PLUNC is not known, but structural similarities to BPI family proteins suggest that PLUNC may be involved in defense responses in the airways. Chu et al. reported that human and mouse bronchial epithelial cells expressed PLUNC (hSPLUNC1 and mSPlunc1), and that exogenously applied PLUNC decreased Mycoplasma pneumoniae levels and lessened interleukin 8 (IL8) production in vitro (11). Conversely, PLUNC siRNA enhanced M. pneumoniae growth and IL8 production. IL13 significantly decreased PLUNC and M. pneumoniae clearance in epithelial cell cultures suggesting that decreased PLUNC expression during allergic responses may contribute to asthma pathobiology. Zhou et al. found that PLUNC protein binds LPS and inhibits the growth of Pseudomonas aeruginosa in vitro (12). Garcia-Caballero et al. reported PLUNC may serve as an airway surface liquid volume sensor by regulating epithelial sodium ion-channel (ENaC) activity (13). Gakhar demonstrated that PLUNC has surfactant activity and that this activity may account for the low surface tension of airway secretions (14). What was lacking in these studies is a demonstration that PLUNC has antimicrobial activity in vivo and that additional PLUNC may be protective during infection. To determine whether PLUNC can limit Pseudomonas aeruginosa infection in the lung, transgenic mice (Scgb1a1-hPLUNC) were generated in which human PLUNC was directed to the airway epithelium with the Scgb1a1 promoter and these mice were challenged with P. aeruginosa.
Materials and Methods
Generation of Scgb1a1-hPLUNC transgenic mice
Constitutive expression of human PLUNC (hPLUNC) protein in the mouse respiratory epithelium was achieved through generation of mice harboring a transgene composed of the human PLUNC cDNA under the transcriptional control of the mouse Scgb1a1 (also known as Clara cell secretory protein, CCSP) promoter (Figure 1A). Total human airway epithelial cell RNA was used as a template for RT-PCR amplification of PLUNC cDNA (0.78kb). The forward primer included the protein translational start codon and 12 nucleotides of flanking sequence: 5′-ATA AGA ATG CGG CCG CCT AAG AGC AAA GAT GTT TC-3′; while the reverse primer covered the translational stop site: 5′-ATA AGA ATG CGG CCG CAC CTT GAT GAC AAA CTG-3′. This cDNA coded for the active and mature PLUNC protein. The PCR product was gel purified, cloned using the pCR2.1-TOPO cloning system (Invitrogen, Carlsbad, CA), and sequenced. A 9.6kb Scgb1a1 genomic fragment in pUC19 (kindly supplied by Dr. Magnus Nord, Karolinska Institute, Stockholm, Sweden) was modified by site directed mutagenesis to include a NotI site upstream of the Scgb1a1 translational start site and resulted in generation of pNotI-9.Scgb1a1. In contrast to the short (2.1 kb) Scgb1a1 promoter, the 9.6 kb construct included approximately 3kb and 2kb of 5′-and 3′ gene-flanking sequence, respectively. The human PLUNC cDNA was cloned into the NotI site of pNotI-9.6ScgbB1a1 and the construct verified by restriction enzyme digestion and sequencing. The 10.38 kb Scgb1a1-hPLUNC cDNA fragment was isolated by digestion with SphI and transgenic generated by microinjection into FVB/N mouse oocytes. One of 19 offspring was positive for the transgene, as assessed by PCR genotyping and confirmed by Southern blot analyses of tail DNA. Subsequently, genotype was determined by PCR. The endogenous Scgb1a1 gene and the Scgb1a1-hPLUNC transgene were distinguished as 450bp and 545bp amplicons using a mouse Scgb1a1 promoter-specific forward primer: 5′-GTT GGC AAG TCT ACA GTT GC-3′, a PLUNC coding region forward primer: 5′-GAC GTC AGT GAT TCC TGG CC-3′, and in combination with a Scgb1a1 intron 1-specific reverse primer: 5′-GAA AGA GAC CCT GGG CAC TCA-3′.
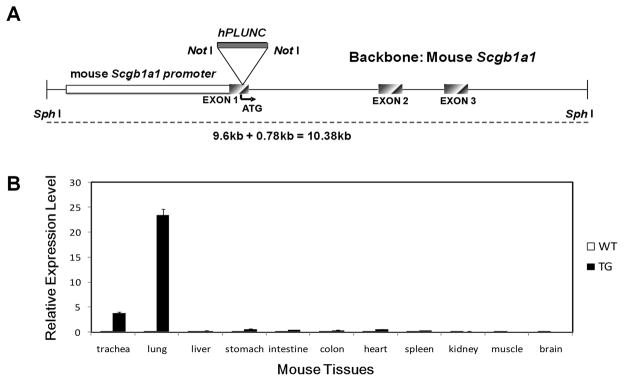
Fig. 1. Generation and assessment of mouse and human PLUNC gene expression in Scgb1a1-PLUNC transgenic mice.
A: Transgene construct. B: Real time Taqman RT-PCR analysis of human PLUNC mRNA abundance in tissues from wild type and transgenic mice. Relative expression was determined by the ΔΔCt method using mouse GUS-B RNA as a control. (mean ± S.D., n = 10). Human PLUNC was highly expressed in intrapulmonary airways of transgenic mice.
Animal Husbandry
The Scgb1a1-hPLUNC transgene was maintained in a hemizygous state by breeding with wild-type FVB/N mice. Mendelian transmission frequencies were observed and no overt consequences of the transgene on growth, breeding, or survival were noted. Mice were maintained in a specific pathogen free status in a 12h light/dark cycle conditions. All procedures were conducted using mice 8–12wk of age maintained in ventilated microisolator cages housed in an American Association for Accreditation of Laboratory Animal Care (AAALAC)-accredited animal facility. Protocols and studies involving animals were conducted in accordance with National Institutes of Health guidelines and approved by Institutional Animal Care and Use Committee (IACUC) at the University of Pittsburgh.
Real time PCR analysis
Total RNA was isolated from various tissues of wild type and transgenic mice by a single-step acid guanidium thiocyanate extraction method (15). qRT-PCR (ABI7700; Applied Biosystems, Foster City, CA) was performed using human specific PLUNC primers (forward 5′-TTC AGG GCA ACG TGT GCC-3′, reverse 5′-TAG TCC GTG GAT CAG CAT GTT AAC A-3′, probe 5′-/56-FAM/CTG GTC AAT GAG GTT CTC AGA GGC TTG G TAMTph/-3′) or mouse specific plunc primers (forward 5′-TGG GAT TCT CAG CGG TTT GGA TGT -3′, reverse 5′-TCA GCC AAG ATA GCC TTC CTT CCT -3′, probe 5′-/56-FAM/CAC CCT GGT GCA CAA CAT TGC TGA AT/TAMTph/-3′). Validation tests were performed to confirm equivalent PCR efficiencies for the target genes. Test and calibrator lung RNAs (Ambion, Austin, TX) were reverse transcribed using a Superscript III kit (Invitrogen), and PCR amplified as follows: 95°C for 12min, 40cycles; 95°C for 15s; 60°C for 1min. Three replicates were used to calculate the average cycle threshold for the transcript of interest and for a transcript for normalization [glucuronidase, beta (GUS-B)] (Assays on Demand, Applied Bioystems). Relative mRNA abundance was calculated by the delta delta CT method.
Gram negative bacteria Pseudomonas aeruginosa
The P. aeruginosa strain (PAO1, ATCC BAA-47) was used for all experiments. P. aeruginosa obtained from a single colony was stored in aliquots at −80°C in 20% glycerol/Luria-Bertoni (LB) broth. For each experiment, an aliquot of bacteria was thawed, inoculated into 10ml LB, and incubated (6h;37°C) with shaking. An aliquot was then diluted 1:100 into 100ml LB broth and incubated (16h; 37°C). Bacteria were washed twice and resuspended in 10ml phosphate-buffered saline (PBS) containing 10mM magnesium chloride.
