Abstract
Despite their importance for the functioning of the immune system, thymic development and peripheral maintenance of Foxp3+ regulatory (TR) cells are poorly understood. We have found that connexin 43 (C×43), expressed by thymic TR cells progenitors, supports TR development. Mice with deletion of the C×43 gene induced in T cells produce only very few TR cells and had elevated proportion of activated T cells in the lymph nodes suggesting impaired peripheral tolerance. Reduction of the TR cell numbers was accompanied by increased presence of CD4+CD25+GITR+Foxp3− T cells which did not produce inflammatory cytokines and lost suppressor function. These results strongly argue that we have discovered a novel signaling pathway, controlled by C×43, that enhances the generation of TR cells. We propose that a possible mechanism of C×43 activity is by regulating Foxp3 expression in TR lineage cells.
Keywords: T cells, Tolerance, Transcription Factors, Foxp3, Connexin 43
Introduction
CD4+ regulatory T cells expressing the transcription factor Foxp3 (TR cells) constitute a major dominant mechanism of peripheral tolerance and homeostasis of the immune system (1). TR cells also control immune responses to various microorganisms, cancer and transplantation antigens. It has been proposed that TR precursors are instructed to develop into TR lineage by a strong, agonist-mediated signal delivered through the TCR (2, 3). However, other reports have suggested that cues other than recognition of self antigens induce the development of TR cells and involve selective survival or interactions with progenitors of double positive thymocytes (4, 5).
In addition to TCR/MHC interaction, signals necessary for thymic TR development include costimulatory molecules and cytokines. Signals mediated by CD28 and its ligands CD80 and CD86, CD40 and LFA1 and cytokines IL-2 and TGF-β are crucial for TR development which is impaired in mice lacking the respective surface molecules, cytokines or their signaling components (6–10).
Binding of TCR, costimulatory molecules and cytokine receptors activates the signaling circuitry which regulates TR development. Molecular events associated with TR lineage commitment and the stages of thymocyte development when they occur, are not fully known. It is however generally accepted that expression of the transcription factor Foxp3 is pivotal for TR lineage differentiation. Transcription factors activated by signaling pathways downstream of the TCR, cytokine receptors and still unknown. TR developmental signals bind promoter and regulatory elements of the Foxp3 gene (11–16). Once induced, Foxp3 regulates transcription of a number of TR-specific genes which are essential for TR phenotype and suppressor function (17, 18). Mice deficient for Foxp3 develop aggressive, fatal autoimmune disease (19, 20). However, these mice do have a population of TR lineage cells lacking suppressor function which suggests that Foxp3 expression can be dissociated from the TR cell development per se (21–23). Recent reports that Foxp3 expression is controlled by a higher order network of transcription factors opens the possibility that its expression is a concluding, rather than initiating event in TR lineage commitment (24, 25).
Cellular and molecular studies show that TR lineage commitment is a multistage process often controlled by molecules expressed in other cell types which have a unique role in regulating TR cell development. Here we report that connexin 43 (C×43, Gja1) supports the development of TR thymocytes and their precursors before the onset of Foxp3 expression. Connexins are small, transmembrane proteins that allow cells of a multicellular organism to communicate with their surrounding through small channels called gap junctions (26). Over 20 connexin genes have been found in human and mouse genomes and disruption of connexin expression often results in developmental defects and lethality (27). Besides intercellular communication, connexins, especially C×43, which has a relatively long C-terminal cytoplasmic domain, may serve as adaptor protein and interact with cytoskeleton and signaling molecules like β-catenin, src, PKC, MAP kinase (28).
Multiple cell types of the immune system, which express C×43, include monocytes, dendritic cells, and NK, B, and T cells (29). C×43 is expressed in thymocytes and thymic epithelial cells and thymocyte precursors and stromal cells communicate through the gap junctions formed by C×43 (30, 31). C×43 is essential for terminal differentiation of B and T cells when tested in C×43-deficient mice, however, when C×43 was missing in bone marrow cells alone, T cell development was normal (32, 33). Antigen activation upregulates C×43 in T cells and it has been suggested that C×43 is a component of the immunological synapse (34). Gap junction signaling mediated by C×43 contributes to antigen-mediated clonal expansion but not cytokine production of activated peripheral T cells (35). In TR cells C×43 contributes to the formation of gap junctions with target cells which is necessary for TR-mediated suppressor function (36).
