Abstract
CRISPR-Cas systems are recently discovered, RNA-based immune systems that control invasions of viruses and plasmids in archaea and bacteria. Prokaryotes with CRISPR-Cas immune systems capture short invader sequences within the CRISPR loci in their genomes, and small RNAs produced from the CRISPR loci (CRISPR (cr)RNAs) guide Cas proteins to recognize and degrade (or otherwise silence) the invading nucleic acids. There are multiple variations of the pathway found among prokaryotes, each mediated by largely distinct components and mechanisms that we are only beginning to delineate. Here we will review our current understanding of the remarkable CRISPR-Cas pathways with particular attention to studies relevant to systems found in the archaea.
Introduction
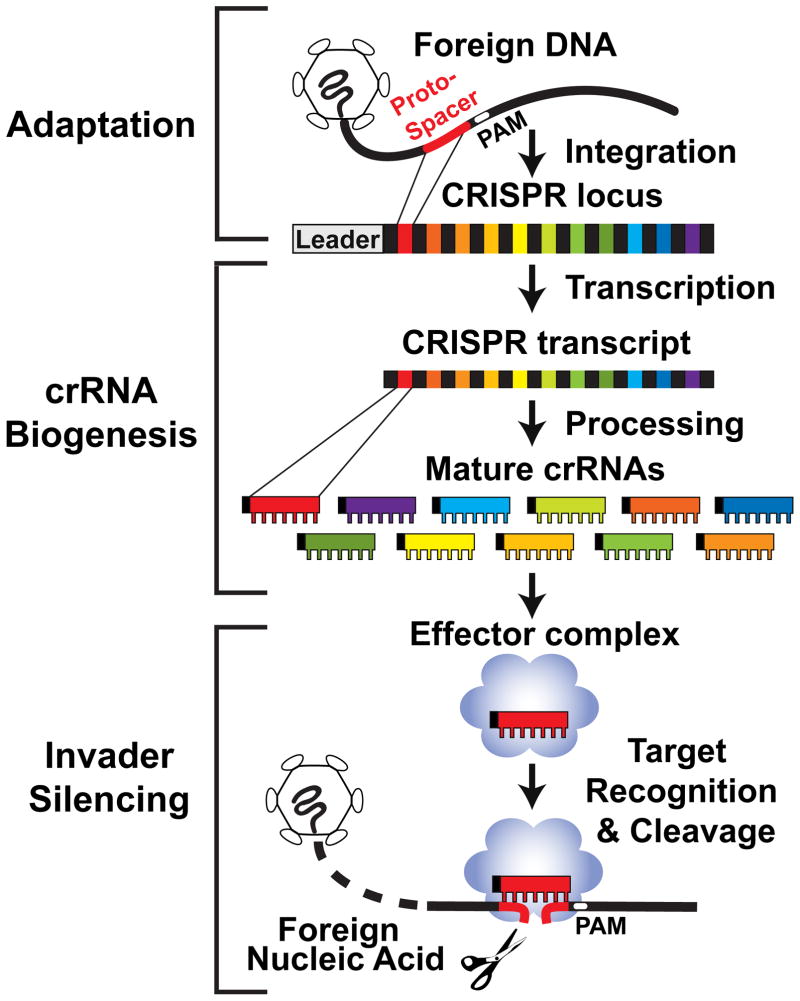
Small RNA-based defense systems that provide adaptive, heritable immunity against viruses, plasmids and other mobile genetic elements have recently been discovered in archaea and bacteria. The RNA and protein components of these immune systems arise from the CRISPR (clustered regularly interspaced short palindromic repeat) and Cas (CRISPR-associated) genes, respectively. The CRISPR-Cas pathway functions in three phases adaptation of CRISPRs to invaders, crRNA biogenesis, and invader silencing (Figure 1). It appears that nearly all archaea and approximately half of bacteria are equipped with CRISPR-Cas systems [1–3], which have been shown to provide protection from viral predation and plasmid invasion in both laboratory settings [4–9] and natural environments [10–13].