Colony Forming Unit (CFU) Assay
The number of colony forming units (CFU) was determined by serial dilution and quantitative culture on LB agar plates. P. aeruginosa was resuspended in PBS and the optical density (OD470) was adjusted to ~0.74, approximately 2 × 1011 bacteria/ml. Five serial 10-fold dilutions in PBS were prepared immediately before the use. Duplicate bacterial samples were mixed with 50μl of PBS, or BALF from either transgenic or wild-type control mice were prepared and placed on ice. An equal volume of the 2 × 105 bacteria/ml solution was added to the wells. Samples were mixed and incubated (2 h; 37°C) with shaking. Three 50μl aliquots with different concentrations (no dilution, five-fold dilution, and ten-fold dilution) from each well were plated on LB agar plates. Plates were inverted and incubated overnight at 37°C. Colonies were counted and the number of colonies per plate determined. For antibody neutralization studies, mouse BALF samples (50 μl) were pre-incubated (1h; 23°C) with 7.5 μl normal rabbit serum (NRS) or with anti-PLUNC serum hour with gentle agitation and CFU were calculated.
In vivo exposure of mice to Pseudomonas aeruginosa
Gender-matched 6- to 8-week-old wild-type and transgenic mice were co-exposed to P. aeruginosa aerosol using an inhalation exposure system (model A42X, Glas-Col, Terre Haute, IN) as previously described (16). Mice were placed in a compartmentalized mesh basket (five chambers, 20 mice/chamber capacity) and exposed (45min); followed by cloud decay (15 min) and decontamination (5 min; UV irradiation). Preliminary studies demonstrated that exposure to 1010 bacteria/ml for 45min resulted in a bacterial deposition of about 2 × 106 CFU/lung immediately after exposure (5 independent experiments, n = 5~6/experimental group, 1.6 2.5 × 106, median and 25th–75th percentile). For survival test, female 6- to 8-week-old wild-type and transgenic mice were infected with by an intra-tracheal instillation of P. aeruginosa at a concentration of 1×109 cfu/mouse.
Bronchoalveolar lavage and cell differential counts
At 4 h or 24 h after aerosol exposure, mice (5~6 mice/group) were anesthetized with 2.5% Avertin. The trachea was cannulated,, the lungs were lavaged (1 ml PBS twice), and the BALF samples pooled (pool 1). The lungs were lavaged an additional five times with 1 ml PBS and the recovered fluid pooled (pool 2). Cells from the two pools were recovered through centrifugation at 300 g and resuspended in 0.5ml PBS. A 50μl aliquot was stained with an equal volume of 0.4% trypan blue (Invitrogen, San Diego, CA) and cells counted with a hemocytometer. An additional aliquot placed onto glass microscope slides (Shanon Cytospin, Thermo Fisher, Pittsburgh, PA), stained with Diff-Quick, and cell differential determined microscopically. Protein concentration in pool 1 was determined by DC Protein assay using bovine serum albumin standards (Bio-Rad, Hercules, CA).
Bacterial clearance
Bacterial deposition was determined by harvesting the right lungs immediately after infection (n = 4 mice group). Bacterial clearance was assayed 4 and 24 hours post infection (n = 5~6 mice/group). The right lungs or spleen were placed into 1ml sterile PBS and kept on ice prior to homogenization. Six serial 10-fold dilutions were prepared and 50μl aliquots plated on LBagar plates. Each dilution was plated in triplicate. Plates were inverted and incubated overnight at 37°C. The number of viable bacteria in the lung and spleen was determinedand expressed as CFU per lung.
Immunohistochemistry
Lungs were inflation fixed in situ with 4% paraformaldehyde (10cm H2O; 10min) with chest cavity open. The lower right lobe was embedded in paraffin and 5μm sections prepared. Sections were cleared with xylenes, hydrated to water, and quenched in 3% H2O2 (10min). Antigen retrieval was performed in 10 mMcitrate buffer (20min; 100°C; pH 6.0). Sections were blocked in 0.03% casein in 0.05% PBS/Tween-20 solution (1h) and incubated (18h, 4°C) with hPLUNC antibody (R&D Systems, Minneapolis, MN). Normal goat serum was used as a control. Following extensive washing with PBS, sections were incubated (2h; 23°C) with biotinylated donkey anti-goat antibody (Pierce Biotechnology, Rockford, IL.) diluted in blocking solution. Sections were washed (30min; 23°C) and incubated with streptavidin-HRP (Pierce Biotechnology) in PBS. Antigen-antibody complexes were detected with Metal Enhanced DAB solution (Pierce Biotechnology). Tissue was counterstained with Mayer’s hematoxylin solution (Sigma-Aldrich Co., St Louis, MO).
Dual Immunofluorescence
Cells co-expressing PLUNC and SCGB1A1 were identified using dual immunofluorescence techniques. Primary antibodies were goat anti-PLUNC (R&D Systems, Minneapolis, MN) and rabbit anti-SCGB1A1 (17). Secondary antibodies (Invitrogen, San Diego, CA) were used in the followingcombinations: Alexa Fluor 488 donkey anti-goat IgG for PLUNCand Alexa Fluor 594 donkey anti-rabbit IgG for SCGB1A1. All secondary antibodies were used at a dilution of 1:500. Stained sections were mounted with Fluoromount-G containing 2 μg/ml 4,6-diamidino-2-phenylindole (DAPI, Sigma). Images of representative fields were acquired with a Provis AX70 microscope (Olympus, Center Valley, PA) equipped with a DAPI/Texas red dual optical excitation filter cube and an FITC optical excitation filter cube (Olympus) and a Spot RT color digital camera (Diagnostic Instrument, Sterling Heights, MI). Images were processed with Image-Pro Plus software (Media Cybernetics, Bethesda, MD).
SDS-PAGE and Western blot analysis
BALF (25μl), lung homogenized in PBS (20μg), or recombinant PLUNC (rPLUNC, 0.1μg) (R&D Systems, Minneapolis, MN) were diluted with sample loading buffer, heated (5min; 95°C), and separated on a NuPAGE 12% Bis-Tris Gel (Invitrogen). Electrophoresis was performed at 70 volts (20min) and continued at 140 volts (80min). For Western blots analysis, proteins were electroblotted onto nitrocellulose membrane (Pierce Biotechnology) in transfer buffer containing 25mM Tris base, 0.2M glycine, 20% methanol (pH~8.5). Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline with 0.1% Tween-20 (TBST)) (1h; 23°C) washed thrice with TBST (5min), and incubated (18h; 4°C) with goat anti-PLUNC antibody (1:1000 in blocking solution, R&D Systems). Membranes were washed thrice with TBST and incubated (1h; 23°C) with mouse anti goat-HRP antibody (1:10,000, Pierce Biotechnology, Inc.). Following 3 washes with TBST, the membranes were developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnology). Densitometry was performed and ImageQuant software (GE Healthcare, Piscataway, NJ) was used to quantify PLUNC protein levels.
Cytokine assay
Cytokine levels in BAL were quantified using mouse Cytokine Multiplex Panel assay (Bio-Rad Inc.). Levels of IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-10, IL-12(p40), IL-17, IFN-γ, TNF-α, GM-CSF, KC, and MIP-1β. Standard recombinant protein solution was used to generate standard curves. Anti-cytokine antibody conjugated xMAP beads and standard or test BALF proteins were added to wells of a 96-well filter plate. The plate was incubated with shaking at room temperature for 30 minutes, washed thrice with Bioplex wash buffer, and 25μl detection antibody was added. After incubation (30min; 23°C), the plate was washed and streptavidin solution added to each well. The plate was incubated (10min) with shaking, washed, and the beads resuspended in Bioplex assay buffer A. The plate was agitated (30s) and absorbance determined with a Bioplex Suspension Array System. Absolute cytokine concentrations were calculated from the standard curve for each cytokine.