Here we report that C×43 supports lineage commitment of double positive and single positive TR precursors. We found that C×43 is expressed in double positive and single positive thymocytes at the developmental stages preceding Foxp3 expression and T cell specific deletion of C×43 gene resulted in a profound deficiency of TR cells in the thymus and in the periphery. Aging C×43 mutant mice had progressively increasing numbers of activated CD4+ T cells in the lymph nodes and spleen. Surprisingly, mice deficient in C×43 in T cells had an expanded population of non-suppressive CD4+CD25+GITR+Foxp3GFP− cells, whose phenotype suggests that they are related to TR cells deficient in Foxp3 expression. C×43-deficient thymocytes were less sensitive to signaling by IL-2 which likely contributed to impaired generation of TR cells in the thymus. In conclusion, our data show that C×43 supports TR lineage development and we postulate that it occurs by a mechanism involving regulation of Foxp3 expression.
Materials and Methods
Mice
Mice with conditional knockout of the C×43 gene (Jackson Labs)(C×43loxP mouse) were crossed to CD4-cre (Taconic) mice and Foxp3GFP reporter mice (23, 37, 38). All mouse strains used for crossing were on the C57BL6 background. Scurfy, TCRα−, RAG−/− mice were purchased from Jackson Labs. Mice were housed under specific pathogen-free conditions and used according to the guidelines of the Institutional Animal Care and Use Committee of the Medical College of Georgia.
T cell activation, proliferation and TR cell suppression assay
T cells were activated in vivo by injecting Foxp3GFP mice with 50 μg of Staphylococcal enterotoxin B (SEB) in a footpad and T cells were isolated after 4 days from popliteal lymph nodes. Naive CD4+ T cells were activated with plate-bound anti-CD3ω (10 μg/ml) and anti-CD28 (1 μg/ml) antibodies. After culturing cells for 3 days, proliferation was measured by adding 1 μCi/well of 3H-thymidine. IL-2 (50 u/ml) was added to indicated wells in some experiments. Adaptive TR cells were produced by activating naive CD4+ T cells in the presence of TGF-β (3 ng/ml) for 4 days and by sorting Foxp3GFP+ cells. Activated TR cells were produced by incubating Foxp3GFP+ cells with plate-bound anti-CD3ω and anti-CD28 antibodies in the presence of 100 u/ml of IL-2. For Th1 differentiation cells were stimulated in the presence of anti-IL-4 antibody (10 μg/ml) and IL-12 (10 ng/ml). For Th2 differentiation cells were stimulated in the presence of IL-4 (1000 U/ml), anti-IFN-γ (10 μg/ml) and anti-IL-12 (10 μg/ml) antibodies. Finally, for Th17 priming cells were be stimulated in the presence of TGF-β (3 ng/ml) and IL-6 (20 ng/ml). Cells were cultured for 4 days.
Lymph node proliferation and TR cell suppression assays were performed as previously described (39).
Cell purification, flow cytometry and cell sorting
Single-cell suspensions were stained with antibodies available commercially (BD Biosciences, eBioscience and BioLegend). Cells were analyzed using FACSCanto flow cytometer (Becton Dickinson) and sorted on a MoFlo cell sorter (Cytomation). Purity of sorted populations exceeded 98.5%.
Production of mixed bone marrow chimeras
Bone marrow from C×43Twt and C×43T−/− mice (expressing congenic markers Ly5.1− and Ly5.1+) was mixed 1:1 and total of 6×106 cells were injected i.v. into sub lethally irradiated (600 Rad) RAG−/− mice. Mice were analyzed 8 weeks after transfer.
In vitro cultures of T cells and cytokine detection
Cytokine production was measured in supernatants of sorted T cell subsets activated and cultured in vitro. CD4+CD25−GFP−, CD4+CD25+GFP− and CD4+CD25+GFP+ cells (2×105 cells per well of 24-well plate) from C×43Twt and C×43T−/− mice were sorted and stimulated with plate-bound anti-CD3 (10 μg/ml) and anti-CD28 (1 μg/ml) antibodies for 40 hours. Supernatants were then collected and used for cytokine detection using commercially available kits (eBioscience). All samples were done in triplicate using pooled cells from two mice. Experiments were repeated three times.