Figure 1.
Overview of the CRISPR-Cas invader defense pathway. In the adaptation phase, a short fragment of foreign DNA (protospacer) is acquired from the invader and integrated into the host CRISPR locus adjacent to the leader. Protospacer adjacent motifs (PAMs) are found near invader sequences selected for CRISPR integration. The CRISPR locus consists of short direct repeat sequences (black) that separate similarly-sized, invader-derived sequences (multiple colors). In the biogenesis phase of the pathway, CRISPR locus transcripts are processed to release individual mature crRNAs (each targeting a different sequence). Mature crRNAs typically retain some of the repeat sequence, which is thought to provide a recognizable signature of the crRNAs. In the silencing phase, crRNA-Cas protein effector complexes recognize foreign DNA or RNA through basepairing of the crRNA. The Cmr and Csn systems affect cleavage of target RNA and DNA, respectively. PAMs provide important auxiliary signals for the recognition of invaders for some DNA-targeting systems.
The discovery of these prokaryotic immune systems has generated considerable excitement, and several excellent reviews are available [2,14–20]. Here, we describe the components and mechanisms of CRISPR-mediated immunity with emphasis on the systems found in archaea. Advances in understanding the three key steps in the CRISPR-Cas pathway are described, including important contributions from studies done in archaea. Finally, we summarize the significant gaps that remain in our knowledge of the molecular mechanisms of CRISPR-Cas-based invader defense.
CRISPRs: Genetic memory banks of past invasions and source of small invader-targeting RNAs
The hallmark feature of the CRISPR-Cas system is the CRISPR locus (see Figure 1). CRISPR loci are characterized by short, direct repeat sequences (typically 30–40 nts) that separate variable sequences of similar size. There are 12 families of CRISPR repeats based on sequence and predicted secondary structure [21]. The variable sequences (called spacers or guide sequences) are derived from viruses, plasmids, and other invaders [17,22–25] and, remarkably, confer immunity against the corresponding invader [4–9,26]. CRISPR locus transcripts are processed to generate small crRNAs that contain individual invader-derived sequences and target invading nucleic acids for silencing ([5,9,26–28], and see Figure 1). Thus, CRISPRs capture and store fragments of invader sequence and give rise to small RNAs that impart heritable immunity against the invaders.
Cas Proteins: Hubs of CRISPR-Cas diversity
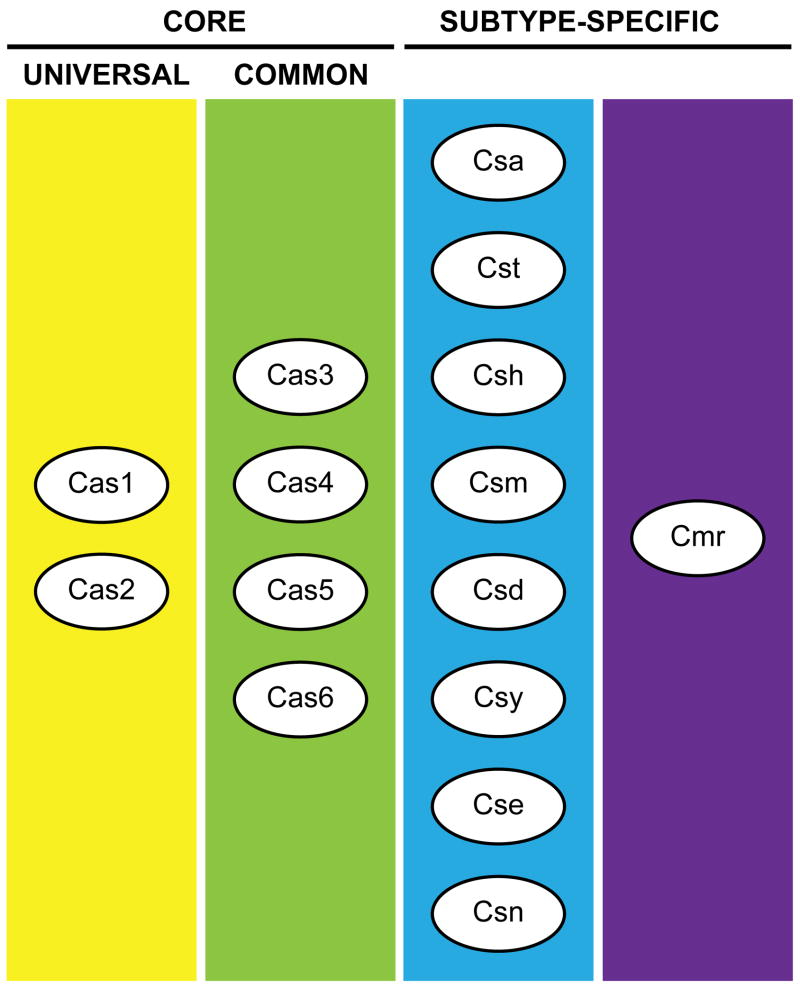