Data analysis
Data are expressed as mean ± S.D. Statistical comparisons between the groups of mice were made using ANOVA (continuous and normally distributed data) or Poisson regression (discontinuous data). A p < 0.05 was considered to be statistically significant. Determinations of the significance of the differences in survival outcomes between Scgb1a1-hPLUNC TG mice and WT control littermates after P. aeruginosa infection were calculated by Kaplan-Meier survival curve comparisons and the p values derived from a log-rank test.
Results
Expression of hPLUNC protein in transgenic mouse
A schematic diagram of PLUNC transgene construct using Scgb1a1 promoter is represented in figure 1A. The tissue specificity of hPLUNC transcript expression was assesses by qRT-PCR analysis (Fig 1B) and was detected exclusively in trachea and lung from transgenic mice. This distribution paralleled that of SCGB1A1 mRNA and was consistent with previously reported specificity of the mouse Scgb1a1 promoter.
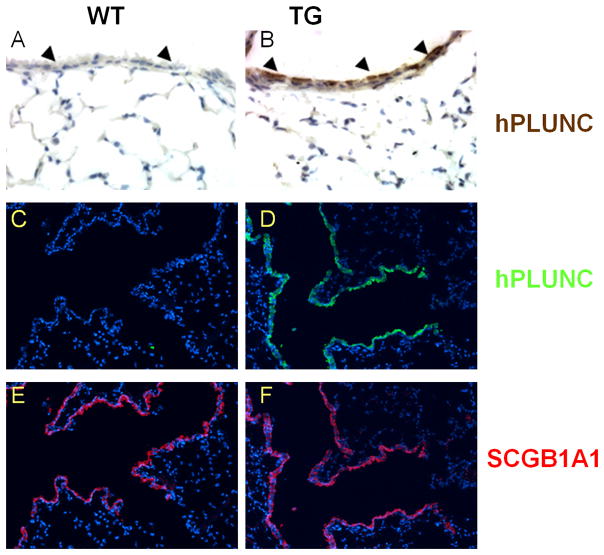
Expression of hPLUNC protein in mouse airways was assessed by immunohistochemical staining of paraffin-embedded trachea and lung sections from wild type control (WT) and transgenic (Scgb1a1-hPLUNC, TG) littermates using an antibody that detects both human and mouse PLUNC. Expression of endogenous mouse PLUNC (mPlunc) was detected in the tracheal and bronchial epithelium (data not shown) but was absent in bronchiolar cells and alveolar epithelial cells of WT control mice (Fig. 2A and Fig. 2C). In transgenic Scgb1a1-hPLUNC mice, PLUNC was detected in tracheal (data not shown), bronchial, and bronchiolar epithelial cells (Fig. 2B and Fig. 2D). The distribution of SCGB1A1 protein was not altered (Fig. 2E and Fig. 2F).
Fig. 2. Human PLUNC protein is expressed in bronchiolar airway epithelial cells in Scgb1a1-PLUNC transgenic mice.
Cellular localization of PLUNC protein in mice was assessed by immuno-histochemistry (A,B) and immuno-fluorescence microscopy (C–F) on lung sections of wild-type (A,C,E) and transgenic (B,D,F) mice using anti-PLUNC antibody. Signal was not detected when parallel sections from transgenic mice were incubated with pre-immune goat serum (data not shown). PLUNC was detected in airway epithelial cells (arrowheads) of transgenic but not WT mice. Original magnification, 400X (A,B) and 100X (C–F).
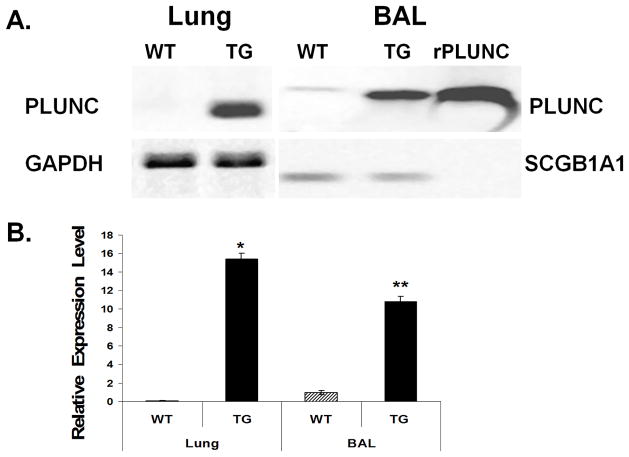
It has been demonstrated that PLUNC is secreted into lumen of human airway and can be detected in sputum (6). To determine whether hPLUNC is similarly secreted in transgenic mice, we used western blot analysis to examine BALF and lung homogenate from WT and TG littermates. SCGB1A1 protein, also known as Clara cell secretory protein (CCSP), was used as a loading control for secreted BALF proteins and GAPDH was used to normalize tissue proteins in lung homogenates. Murine Plunc (mPlunc) is not normally expressed in intrapulmonary airway epithelium and very little protein could be detected in lung homogenate from control littermates. Total PLUNC protein (mouse and human) in BALF was >10-fold greater in transgenic mice as compared to control littermates (Fig. 3). These results indicate that hPLUNC was highly expressed in intrapulmonary airways and that it was effectively secreted into the luminal space.
Fig. 3. Expression and secretion of human PLUNC is significantly increased in Scgb1a1-PLUNC transgenic mice.
A. Western blot analysis was used to assess protein expression of PLUNC in BAL and total lung homogenate from 6-wk-old wild-type (WT) and Scgb1a1-PLUNC transgenic (TG) mice. Proteins were separated by SDS-PAGE, blotted onto PVDF membranes. Blots were incubated with anti-PLUNC antibody, which detected both mouse and human PLUNC. Recombinant protein (rPLUNC) was used as a positive control. GAPDH was used as a loading control for lung homogenates. Mouse SCGB1A1 was used as a control for blots of BAL samples. B. Quantitative results that represent PLUNC protein expression levels detected by Western blot analysis after normalization with GAPDH (lung) or SCGB1A1 (BAL). *p and **p < 0.05 when the transgenic mice group compared with the wild-type mice group with each respective sample type. (mean ± S.D., n = 8).
Enhanced antibacterial activity in BALF from Scgb1a1-hPLUNC transgenic mice
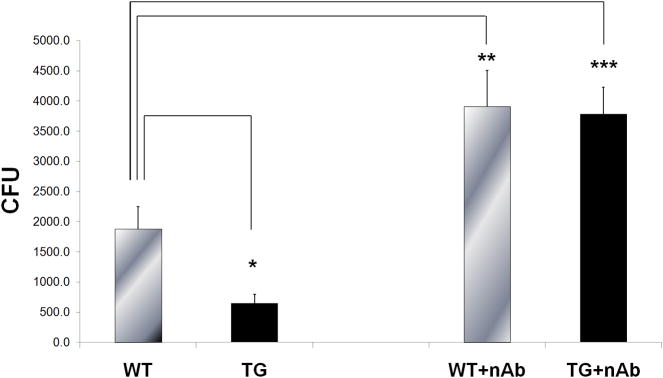
To determine whether BALF from transgenic TG mice had increased anti-bacterial activity, we assessed P. aeruginosa (PAO1, ATCC BAA-47) colony formation in BALF obtained from WT control and Scgb1a1-hPLUNC transgenic littermates. CFU decreased 2.9-fold in cultures treated with BALF from transgenic mice compared to that from control littermates (Fig. 4). To determine the contribution of human and mouse PLUNC to this bactericidal activity, pre-immune normal rabbit serum immunoglobulin G (IgG) or rabbit anti-PLUNC IgG were pre-incubated with BALF and antibacterial assay was performed. Antibody neutralization by neutralizing anti-PLUNC IgG decreased total bactericidal activity of WT control BALF indicating that endogenous mouse Plunc was responsible for a portion of the activity (Fig 4). In addition, the bactericidal activity of antibody treated Scgb1a1-hPLUNC BALF was not different from antibody treated BALF from WT control littermates. These results demonstrated that murine Plunc (mPlunc) is active in the airway, and that the additional hPLUNC is secreted to and is active in the airway lumen in Scgb1a1-hPLUNC transgenic mice.