Adoptive transfer
Total CD4+ cells from Cx43Twt, Cx43T−/− and 3 weeks old scurfy males were sorted on AutoMacs (Miltenyi). Purity of sorted cells exceeded 97%. 5×105 cells from Cx43Twt or Cx43T−/− were mixed with 5×105 scurfy cells and transferred (i.v.) into TCRα− recipient mice. Recipient mice were weighted every day with weight at the day of transfer marked as 100%. 5 mice of each kind were analyzed.
CD4+Foxp3GFP− effector cells from Cx43Twt and Cx43T−/− mice, CD4+CD25+Foxp3GFP− T cells from Cx43T−/− mice and CD4+Foxp3GFP+ TR cells from Cx43Twt mice were isolated by flow cytometry. 5×105 effector cells were transferred (i.v.) into RAG knockout mice alone or mixed with 1.5×105 CD4+CD25+Foxp3GFP− T cells from Cx43T−/− mice or CD4+Foxp3GFP+ cells from Cx43Twt mice. Recipient mice were sacrificed after 10 weeks and colon sections were examined for inflammatory bowel disease as described (40).
RT-PCR
RNA was isolated from sorted cells with RNeasy Mini Kit (Qiagen) and reverse transcribed using Superscript kit (Invitrogen). β-actin was used to normalize cDNA quantities. PCR products were resolved on agarose gels, with the gel being scanned. PCR primers used to detect Cx43, Foxp3 and β-actintranscripts were: Cx43 sense: 5'CAGACAGGTCTGAGAGCCCGAACTCT3', antisense: 5'AAGGACCCAGAAGCGCACGTGAGAG3', Foxp3 sense: 5'ATCCAGCCTGCCTCTGACAAGAACC 3', antisense: 5'GGGTTGTCCAGTGGACGCACTTGGAGC3', β-actin sense: 5'CCTTCTACAATGAGCTGCGTGTGGC3', antisense: 5'CATGAGGTAGTCTGTCAGGTCC3. Foxp3-specific primers distinguish between amplification product of the endogenous Foxp3 gene (401 bp) and the transgenic transcript (1357 bp). The PCR conditions were: 94°C for 2 min., 30 cycles of: 94°C for 30 sec., 56°C for 20 sec., 72°C for 50 sec; 72°C for 2 min
Western blotting
Foxp3 protein was detected in sorted (105 cells/sample) CD4+CD25−Foxp3GFP−, CD4+CD25+Foxp3GFP− and CD4+CD25+Foxp3GFP+ cells. Cells were lysed in the gel-loading buffer and resolved on 10% polyacrylamide gel. Proteins were transferred onto a PVDF membrane (Millipore). Membranes were probed with anti-Foxp3 antibody eBio7979 (1:500, eBioscience) followed by goat anti-mouse polyclonal antibody coupled with horseradish peroxidase (1:5000, BioRad). Membranes were developed with ECL chemiluminescence kit (Amersham) according to the manufacturer's instructions. Detection of β-actin was performed to ensure equal loading. Primary antibody (1:8,000) was from Sigma (clone AC-74). For detection of Foxp3 in EL-4 cells, lysates from 5×105 cells were run per a lane.
Production of Cx43 expression constructs in retroviral vectors and analysis of EL-4 cells
Cx43 full-length cDNA was purchased from Open Biosystems (Huntsville, AL). cDNA lacking the stop codon was amplified by PCR and cloned into pEYFP-N1 vector (Clontech) between EcoRI and BamHI in frame with the YFP. Cx43 consisting of amino acids 1–228 (named Cx43NT where NT stands for N-terminal portion) was prepared as described above. All constructs were sequenced to ensure no mutation was created by the PCR. Full-length wild-type and Cx43NT sequences tagged with YFP were then inserted into retroviral plasmid pMSCV-IRES-rCD2 expressing rat CD2 extracellular domain driven by IRES. Retroviral particles were created by calcium phosphate mediated transfection of Phoenix-Eco packaging cell line as described (41). EL-4 cell line obtained from ATCC was then transduced and infected cells were sorted on magnetic sorter (Miltenyi) using biotinylated antibody against rat CD2 (Cedarlane, Burlington, NC) and streptavidin beads (Miltenyi). EL-4 cells expressing Cx43 that were used for all experiments were >98% rCD2+YFP+ as assessed by flow cytometry.
Statistical analysis
Differences between cell populations or cytokine concentrations were analyzed by two sample t test. Values of p < 0.05 were considered statistically significant. Colitis scores were compared using two sample permutation test which is a non-parametric analogue of the two sample t-test (42).