The cas genes are very tightly linked to CRISPR loci, both physically (location within genomes) and evolutionarily (co-segregation among genomes), consistent with the co-function of crRNAs and Cas proteins. Over 45 cas gene families have been identified, but a given organism only possesses a subset of these [2,29–31]. A few “core” cas genes (cas1-6) are present in a wide array of organisms [2,29–31]; however, most organisms have only some of these six genes, and only cas1 and cas2 appear to be universal (Figure 2). In a given organism, the core cas genes are supplemented by one or more of the nine sets of subtype-specific cas genes (Figure 2). These sets of two to six genes co-segregate among genomes as distinct cas gene modules. Eight of the modules are named for a prototypical organism where they are the only additional cas genes found [29]. For example, the cas subtype Aeropyrum pernix or csa genes are a set of 6 non-core cas genes found together in A. pernix as well as other organisms. The other Cas subtypes include: Thermotoga (cst), Haloarcula (csh), Mycobacterium (csm), Desulfovibrio (csd), Yersinia (csy), Escherichia (cse) and Neisseria (csn). Each of these eight modules is associated with particular subsets of the core cas genes [18,21]. The ninth module, cas module RAMP (cmr), is only found in conjunction with other subtype-specific modules [29,31,32]. As more genomes are sequenced, new cas gene modules continue to emerge and relationships between components of the modules are recognized [2]. The limited available information confirms that cas genes are essential for the function of CRISPR-Cas systems [4–6,33,34].
Figure 2.
Combinations of Cas proteins create diverse CRISPR-Cas systems. Cas1-6 are core Cas proteins found in many and diverse organisms. In addition, there are eight primary modules of subtype-specific Cas proteins (consisting of 2–6 proteins each), and the auxiliary Cmr module. A typical CRISPR-Cas system is comprised of the nearly universal Cas1 and Cas2 proteins, a specific combination of the other core Cas proteins and a set of sub-type specific Cas proteins. A given organism may possess more than one CRISPR-Cas system, and may also have the Cmr module. See Haft et al. [29].
The Csa, Cst, Csh, and Csm subtype Cas systems are common in archaea [2,29,31,32]. All of the Cas systems are found in bacteria with the exception of Csa, which may be exclusive to archaea. In general, archaea tend to have multiple (or mixed) Cas systems. The diversity of Cas systems found among prokaryotes is illustrated in the cas genes present in the some of the currently studied model organisms (Table 1). It is thought that the Cas systems are disseminated by horizontal gene transfer [32,35–38] with the result that closely related species can have completely different systems and highly divergent organisms can have very similar Cas systems.
Table 1.