Fig. 4. BALF from transgenic mice exhibits enhanced antimicrobial activity.
Left. BALF from unchallenged WT and TG littermates was mixed with P. aeruginosa (PAO1) as described in Materials and Methods and colony forming units (CFU) were assessed. Right. Neutralizing antibody against PLUNC was pre-incubated with WT or TG BALF for 1h prior to mixing with bacteria and CFU counts were assessed. Results are the mean and S.D. from three independent experiments (n = 8 for each experiment), *p < 0.05 when the transgenic mice group compared with the wild-type mice group.
Murine Plunc enhanced host defense in mouse lung against inhaled P. aeruginosa
To evaluate the effect of inhaled P. aeruginosa on mPlunc expression, lung mRNA levels were assessed by qRT-PCR analysis. PAO1 bacterial exposure significantly increased Plunc expression at both 6h (5.1 fold, p=0.0002) and 24h (2.5 fold, p=0.021) after the exposure (Fig. 5) but the induction of Plunc at 6h after exposure was higher than 24h post exposure. To determine the functional significance of increased plunc in mouse lung after bacterial infection, the mice were pre-treated with specific anti-PLUNC antibody to neutralize the antimicrobial activity of PLUNC in mouse airways. The antibody concentration at either 10 or 25 μg did not show any noticeable difference in affecting bacterial susceptibility. However, a pre-treatment on mice with either concentration of neutralizing antibody significantly increased the mouse susceptibility to PAO1 induced bacterial infection when compared with the control mice that were pre-treated with non-specific IgG (Fig. 6). There was at least three-fold increase in bacterial burden of mouse lung when mice were pre-treated with the neutralizing antibody. These data further supported the important role of PLUNC in host defense against P. aeruginosa induced lung infection.
Fig. 5. Exposure to P. aeruginosa increases the mRNA expression of mPlunc.
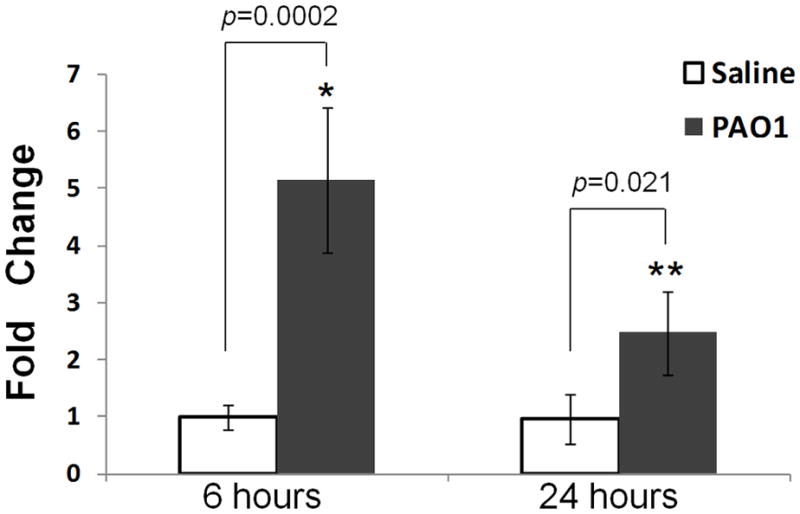
Murine Plunc expression was analyzed 6h and 24h after aerosolized exposure of P. aeruginosa PAO1 strain to FVB/N wild-type mice and all data expressed as fold induction over saline-exposed mice for time point at 6h post exposure. Levels of mPlunc was quantified in lung homogenates by real-time RT-PCR and determined by the ΔΔCt method using mouse GUS-B RNA as a control. Results are mean ± SEM from two experiments; n = 6~8 mice for each group. *p < 0.001 and **p < 0.05 when the PAO1 exposed mice group compared with the saline exposed mice group at 6h and 24h after exposure.
Fig. 6. Pre-treatment with anti-PLUNC antibody increases susceptibility of wild-type mice to bacterial infection.
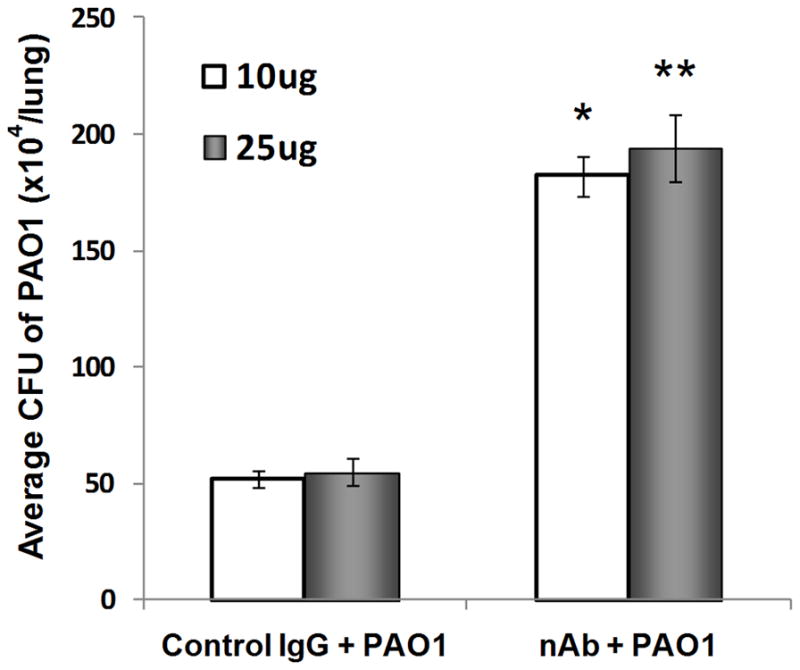
Wild-type FVB/N mice were intranasally instilled (10 & 25 μg/mouse) with control rabbit IgG or neutralizing mouse anti-PLUNC antibody (nAb) 2h prior to exposure to ~2 × 106 CFU/lung of aerosolized P. aeruginosa (PAO1). The number of bacteria in lung homogenates was determined 24h after bacterial exposure. Anti-PLUNC Ab treatment resulted in a significant increase in CFU counts compared with IgG control. Results are mean ± SEM from two experiments; n = 5 mice for each group. *p < 0.05 and **p < 0.05 when two different neutralization antibody concentrations were compared with their respective IgG control group.
Improved bacterial clearance in Scgb1a1-hPLUNC transgenic mice against inhaled P. aeruginosa
To determine whether hPLUNC expression could alter bacterial infection in vivo, WT control and Scgb1a1-hPLUNC transgenic littermates were co-exposed to saline or P. aeruginosa aerosols and deposition and clearance monitored. The aerosolized exposure protocol resulted in a deposition of 2.26±0.39 × 106 CFU/lung P. aeruginosa and was consistent among exposures. Immediately after exposure, bacterial deposition was equivalent between transgenic mice and WT control littermates (Fig. 7). This exposure concentration enabled us to examine PLUNC-mediated antimicrobial activity after a reversible respiratory infection. Because a whole body exposure method was utilized, a systemic bacteremia was also assessed by determining the CFU in spleen homogenates 24h post infection. Less than 500 CFU were detected in spleens of mice and no statistical difference was detected between the transgenic mice as compared to their control littermates.
Fig. 7. Scgb1a1-PLUNC transgenic mice are resistant to P. aeruginosa induced lung infection.