Results
Expression pattern of Cx43 in peripheral CD4+ T cells
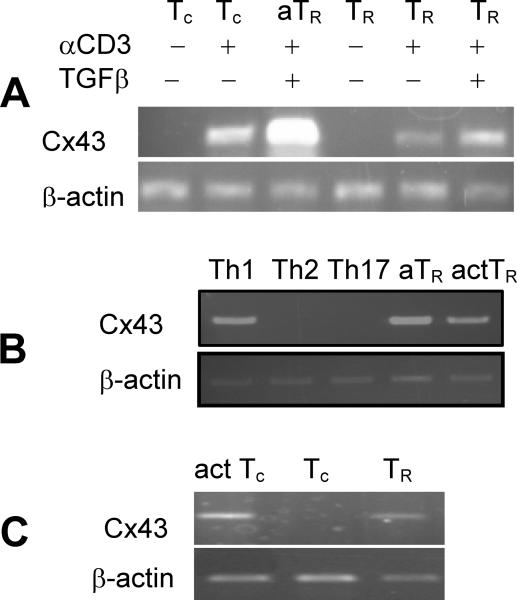
To gain insight into the molecular mechanisms of TR cell function, we compared global gene expression profiles of resting and activated conventional CD4+ T cells (Tc), adaptive TR cells and resting and activated natural TR cells using GeneChip arrays. We have identified Cx43 as a gene differentially expressed in resting and activated effector CD4+ and TR cells. To corroborate microarray data, we investigated Cx43 expression in resting CD44−CD62L+Foxp3GFP− and activated CD44+CD62L−Foxp3GFP− conventional CD4+ T cells and CD4+Foxp3GFP+ TR cells activated in vitro without and in the presence of TGF-β (Fig. 1A). Only CD4+ T cells activated in conditions promoting generation of Th1 cells but not Th2 and Th17 cells express Cx43 (Fig. 1B). To corroborate in vitro data, Cx43 expression was investigated in naive, activated and TR cells sorted from Foxp3GFP reporter mice immunized with SEB (Fig. 1C). In summary, Cx43 was expressed in Tc and TR cells only after activation, and its expression is significantly augmented by TGF-β. Though, Cx43 is greatly upregulated in activated TR cells, its expression was independent of Foxp3 because it was found in Foxp3-deficient T cells from scurfy mice (data not shown). This expression pattern suggested that Cx43 is dispensable in naive conventional and TR cells, but could be important in T cell developmental processes requiring TCR stimulation and T cell activation.
FIGURE 1.
C×43 expression in various T cell subsets assessed by RT-PCR. (A) C×43 expression in naive conventional CD4+ T cells (Tc), CD4+ T cells activated in vitro, adaptive TR cells (aTR, Tc cells activated in the presence of TGF-β), resting natural TR (sorted Foxp3GFP+ cells) and activated nTR cells (sorted Foxp3GFP+ cells activated in vitro). Where indicated, populations of T cells were activated in vitro with plate-bound anti-CD3/anti-CD28 antibodies and TGF-β. Experiment was repeated on cell populations sorted from three independent mice. (B) C×43 expression in CD4+ T cells stimulated in conditions promoting generation of Th1, Th2, Th17, aTR and activated TR cells. (C) C×43 expression in populations of activated CD4+ T cells (act. Tc), naive (Tc) and TR cells (Foxp3GFP+) sorted from mice immunized with SEB. Upper panels show C×43 expression, lower panels show β-actin expression.