Core and sub-type specific Cas protein genes in model organisms being employed to understand CRISPR-Cas systems. Information from [1] except for S. thermophilus [15]. Cas nomenclature is from Haft et al. [29].
| ORGANISM | CORE | SUBTYPE-SPECIFIC | TOTAL # |
|---|---|---|---|
| ARCHAEA | |||
| Pyrococcus furiosus (DSM 3638) | Cas 1, 2, 3, 4, 5, 6 | Csa, Cst, Cmr | 27 |
| Sulfolobus sulfataricus (P2) | Cas 1, 2, 3, 4, 5, 6 | Csa, Csm, Cmr | 53 |
| BACTERIA | |||
| Escherichia coli (K-12) | Cas 1, 2, 3 | Cse | 8 |
| Psuedomonas aeruginosa (PA14) | Cas 1, 2/3 fusion | Csy | 6 |
| Staphylococcus epidermidis (RP62a) | Cas 1, 2, 6 | Csm | 8 |
| Streptococcus thermophilus (DGCC7710) | Cas 1, 2, 3, 6 | Csn, Csm, Cse | 25 |
The diversity of Cas proteins that populate CRISPR-Cas systems would suggest that there are multiple variations of the CRISPR-Cas pathway to be delineated and available information substantiates the expected diversity in the pathways (for example in the targeting of DNA versus RNA targeting, see below). Cas protein sequences indicate potential functions as nucleases, helicases, RNA binding proteins, etc. [29,31]; however, most of the proteins have not yet been biochemically characterized or assigned functions in the pathways. Exceptions are described below.
Three steps in the CRISPR-Cas invader defense pathway
Cas proteins function in each of the three steps required for CRISPR-Cas system function: 1) adaptation of CRISPRs, 2) crRNA biogenesis, and 3) invader silencing (Figure 1).
1. Adaptation: Acquisition of new invader sequences in the CRISPR loci
In adaptation, a copy or fragment of invading nucleic acid termed a protospacer is generated and integrated into the CRISPR locus (Figure 1). Protospacers are typically inserted immediately adjacent to the leader sequence at one end of the CRISPR ([4,10,15,24,25]; see Figure 1) providing an approximately chronological record of past infections. Arguably, adaptation is the most unique and fascinating aspect of CRISPR-Cas biology, but there is scant information regarding the molecular mechanism.
Short (3–6 nt) sequence elements found adjacent to the protospacer in the foreign nucleic acid, termed PAMs [14,23,28], are critical in generation and/or integration of protospacers into CRISPR loci [4,6,39]. The PAM is presumably recognized by the adaptation machinery. The trans-acting factors involved in novel spacer acquisition remain largely unknown. A central role for the universal Cas1 in invader DNA cleavage has been suggested based on observed cleavage of dsDNA in vitro [40]; however, Cas1 generates ~80 bp DNA fragments without a requirement for a flanking PAM. Compelling genetic evidence implicates Csn2 (also called Cas7) in acquisition in Streptococcus thermophilus [4,6], suggesting that sub-type specific Cas proteins may act with Cas1 in adaptation. Moreover, at least in E. coli, there is evidence that Cas1 may function with non-Cas proteins [41]. Mechanisms that limit the size of CRISPR arrays are also now coming to light. Loss of CRISPR length appears to occur via spontaneous homologous recombination between repeat sequences [7,13,24,39].
2. CRISPR RNA biogenesis
Biogenesis entails production of numerous individual crRNAs from CRISPR locus transcripts ([5,9,26–28,33], and see Figure 1). “RNomic” (RNA profiling) studies of the small RNAs from two thermophilic archaea, A. fulgidus and S. solfataricus, revealed that CRISPR loci were transcriptionally active and yielded elaborately processed RNAs (prior to our understanding of CRISPR function) [42,43]. CRISPR loci are predominantly transcribed from promoters located at the leader ends [5,9,26–28,33]. In most organisms analyzed thus far, CRISPR RNAs and Cas proteins are constitutively expressed, consistent with an immune system operating in “surveillance mode.” However, there is evidence that the expression of the CRISPR-Cas system of specific E. coli strains is highly regulated [44] and that in T. thermophilus, the presence of an invader triggers elevations in the expression of CRISPR-Cas components [45].