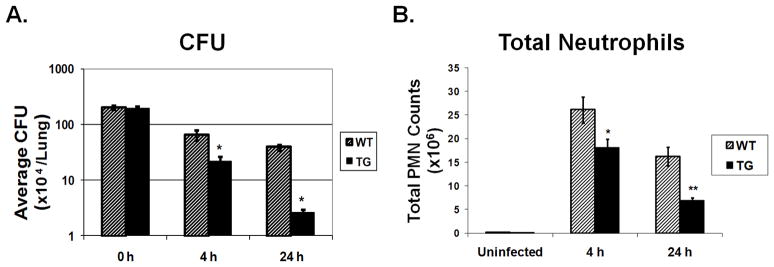
Age-matched WT and TG littermates were infected with ~2 × 106 CFU P. aeruginosa (PAO1) per lung by inhalation. A. Colony forming units (CFU) in lung homogenates were determined at the indicated recovery time points. *p < 0.01 for WT to TG comparisons. B. Total neutrophils in BAL fluid from WT compared to TG mice 4h and 24 h after bacterial exposure (*p<0.05). Total neutrophils were significantly lower in TG mice 24 h following infection (**p < 0.01). Results are mean ± SD (n = 5~6) from three independent experiments.
Four hours after exposure, WT control mice became slightly lethargic and demonstrated signs of infection (e.g., ruffed fur and hunched back), whereas Scgb1a1-hPLUNC transgenic littermates displayed more active behavior and little observable signs of infection. The bacterial clearance was ~3-fold greater (p<0.05) in lung homogenates from Scgb1a1-hPLUNC transgenic mice (2.31±0.41×105CFU/lung/ml) as compared to that from WT control mice (6.83±0.57×105CFU/lung/ml) (Fig. 7A). At 24h after exposure, WT control and Scgb1a1-hPLUNC transgenic mice exhibited similar activity levels. However, bacteria clearance was ~15-fold greater (p<0.05) in lung homogenates from Scgb1a1-hPLUNC transgenic mice (2.6±0.29×104CFU/lung/ml) as compared to that from WT control mice (3.9±0.34×105CFU/lung/ml) (Fig. 7A). Thus, control mice had cleared 80% whereas transgenic mice had cleared 98% of the initial bacterial burden. These data suggest that PLUNC acts early in infection and promotes efficient clearance of the initial bacterial inoculum.
Decreased inflammatory cell infiltration in Scgb1a1-hPLUNC transgenic mouse lung following inhaled P. aeruginosa
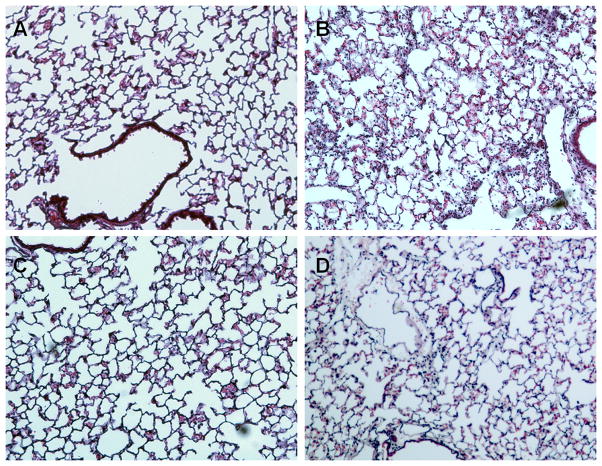
BALF total cell count (2~4×105cells/lung) or differentials (≥99% macrophages) from unexposed Scgb1a1-hPLUNC transgenic mice were not different than those from WT control mice. Four hours after bacterial exposure, the total inflammatory cell counts in BALF increased in both TG and WT control mice but the increase was higher in WT mice than in TG mice. The increased macrophage number between WT (3.1±1.4×106 n=18) and TG (3.6±1.6×106 n=18) mice was not statistically different but the total neutrophils in WT control mice (26.1±2.8×106 n=18) were slightly greater (~1.44-fold, p<0.05) than from Scgb1a1-hPLUNC transgenic mice (18.1±2.3×106 n=18) (Fig. 7B). However, 24h after exposure the total neutrophils were more than 2-fold (p<0.01) greater in BALF from WT control mice (16.2±2.0×106 n=18) as compared to BALF from Scgb1a1-hPLUNC transgenic mice (6.90±0.6×106 n=18) (Fig. 7B). There were no significant differences in the total macrophage number between TG and WT mice at 24h after PAO1 induced bacterial infection. These observations were supported by histological analysis of lung tissue. Lungs from uninfected WT control mice (Fig. 8A) were not different than those from Scgb1a1-hPLUNC transgenic mice (Fig. 8C). However, 24h after bacterial exposure, lung tissues from WT control mice (Fig. 8B) contained more areas of focal peribronchial and alveolar neutrophilic infiltrates than lungs from Scgb1a1-hPLUNC transgenic littermates (Fig. 8D).
Fig. 8. Decreased neutrophilic infiltration Scgb1a1-PLUNC mice after bacterial exposure.
Wild-type and Scgb1a1-PLUNC mice were exposed to saline (A, C) or aerosolized P. aeruginosa (B, D). Lung tissues were harvested at 24h after exposure and fixed for histological evaluation. No histological differences were observed between WT and TG mice prior to bacterial exposure. Following bacterial exposure, TG mice showed less neutrophil infiltration. Original magnification, 40X as indicated. Sections are representative of at least five mice studied at each time point from each infection.
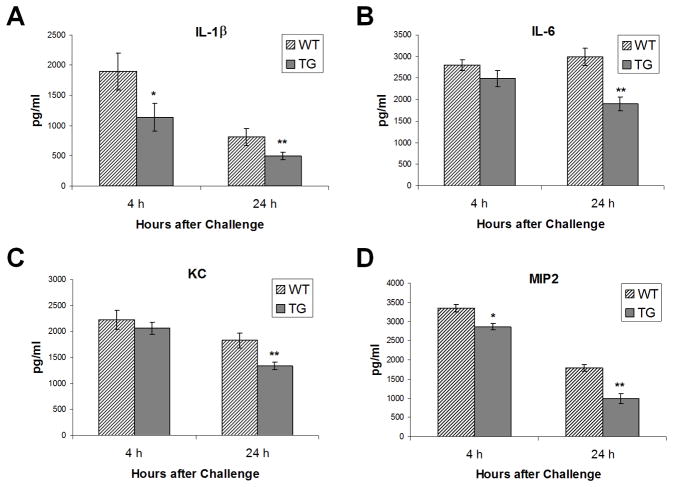
Decreased pro-inflammatory cytokines in Scgb1a1-hPLUNC transgenic mice after P. aeruginosa exposure
To determine whether production of inflammatory cytokines varied 4h or 24h after exposure, BALF samples were collected and cytokine concentrations were determined by BioPlex assay. Without bacterial exposure, BALF cytokine levels from WT control mice were not different than those from Scgb1a1-hPLUNC transgenic mice and most cytokine levels were not detected (data not shown). After exposure, the spectrum of BALF cytokines detected did not vary by genotype, but the magnitude of the response was consistently lower in BALF from Scgb1a1-hPLUNC transgenic mice as compared to that from WT control mice (Fig. 9). At 4h after exposure, interleukin 1, beta (IL1β) levels were lower in BALF from Scgb1a1-hPLUNC transgenic mice as compared to that from WT control mice (Fig. 9A). At 24 h after exposure, the difference was greater and additional cytokines including IL6 (Fig. 9B) and chemotatic cytokines, chemokine (C-X-C motif) ligand 1 (a.k.a. KC) (Fig. 9C) and chemokine (C-X-C motif) ligand 2 (a.k.a. MIP-2) (Fig. 9D), were significantly lower in BALF from Scgb1a1-hPLUNC transgenic mice as compared to that from WT control mice. We did not observe significant differences in the production of IL-2, IL-3, IL-10 and IL-12(p40) in steady state or PA-challenged WT and Scgb1a1-hPLUNC transgenic mice (data not shown).