Cx43 is necessary for thymic generation of TR cells but not conventional T cells
To further evaluate possible role of Cx43 in T cells, we generated mice with T-cell specific deletion of Cx43. Cx43 conditional knockout mice, prepared by flanking the only protein coding exon of the Cx43 gene with loxP sites (Cx43loxP mouse), were crossed to CD4-cre mice expressing cre recombinase in T cells starting at the stage of double positive thymocytes (37, 38). In addition, we introduced Foxp3GFP reporter transgene into CD4-cre/Cx43loxP/loxP mice to tag TR cells with the GFP (23). Mice transgenic for CD4-cre and Foxp3GFP and homozygous for the Cx43loxP/loxP allele (Cx43T−/− mice) had normal numbers and proportions of double and single positive thymocytes and peripheral CD4+ and CD8+ T cells, but a greatly decreased population of TR cells (Fig. 2A–F). Younger mice had more pronounced deficiency of TR cells than older mice rising a possibility that TR cells are differentially sensitive to Cx43 deficiency (Fig. 2E, F). Control mice (Cx43Twt) used for the experiments expressed FoxpGFP reporter transgene and both alleles of Cx43 flanked by loxP sites, but these mice lacked CD4-cre transgene allowing for normal expression of Cx43. Young (3 week) Cx43T−/− mice had the same proportion of activated/memory cells in the lymph nodes as control mice, while older mice (8 week) had elevated proportion of activated T cells (Fig. 2G, H). CD4+ T cells from Cx43T−/− mice had higher expression of activation markers CTLA-4, GITR, ICOS and higher proportion of proliferating peripheral CD4+ T cells (Fig. 2I). In contrast, Cx43T−/− mice had much lower proportion of FoxpGFP+ TR cells both in the thymus and in the peripheral lymph nodes (Fig. 2B, D–F). The remaining Foxp3GFP+ cells in Cx43T−/− mice had lower expression of Foxp3 and decreased suppressor function compared to TR cells isolated from normal mice (see below). Altogether, phenotypic analysis of T cell populations in the thymus and peripheral lymph nodes showed that Cx43 is required for proper differentiation of TR cells. Decreased numbers of TR cells in Cx43T−/− mice corresponded with higher proportion of cycling conventional CD4+ T cells with activated phenotype and suggested impaired peripheral tolerance (Fig. 2G–I and data below).
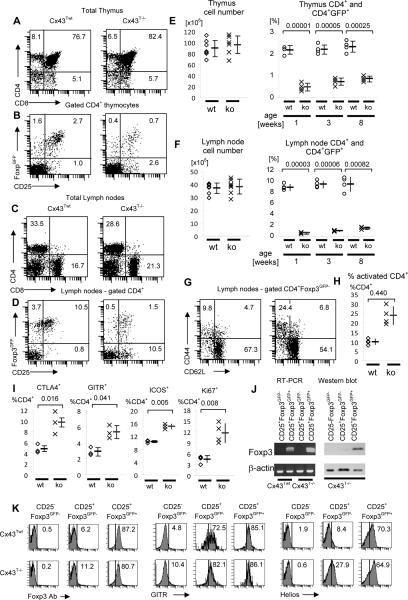
FIGURE 2.
Flow cytometry and Foxp3 gene expression analysis of T cell subsets in the thymus and lymph nodes of an 8 week old C×43Twt and C×43T−/− mice. (A–D) Representative flow cytometry analysis of thymocytes (A, B) and lymph node cells (C, D) of C×43Twt and C×43T−/− mice. (A, C) Total thymocyte and lymph node populations were stained with antibodies specific for CD4 and CD8. (B, D) Flow cytometry analysis of CD25 and Foxp3GFP expression on gated single positive CD4+ thymocytes (B) or lymph node cells (D). (E, F) Total number of thymocytes and lymph node cells (left panels) and percentage of Foxp3GFP+ cells in the populations of CD4+ cells in the thymus and lymph node cells (right panels) in 1, 3 and 8 week old mice. Values for individual C×43Twt (wt) mice are shown as (o) and for C×43T−/− (ko) mice as (x). Average values and standard deviations are shown to the right of individual mice data. Statistical differences between data are indicated by brackets and p values are shown above brackets. Seven mice of each type were analyzed for cell number comparison and four mice for each time point to compare percentages of Foxp3GFP+ cells. (G, H) Representative flow cytometry analysis of CD44 and CD62L expression on gated CD4+Foxp3GFP− lymph node cells (G), and the corresponding statistical analysis showing percentage of activated cells in the population of gated CD4+ T cells (H). Four mice of each type was analyzed. Analysis description and symbols used are as in panels E and F. (I) Expression of CTLA4, GITR, ICOS and Ki-67 by peripheral CD4+ T cells in C×43Twt (wt) and C×43T−/− (ko) mice. Analysis description and symbols used are as above. Data from at least three mice are shown. (J) RT-PCR (left panel) and Western blot analysis (right panel) of Foxp3 expression. For RT-PCR, CD4+CD25+Foxp3GFP− and CD4+CD25+Foxp3GFP+ cells were sorted from C×43Twt and C×43T−/− mice. For Western blot populations of naive CD4+CD25−Foxp3GFP− and CD4+CD25+Foxp3GFP− and CD4+CD25+Foxp3GFP+, cells were sorted from C×43T−/− mice and used for analysis. Experiment was repeated three times. (K) Flow cytometry analysis of Foxp3 (left set of panels), GITR (middle set of panels) and Helios (right set of panels) expression in populations of CD4+CD25−Foxp3GFP−, CD4+CD25+Foxp3GFP− and CD4+CD25+Foxp3GFP+ cells sorted from C×43Twt and C×43T−/− mice. One of two independent experiments is shown.