Considerable progress has been made identifying and characterizing the primary crRNA biogenesis enzymes. In several CRISPR-Cas systems, RAMP [31] superfamily Cas proteins catalyze cleavage of CRISPR transcripts. For example, the core Cas protein, Cas6 (from P. furiosus) [46–48] and sub-type-specific Cas proteins, Cse3 (E. coli) [5] and Csy4 (P. aeriginosa) [49] each recognize specific crRNA repeat sequences/structures and catalyze a single-cut within each repeat which liberates unit crRNAs (known as 1× processing intermediates [47]) containing 5′ and 3′ flanking repeat-derived sequences. The repeat sequences can be further trimmed by unknown mechanisms [26,33]. An 8-nucleotide, repeat-derived sequence is retained at the 5′ ends of CRISPR RNAs from several archaea and bacteria [5,9,26,47] and likely plays an important role as a crRNA “identity tag” that serves as a Cas protein binding site. Detailed biochemical and structural studies of the pre-crRNA biogenesis enzymes have provided a wealth of information on modes of cleavage and recognition of both palindromic and unstructured CRISPR repeat RNAs [46–50]. At the same time, a very distinct mechanism of pre-crRNA biogenesis has been found in bacteria with Csn subtype systems. A trans-acting RNA encoded within the CRISPR-Cas region forms a duplex with the repeat sequence of the pre-crRNA that is processed by RNase III and perhaps Csn1 [33].
3. Invader Silencing
crRNAs are incorporated into effector complexes and guide the complexes to invading nucleic acid (via base-paired interactions). Silencing can occur at the DNA or RNA level, and DNA targeting requires a PAM in the DNA target for at least a subset of CRISPR-Cas systems [4–9,26,51].
CRISPR-Cas systems that target invader DNA
Evidence indicates that Cse [5], Csn [6] and Csm [9] subtype systems directly or indirectly target the DNA of invaders. Cleavage of invader DNA has been observed in the case of the Csn system of S. thermophilus (but not yet in the others; [9]). It is not known whether Cas systems that target DNA employ silencing mechanisms other than cleavage or can also target RNA (DNA targeting can obscure identification of RNA targeting unless this is accounted for in the experimental design (e.g., as in [9])). The core Cas3 protein has been shown to be essential for invader defense in vivo [5], to degrade double-stranded DNA or RNA substrates in vitro [52], and likely catalyzes DNA cleavage in several CRISPR-Cas systems. In contrast, the predicted invader DNA nuclease in the Csn system is Csn1 (also called Cas5 [6] or Cas 9 [2]).
Systems to investigate silencing mechanisms in vivo have recently been developed in the crenarchaea Sulfolobus solfataricus and Sulfolobus islandicus [7,8]. Results from studies in which artificial invaders containing sequences recognized by existing CRISPR spacers (i.e. protospacers) were introduced into cells indicate that invaders are targeted at the DNA level, though more work is required to understand which CRISPR-Cas system(s) (Csa, Csm or Cmr) are responsible for the observed activity and whether the targeted DNA is cleaved.
For DNA targeting systems, it is critical that the pathway not target the corresponding spacer in its own CRISPR (which matches the protospacer in the invader). PAMs provide one mechanism for distinction of self vs. non-self. PAMs are critical for silencing by several systems [6,7,51], and the PAM recognized by the CRISPR-Cas system in the invader is not present in the repeat sequence that flanks the potential target in the CRISPR. Another mechanism described in Staphlococcus epidermidis, which harbors a Csm-type system, is the requirement for mismatches between the target DNA and the 5′ repeat tag sequences of the crRNA (which base-pair perfectly with the potential target in the CRISPR) [53]. Interestingly, it appears that host sequences are occasionally integrated into CRISPR loci resulting in self-targeting and cell death [54], and when self-targeting crRNAs were expressed from a plasmid-encoded CRISPR locus, Sulfolobus solfataricus host cell survival was associated with homologous recombination that replaced the self-targeting CRISPR locus with the homologous chromosomal copy [8].