Fig. 9. Decreased pro-inflammatory cytokine production in Scgb1a1-PLUNC mice following PAO1 exposure.
WT and TG mice were infected with P. aeruginosa as described. Cytokines concentrations in BALF were measured using a Bioplex assay and are reported in pg/ml. Values were determined for 4h and 24h after bacterial exposure. Striped bars, WT mice; solid bars, Scgb1a1-PLUNC TG mice. Results are mean ± SD from three experiments (n = 5~6 mice). *p < 0.05 and **p < 0.05 for WT to TG comparisons at each time point.
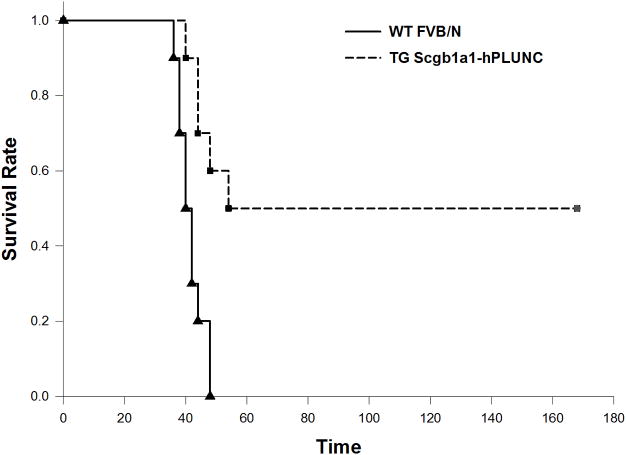
Enhanced survival in Scgb1a1-hPLUNC transgenic mice after P. aeruginosa exposure
To determine if PLUNC affects susceptibility to P. aeruginosa induced lung infection, groups of female 6- to 8-week-old WT and Scgb1a1-hPLUNC transgenic mice were challenged through intra-tracheal instillation with various doses (107, 108, and 109 CFU/mouse) of P. aeruginosa PAO1. We did not observe any mortality when concentrations of PAO1 at 107 and 108 CFU/lung were administered (data not shown). When mice were challenged with 109 CFU/animal of PAO1, Scgb1a1-hPLUNC transgenic mice displayed enhanced survival and decreased lethality (log-rank test, p = 0.001) (Fig. 10). All WT mice succumbed to PAO1 induced lung infection within 48h (mean survival time 41.6±1.3h), while five out of ten Scgb1a1-hPLUNC transgenic mice survived the challenge and were still alive one week after infection. These results indicate that the over-expressed PLUNC is essential to an enhanced survival and resistance to P. aeruginosa infection.
Fig. 10. Over-expressed hPLUNC in Scgb1a1-PLUNC mice enhances the survival from PAO1 induced acute pneumonia.
WT and TG mice were infected with P. aeruginosa as described at a concentration of 1×109cfu/mouse of PAO1 strain. Survival is represented by Kaplan-Meier Survival Curves and there was a statistically significant difference between survival curves of WT and TG groups (P = 0.001, Log Rank Test).a
Discussion
Effective host defense against microbial invasion requires an innate immune system whose response is both rapid and independent of prior exposure (18). Several secreted proteins participate in this innate immune response (19) and data presented here indicate that PLUNC is a member of this group. In this study, we successfully expressed hPLUNC protein in the airway epithelium of transgenicmice using the mouse Scgb1a1 promoter. Scgb1a1-hPLUNC transgenic mice expressed hPLUNC mRNA and hPLUNC protein in the airway epithelium and secreted hPLUNC was detected in BAL fluid. Expression of hPLUNC did not alter lung structure or function, nor did it change the expression and secretion of the major mouse airway secretory protein SCGB1A1. However, hPLUNC was associated with enhanced bacterial killing and decreased inflammation following the exposure with a major human airwaypathogen, the gram-negative bacteria P. aeruginosa (PAO1). Our results provide evidence of a role for PLUNC in mitigation of bacterial infection both in vitro and in vivo. While other pathogens were not tested in the study, the transgenic mice generated in this study represent a useful animal model for future investigations to identify PLUNC mediated antimicrobial activityin the airway.
Several lines of evidence suggest that PLUNC mediated antibacterial activity is an important aspect of the innate immune response in the airway. First, PLUNC is structurally related to BPI and LBP, proteins that have been demonstrated to be important to host defense against gram negative bacteria (20–22). Second, PLUNC belongs to the short-peptide subfamily of PLUNC family proteins and has homology to the N-terminal domains of BPI (23–25). Peptides derived from this region of BPI have previously been shown to possess direct bactericidal activity (23, 26). Using a bio-activity assay to test antimicrobial functions of secretory proteins in BALF, we demonstrated a significant decrease in CFU counts of P. aeruginosa when bacteria were co-incubated with BALF from Scgb1a1-hPLUNC transgenic mice. Furthermore, both in vitro and in vivo antibody neutralization studies indicated that the increased antibacterial activity was due to PLUNC. Therefore, the decreased sensitivity in Scgb1a1-hPLUNC transgenic mice may have partially attributed to the direct killing activity. These data suggested that expression of hPLUNC in mouse airways increased mouse resistance to pathogens.
Because BPI is normally expressed in inflammatory cells but not airway epithelial cells, these data may also suggest that critical aspects of the innate response can be mediated by similar proteins that are secreted by different cell types. Moreover, the augmentation of protection by the additional hPLUNC suggests that PLUNC or BPI congeners could provide a novel anti-infection therapy. PLUNC mediated antibacterial activity highlight its functional significance and support the notion that PLUNC plays an important role in innate immunity against respiratory infection.
PLUNC is one of the most abundant secretory proteins in nasal drainage, BALF, and in primary epithelial cell cultures (6, 27–28). The significant amount of PLUNC secreted into the airway lumen and its antibacterial activity indicated that PLUNC plays a critical role in airway epithelial cell-based innate immunity. The enhanced antibacterial activity in Scgb1a1-PLUNC transgenic mice correlated with expression of hPLUNC in mouse airways. The magnitude of enhanced antibacterial activity (about 3 fold) was similar to previously reported antibacterial activity for lysozyme, a known anti-microbial protein in airway secretions (29). Because PLUNC is: 1) an abundant component of the antimicrobial fraction of nasal secretions (30), 2) present at high levels in airway secretions (6, 27–28, 31–32), 3) expressed at levels similar to that of lysozyme in tracheobronchial submucosal glands of, and 4) has anti-microbial activity comparable to that of lysozyme, it is reasonable to propose that PLUNC is an important component of the innate response to airway pathogens that originates from airway epithelial cells. It has been suggested that PLUNC is a novel airway secretory protein with a surfactant protein function that modulates surface tension and inhibits biofilm formation (14). Therefore, one of the potential PLUNC mediated antimicrobial mechanisms may be over-expressed hPLUNC in Scgb1a1-PLUNC transgenic mice enhanced the mucociliary clearance and disrupted the biofilm formation of P. aeruginosa after PAO1 challenged respiratory infection. The enhanced survival of Scgb1a1-PLUNC transgenic mice compare to their WT littermates after a high dose of P. aeruginosa challenge further indicates an important antimicrobial activity of PLUNC to P. aeruginosa induced lung infection and supports the notion that PLUNC plays a critical role in airway epithelium mediated innate immune response.