Target RNA cleavage by the CRISPR-Cmr complex
In P. furiosus, a complex comprised of the six Cmr subtype proteins and mature crRNAs cleaves complementary RNAs (and not DNAs) [26]. All six Cmr proteins are important for function of the complex and the crRNAs direct cleavage 14 nucleotides upstream of their 3′ ends [26]. Approximately 70% of archaea and 30% of bacteria with CRISPR-Cas systems have the Cmr module in addition to other Cas systems [32] suggesting that this RNA-targeting branch of the CRISPR-Cas immune system plays an important role in the biological warfare against viruses and other mobile genetic elements.
Co-evolving elements of a CRISPR-Cas system
Functional CRISPR-Cas systems include three co-evolved components: the leader region of the CRISPR, the CRISPR repeat, and the cas gene collection (Figure 3). As described above, function of the system very likely requires specific interactions between Cas proteins and both the CRISPR leader (e.g. for integration of new invader-derived sequences) and crRNA repeat sequence (e.g. for crRNA biogenesis and co-function in silencing; Figure 3), and recent studies indicate that these three elements co-segregate [14,21,28,32,39]. Thus, sets of core and subtype-specific cas genes are associated with specific leader and repeat sequence families [21,28,29,32,55]. Likely for the same reason (required Cas protein interaction), CRISPR-Cas systems are associated with specific PAM sequences in the targeted invaders (Figure 3). Extensive analysis of the CRISPR-Cas systems of Sulfolobus species, along with available viral and plasmid sequences, demonstrates the relationships between these elements [28,32,55]. Not surprisingly, it appears that the selective pressure of PAM recognition and silencing by CRISPR-Cas systems is countered by viral evolution of the targeted PAM sequences [51].
Figure 3.
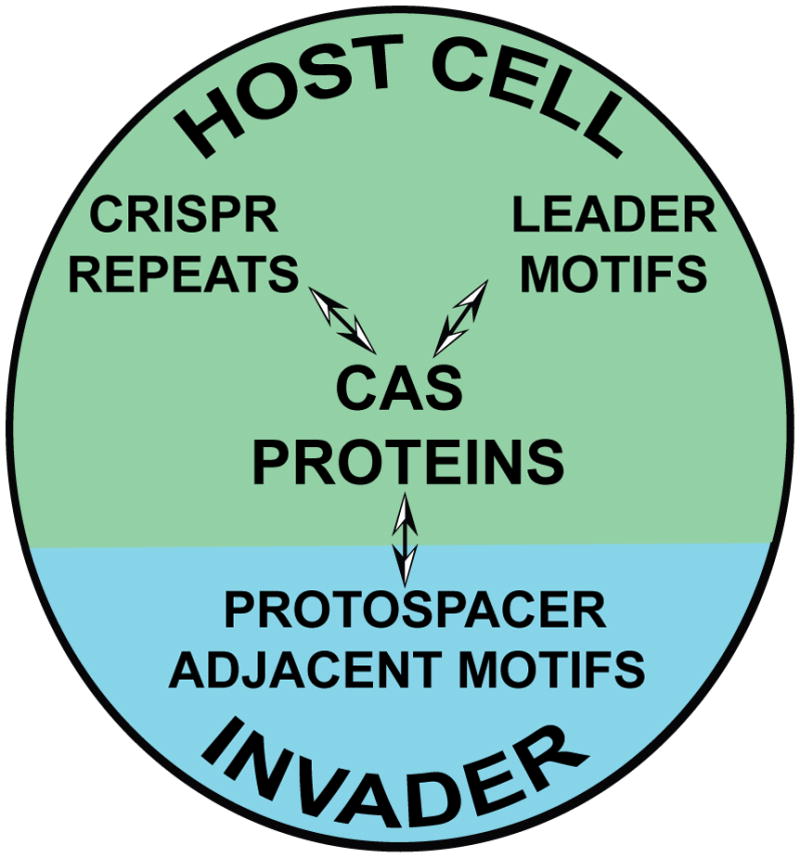
CRISPR-Cas systems of various prokaryotes include three co-evolved components: CRISPR repeats, CRISPR leaders, and associated Cas proteins that function on discrete PAMs (protospacer adjacent elements) present in the viruses and other mobile genetic elements that they encounter. Known and predicted interactions between Cas proteins and the host CRISPR RNA repeat sequences and leader DNA elements plus invader PAMs are indicated (arrows). The specificity of these RNA-protein and DNA-protein interactions likely contribute to the co-evolution of the four components.