PLUNC may possess anti-inflammatory activities that are important in modulating airway innate immune response. Our data demonstrate a lower degree of neutrophil recruitment and less lung inflammation in PA01 exposed Scgb1a1-PLUNC transgenic mice than their WT littermates and suggest that constitutive over-expressed of hPLUNC in mouse airways resulted in more efficient bacterial killing. The decreased inflammation noted in Scgb1a1-PLUNC transgenic mice could have been a direct consequence of more efficient bacterial clearance. The potential binding of PLUNC to LPS may modulate LPS mediated inflammatory response. It has been suggested that PLUNC binds directly to lipopolysaccharide from Escherichia coli (28). A structurally related protein murine parotid secretory protein (PSP), also called SPLUNC2, has been shown to bind to bacterial membranes (33).
Cytokines play an important role in regulation and modulation of immunological and inflammatory processes. Unlike an in vitro cell culture system that represents a single cell type, the use of a transgenic mouse model provided an opportunity to identify coordinated changes in cytokine production between inflammatory and epithelial cells after exposure to aerosolized P. aeruginosa. Normally, following the recognition of microbial products, TLR-mediated signaling results in the production of TNF-α and IL-1β, two early-responsive cytokines that regulate subsequent recruitment of neutrophils (34–36). These substances usually play beneficial roles in the body’s defense systems. However, excess cytokine production may contribute to an exacerbated inflammatory response and could even contribute to severe inflammation and dysfunction in other organs if these inflammatory mediators are released excessively into the bloodstream. Therefore, a well-regulated and balanced production of inflammatory mediators is critical to an effective local and systemic host defense. In our studies, pro-inflammatory cytokines such as IL-1β, IL-6, and TNF-α were nearly undetectable prior to bacterial exposure but were significantly increased after challenge. Enhanced clearance of inhaled bacteria in Scgb1a1-PLUNC mice also correlated with decreased production of pro-inflammatory cytokines (including IL-1β, TNF-α, and IL-6) after bacterial challenge. Another pro-inflammatory chemokine, KC, has been shown to substantially increase the accumulation of neutrophils in the lungs after the intratracheal administration of LPS (37) and we observed significant lower levels of KC secretion in BALF from Scgb1a1-PLUNC transgenic mice when compared to WT control mice after P. aeruginosa exposure. The lower amount of KC in BALF of Scgb1a1-PLUNC transgenic mice might also partially account for decreased neutrophil infiltration and later resulted in lesser damage to the lungs in transgenic mice as compared to their WT littermates. Although the decrease in pro-inflammatory cytokine production noted in P. aeruginosa exposed Scgb1a1-PLUNC transgenic mice is likely to be a consequence of antibacterial activities of hPLUNC, it is also possible that the lower cytokine levels in Scgb1a1-PLUNC transgenic mice could be due to unanticipated roles for hPLUNC in regulation of cytokine production in epithelial and/or inflammatory cells. Further studies to identify potential bioactivity of PLUNC in association with inflammatory response are worthy of future investigation.
Our data suggest that expression of hPLUNC provide an airway epithelial cell specific protection against opportunistic pathogens such as P. aeruginosa. While the major difference between transgenic mice and their wild-type littermates is most likely due to expression of hPLUNC, we could not rule out the possibility that other anti-microbial proteins/peptides may also act synergistically with ectopically expressed hPLUNC to enhance the bacterial killing effect. The enhanced bacterial clearance observed in Scgb1a1-PLUNC transgenic mice may due to interaction of over-expressed PLUNC with other antimicrobial peptides such as defensins and/or antimicrobial proteins such as lysozyme to potentate its antibacterial activities.
In conclusion, this study demonstrated antibacterial activity of PLUNC against airway bacterial pathogens using both in vitro and in vivo approaches. We also demonstrated that Scgb1a1-PLUNC transgenic mice exhibited enhanced survival and improved resistance to P. aeruginosa infection, one of the most common airway infection associated with chronic colonization in cystic fibrosis patients. Given the emergence of highly resistant bacteria pathogens and the increasing population of immuno-compromised patients, the treatment of bacterial infection has and will continue to be challenging. A better understanding of airway epithelial cell-initiated host defense may provide an alternative approach to efficiently combat airway bacterial infection.
Acknowledgments
We thank Dr. Jonathan Chao in making constructs for the generation of Scgb1a1-PLUNC transgenic mice and Dr. Thomas Zhao for his technical assistance in dissecting mice.
Y.P. Di acknowledges support from Public Health Service Grants NIH (ES011033, HL091938) and American Heart Association-Pennsylvania Delaware Affiliate (0365327U).
Abbreviations used in this paper
- WT
wild type
- TG
transgene
- BAL
bronchoalveolar lavage
- PLUNC
palate, lung and nasal epithelium carcinoma associated
- PA
P. aeruginosa
Footnotes
Disclosures
The authors have no financial conflict of interest.
References
- 1.Medzhitov R, Janeway CA., Jr Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 2.Elsbach P, Weiss J. Role of the bactericidal/permeability-increasing protein in host defence. Curr Opin Immunol. 1998;10:45–49. doi: 10.1016/s0952-7915(98)80030-7. [DOI] [PubMed] [Google Scholar]
- 3.Gioannini TL, Teghanemt A, Zarember KA, Weiss JP. Regulation of interactions of endotoxin with host cells. J Endotoxin Res. 2003;9:401–408. doi: 10.1179/096805103225002773. [DOI] [PubMed] [Google Scholar]
- 4.Albers JJ, Wolfbauer G, Cheung MC, Day JR, Ching AF, Lok S, Tu AY. Functional expression of human and mouse plasma phospholipid transfer protein: effect of recombinant and plasma PLTP on HDL subspecies. Biochimica et biophysica acta. 1995;1258:27–34. doi: 10.1016/0005-2760(95)00091-p. [DOI] [PubMed] [Google Scholar]
- 5.Day JR, Albers JJ, Lofton-Day CE, Gilbert TL, Ching AF, Grant FJ, O’Hara PJ, Marcovina SM, Adolphson JL. Complete cDNA encoding human phospholipid transfer protein from human endothelial cells. The Journal of biological chemistry. 1994;269:9388–9391. [PubMed] [Google Scholar]
- 6.Di YP, Harper R, Zhao Y, Pahlavan N, Finkbeiner W, Wu R. Molecular cloning and characterization of spurt, a human novel gene that is retinoic acid-inducible and encodes a secretory protein specific in upper respiratory tracts. The Journal of biological chemistry. 2003;278:1165–1173. doi: 10.1074/jbc.M210523200. [DOI] [PubMed] [Google Scholar]
- 7.Bingle CD, Gorr SU. Host defense in oral and airway epithelia: chromosome 20 contributes a new protein family. Int J Biochem Cell Biol. 2004;36:2144–2152. doi: 10.1016/j.biocel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Leclair EE. Four BPI (bactericidal/permeability-increasing protein)-like genes expressed in the mouse nasal, oral, airway and digestive epithelia. Biochem Soc Trans. 2003;31:801–805. doi: 10.1042/bst0310801. [DOI] [PubMed] [Google Scholar]