Conclusions
Evidence indicates that CRISPR-Cas immune systems play a globally important biological role in host-parasite interactions and collectively shape the evolution and ecology of prokaryotes and viruses [10,11,13,56,57]. The early studies have revealed that there is a diverse series of CRISPR-Cas pathways that function through distinct components and mechanisms, which are dispersed throughout archaea and bacteria. Much of our still very limited knowledge has come from studies with a small set of model organisms that collectively do not encompass the known CRISPR-Cas modules, and further investigation in other organisms will help address this gap. In the near future, the concerted effort of numerous research groups is expected to provide answers to fundamental questions such as how novel protospacers are acquired from invaders and integrated into CRISPRs, what constitutes a functional crRNA and how they are generated, and how silencing is achieved for each of the CRISPR-Cas pathways, and should illuminate the molecular mechanisms governing the astonishing CRISPR-Cas-mediated prokaryotic immune pathways.
Acknowledgments
We are grateful to Claiborne V. C. Glover III for critical reading of this review. This work was supported by NIH grant RO1GM54682 (including American Recovery and Reinvestment Act [ARRRA] funds) to M.P.T. and R.M.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the last two years, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makarova KS, Haft DH, Barrangou R, Brouns S, Charpentier E, Horvath P, Moineau S, Mojica FJ, Yakunin AF, van de Oost J, et al. Evolution and classification of the CRISPR/Cas systems. Nature Reviews Microbiology. 2011 doi: 10.1038/nrmicro2577. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rousseau C, Gonnet M, Le Romancer M, Nicolas J. CRISPI: a CRISPR interactive database. Bioinformatics. 2009;25:3317–3318. doi: 10.1093/bioinformatics/btp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **6.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. This work revealed that the Csn-type CRISPR-Cas system in S. thermophilus can target plasmids bearing antibiotic resistance genes and cleaves DNA targets. [DOI] [PubMed] [Google Scholar]
- *7.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, Garrett RA. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. These studies demonstrated that CRISPR-Cas systems in two Sulfolobus species function to resist plasmid invasion, and that the mechanism does not depend on target transcription (and thus presumably involves DNA targeting). Also provided insights on CRISPR length maintenance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *8.Manica A, Zebec Z, Teichmann D, Schleper C. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol. 2011 doi: 10.1111/j.1365-2958.2011.07586.x. Similar to Gudbergadottir, this work provided evidence for CRISPR-Cas-mediated resistance to viruses at the DNA level in Sulfolobus. [DOI] [PubMed] [Google Scholar]
- 9.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 11.Held NL, Herrera A, Cadillo-Quiroz H, Whitaker RJ. CRISPR associated diversity within a population of Sulfolobus islandicus. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pride DT, Sun CL, Salzman J, Rao N, Loomer P, Armitage GC, Banfield JF, Relman DA. Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time. Genome Res. 2011;21:126–136. doi: 10.1101/gr.111732.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–207. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- 14.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 15.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 16.Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lillestol RK, Redder P, Garrett RA, Brugger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 20.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 23.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 24.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 25.Shah SA, Hansen NR, Garrett RA. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans. 2009;37:23–28. doi: 10.1042/BST0370023. [DOI] [PubMed] [Google Scholar]
- 26.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. First insight on the molecular mechanism of silencing by a CRISPR-Cas effector complex. Defined the components of the crRNA-Cmr protein complex and demonstrated that the complex cleaves RNA targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Lillestol RK, Shah SA, Brugger K, Redder P, Phan H, Christiansen J, Garrett RA. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009;72:259–272. doi: 10.1111/j.1365-2958.2009.06641.x. Identified relationship between cas genes, CRISPR repeat sequences, and leaders that defined three co-evolved families of CRISPR-Cas systems in Sulfolobus. [DOI] [PubMed] [Google Scholar]