- 9.Bingle CD, Craven CJ. Comparative analysis of the PLUNC (palate, lung and nasal epithelium clone) protein families. Biochem Soc Trans. 2003;31:806–809. doi: 10.1042/bst0310806. [DOI] [PubMed] [Google Scholar]
- 10.LeClair EE, Nomellini V, Bahena M, Singleton V, Bingle L, Craven CJ, Bingle CD. Cloning and expression of a mouse member of the PLUNC protein family exclusively expressed in tongue epithelium. Genomics. 2004;83:658–666. doi: 10.1016/j.ygeno.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Chu HW, Thaikoottathil J, Rino JG, Zhang G, Wu Q, Moss T, Refaeli Y, Bowler R, Wenzel SE, Chen Z, Zdunek J, Breed R, Young R, Allaire E, Martin RJ. Function and regulation of SPLUNC1 protein in Mycoplasma infection and allergic inflammation. J Immunol. 2007;179:3995–4002. doi: 10.4049/jimmunol.179.6.3995. [DOI] [PubMed] [Google Scholar]
- 12.Zhou HD, Li XL, Li GY, Zhou M, Liu HY, Yang YX, Deng T, Ma J, Sheng SR. Effect of SPLUNC1 protein on the Pseudomonas aeruginosa and Epstein-Barr virus. Mol Cell Biochem. 2008;309:191–197. doi: 10.1007/s11010-007-9659-3. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Caballero A, Rasmussen JE, Gaillard E, Watson MJ, Olsen JC, Donaldson SH, Stutts MJ, Tarran R. SPLUNC1 regulates airway surface liquid volume by protecting ENaC from proteolytic cleavage. Proc Natl Acad Sci U S A. 2009;106:11412–11417. doi: 10.1073/pnas.0903609106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gakhar L, Bartlett JA, Penterman J, Mizrachi D, Singh PK, Mallampalli RK, Ramaswamy S, McCray PB., Jr PLUNC is a novel airway surfactant protein with anti-biofilm activity. PLoS One. 2010;5:e9098. doi: 10.1371/journal.pone.0009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Yu H, Hanes M, Chrisp CE, Boucher JC, Deretic V. Microbial pathogenesis in cystic fibrosis: pulmonary clearance of mucoid Pseudomonas aeruginosa and inflammation in a mouse model of repeated respiratory challenge. Infect Immun. 1998;66:280–288. doi: 10.1128/iai.66.1.280-288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reynolds SD, Hong KU, Giangreco A, Mango GW, Guron C, Morimoto Y, Stripp BR. Conditional clara cell ablation reveals a self-renewing progenitor function of pulmonary neuroendocrine cells. American journal of physiology. 2000;278:L1256–1263. doi: 10.1152/ajplung.2000.278.6.L1256. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 19.Canny G, Levy O. Bactericidal/permeability-increasing protein (BPI) and BPI homologs at mucosal sites. Trends Immunol. 2008;29:541–547. doi: 10.1016/j.it.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Horwitz AH, Williams RE, Nowakowski G. Human lipopolysaccharide-binding protein potentiates bactericidal activity of human bactericidal/permeability-increasing protein. Infect Immun. 1995;63:522–527. doi: 10.1128/iai.63.2.522-527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marra MN, Wilde CG, Collins MS, Snable JL, Thornton MB, Scott RW. The role of bactericidal/permeability-increasing protein as a natural inhibitor of bacterial endotoxin. J Immunol. 1992;148:532–537. [PubMed] [Google Scholar]
- 22.Tobias PS, Soldau K, Iovine NM, Elsbach P, Weiss J. Lipopolysaccharide (LPS)-binding proteins BPI and LBP form different types of complexes with LPS. J Biol Chem. 1997;272:18682–18685. doi: 10.1074/jbc.272.30.18682. [DOI] [PubMed] [Google Scholar]
- 23.Capodici C, Chen S, Sidorczyk Z, Elsbach P, Weiss J. Effect of lipopolysaccharide (LPS) chain length on interactions of bactericidal/permeability-increasing protein and its bioactive 23-kilodalton NH2-terminal fragment with isolated LPS and intact Proteus mirabilis and Escherichia coli. Infect Immun. 1994;62:259–265. doi: 10.1128/iai.62.1.259-265.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elsbach P, Weiss J. The bactericidal/permeability-increasing protein (BPI), a potent element in host-defense against gram-negative bacteria and lipopolysaccharide. Immunobiology. 1993;187:417–429. doi: 10.1016/S0171-2985(11)80354-2. [DOI] [PubMed] [Google Scholar]
- 25.Ooi CE, Weiss J, Elsbach P. Structural and functional organization of the human neutrophil 60 kDa bactericidal/permeability-increasing protein. Agents Actions. 1991;34:274–277. doi: 10.1007/BF01993301. [DOI] [PubMed] [Google Scholar]
- 26.Elsbach P, Weiss J, Doerfler M, Shu C, Kohn F, Ammons WS, Kung AH, Meszaros KK, Parent JB. The bactericidal/permeability increasing protein of neutrophils is a potent antibacterial and anti-endotoxin agent in vitro and in vivo. Prog Clin Biol Res. 1994;388:41–51. [PubMed] [Google Scholar]
- 27.Campos MA, Abreu AR, Nlend MC, Cobas MA, Conner GE, Whitney PL. Purification and characterization of PLUNC from human tracheobronchial secretions. Am J Respir Cell Mol Biol. 2004;30:184–192. doi: 10.1165/rcmb.2003-0142OC. [DOI] [PubMed] [Google Scholar]
- 28.Ghafouri B, Kihlstrom E, Tagesson C, Lindahl M. PLUNC in human nasal lavage fluid: multiple isoforms that bind to lipopolysaccharide. Biochimica et biophysica acta. 2004;1699:57–63. doi: 10.1016/j.bbapap.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Akinbi HT, Epaud R, Bhatt H, Weaver TE. Bacterial killing is enhanced by expression of lysozyme in the lungs of transgenic mice. J Immunol. 2000;165:5760–5766. doi: 10.4049/jimmunol.165.10.5760. [DOI] [PubMed] [Google Scholar]
- 30.Cole AM, Liao HI, Stuchlik O, Tilan J, Pohl J, Ganz T. Cationic polypeptides are required for antibacterial activity of human airway fluid. J Immunol. 2002;169:6985–6991. doi: 10.4049/jimmunol.169.12.6985. [DOI] [PubMed] [Google Scholar]
- 31.Ghafouri B, Irander K, Lindbom J, Tagesson C, Lindahl M. Comparative proteomics of nasal fluid in seasonal allergic rhinitis. J Proteome Res. 2006;5:330–338. doi: 10.1021/pr050341h. [DOI] [PubMed] [Google Scholar]
- 32.Roxo-Rosa M, da Costa G, Luider TM, Scholte BJ, Coelho AV, Amaral MD, Penque D. Proteomic analysis of nasal cells from cystic fibrosis patients and non-cystic fibrosis control individuals: search for novel biomarkers of cystic fibrosis lung disease. Proteomics. 2006;6:2314–2325. doi: 10.1002/pmic.200500273. [DOI] [PubMed] [Google Scholar]
- 33.Robinson CP, Bounous DI, Alford CE, Nguyen KH, Nanni JM, Peck AB, Humphreys-Beher MG. PSP expression in murine lacrimal glands and function as a bacteria binding protein in exocrine secretions. Am J Physiol. 1997;272:G863–871. doi: 10.1152/ajpgi.1997.272.4.G863. [DOI] [PubMed] [Google Scholar]
- 34.Sherry B, Cerami A. Cachectin/tumor necrosis factor exerts endocrine, paracrine, and autocrine control of inflammatory responses. J Cell Biol. 1988;107:1269–1277. doi: 10.1083/jcb.107.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strieter RM, Remick DG, Ward PA, Spengler RN, Lynch JP, 3rd, Larrick J, Kunkel SL. Cellular and molecular regulation of tumor necrosis factor-alpha production by pentoxifylline. Biochem Biophys Res Commun. 1988;155:1230–1236. doi: 10.1016/s0006-291x(88)81271-3. [DOI] [PubMed] [Google Scholar]
- 36.Larrick JW, Kunkel SL. The role of tumor necrosis factor and interleukin 1 in the immunoinflammatory response. Pharm Res. 1988;5:129–139. doi: 10.1023/a:1015904721223. [DOI] [PubMed] [Google Scholar]
- 37.Tsai WC, Strieter RM, Wilkowski JM, Bucknell KA, Burdick MD, Lira SA, Standiford TJ. Lung-specific transgenic expression of KC enhances resistance to Klebsiella pneumoniae in mice. J Immunol. 1998;161:2435–2440. [PubMed] [Google Scholar]