- 29.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 31.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah SA, Garrett RA. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol. 2011;162:27–38. doi: 10.1016/j.resmic.2010.09.001. [DOI] [PubMed] [Google Scholar]
- **33.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoding small RNA and host factor RNase III. Nature. 2011 doi: 10.1038/nature09886. In Press. This work revealed an unanticipated mechanism of CRISPR RNA biogenesis involving a novel non-coding RNA encoded within the CRISPR-Cas locus and a non-Cas protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O’Toole GA. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakraborty S, Snijders AP, Chakravorty R, Ahmed M, Tarek AM, Hossain MA. Comparative network clustering of direct repeats (DRs) and cas genes confirms the possibility of the horizontal transfer of CRISPR locus among bacteria. Mol Phylogenet Evol. 2010;56:878–887. doi: 10.1016/j.ympev.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 36.Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–729. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 37.Horvath P, Coute-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol. 2009;131:62–70. doi: 10.1016/j.ijfoodmicro.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 38.Portillo MC, Gonzalez JM. CRISPR elements in the Thermococcales: evidence for associated horizontal gene transfer in Pyrococcus furiosus. J Appl Genet. 2009;50:421–430. doi: 10.1007/BF03195703. [DOI] [PubMed] [Google Scholar]
- 39.Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 41.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A, et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol. 2011;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci U S A. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Huttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 44.Mojica FJ, Diez-Villasenor C. The on-off switch of CRISPR immunity against phages in Escherichia coli. Mol Microbiol. 2010;77:1341–1345. doi: 10.1111/j.1365-2958.2010.07326.x. [DOI] [PubMed] [Google Scholar]
- 45.Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J Mol Biol. 2010;395:270–281. doi: 10.1016/j.jmb.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 46.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *48.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. The co-structure of Cas6 (riboendonuclease that functions in crRNA biogenesis) and a bound crRNA revealed the molecular basis for cleavage of the repeat RNA at a site that is distal to the sequence-specific RNA binding site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *49.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. This study showed the structural basis for the binding and cleavage of a structured CRISPR repeat RNA by the CRISPR RNA biogenesis enzyme, Csy4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ebihara A, Yao M, Masui R, Tanaka I, Yokoyama S, Kuramitsu S. Crystal structure of hypothetical protein TTHB192 from Thermus thermophilus HB8 reveals a new protein family with an RNA recognition motif-like domain. Protein Sci. 2006;15:1494–1499. doi: 10.1110/ps.062131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han D, Krauss G. Characterization of the endonuclease SSO2001 from Sulfolobus solfataricus P2. FEBS Lett. 2009;583:771–776. doi: 10.1016/j.febslet.2009.01.024. [DOI] [PubMed] [Google Scholar]
- **53.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. Determined a novel mechanism for the important function of discrimination of invader DNA from similar sequences in the CRISPR locus by the CRISPR-Cas system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrett RA, Shah SA, Vestergaard G, Deng L, Gudbergsdottir S, Kenchappa CS, Erdmann S, She Q. CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochem Soc Trans. 2011;39:51–57. doi: 10.1042/BST0390051. [DOI] [PubMed] [Google Scholar]
- 56.Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol. 2009;11:457–466. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
- 57.Sorokin VA, Gelfand MS, Artamonova Evolutionary dynamics of clustered irregularly interspaced short palindromic repeat systems in the ocean metagenome. Appl Environ Microbiol. 2010;76:2136–2144. doi: 10.1128/AEM.01985-09. [DOI] [PMC free article] [PubMed] [Google Scholar]