Remarkably, Cx43T−/− mice had an expanded population of CD4+CD25+Foxp3GFP− cells in the thymus and in lymph nodes (Fig. 2B, D–F). These cells expressed GITR, CTLA-4 and Helios, markers characteristic of TR cells, but did not express Foxp3 (Fig. 2J, K and data not shown)(43–45). The population of CD4+CD25+Foxp3GFP− cells resembled TR cells which develop in the absence of Foxp3 expression, in scurfy or Foxp3 knockout mice. The presence of such cells argues that Cx43 is involved in regulating Foxp3-dependent, suppressor functions which could be separated from the rest of the TR lineage developmental program. Consistent with the lack of Foxp3 and Cx43, CD4+CD25+Foxp3GFP− cells did not inhibit proliferation of conventional CD4+ T cells in vitro (Fig. 3A). In vivo, total CD4+ T cells from Cx43T−/− mice were less efficient in controlling effector CD4+ T cells from scurfy mice when co-transferred into recipient lymphopenic mice (Fig. 3B). The CD4+CD25+Foxp3GFP− cells proliferated much less than conventional CD4+ T cells when stimulated in vitro, did not produce inflammatory cytokines and produced only small amounts of IL-2 (Fig. 3C, D). The lack of suppressor function of CD4+CD25+Foxp3GFP− T cells in Cx43T−/− mice was most likely due to the deficiency of Foxp3, but also could be due to the lack of Cx43, which has been shown to contribute directly to the suppressor function of TR cells (36). In an adaptive transfer experiment, CD4+CD25+Foxp3GFP− cells from Cx43T−/− mice were not able to protect mice from inflammatory bowel disease (Fig. 4A–F). In summary, the phenotype of CD4+CD25+Foxp3GFP− cells strongly argues that they represent cells committed to TR lineage which did not complete whole differentiation process.
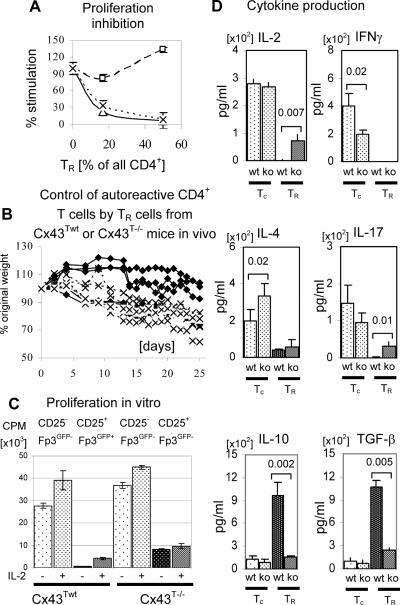
FIGURE 3.
Suppressor function, proliferation and cytokine production of CD4+ T cell subsets from C×43Twt and C×43T−/− mice. (A) Proliferation inhibition assay. Proliferation of CD4+Foxp3GFP− responder cells (expressing wild type C×43 alleles) and stimulated with soluble anti-CD3 antibody was inhibited by adding increasing numbers of CD4+CD25+ TR cells sorted from C×43Twt (triangles, continuous line) and CD4+CD25+Foxp3GFP− (squares, dashed line) or CD4+CD25+Foxp3GFP+ cells (x, dotted line) sorted from C×43T−/− mice. The percentage stimulation was calculated by dividing the proliferation reading from a particular well by the reading from a well with only Foxp3GFP− responder cells. The plot shows a representative experiment of three. (B) TR cells from C×43T−/− mice are less efficient in controlling CD4+ T cells isolated from scurfy mice. CD4+ T cells from scurfy mice were mixed 1:1 with total CD4+ T cells from C×43Twt (◆) or C×43T−/− (x) mice and transferred into TCRα− recipient mice. The plot shows percentage of the original weight of individual mice at various time intervals after cell transfer. (C) Proliferation assay of T cell subsets isolated from C×43Twt and C×43T−/− mice and stimulated with plate bound anti-CD3/anti-CD28 antibodies. CD4+CD25Foxp3GFP− (conventional T cells) and CD4+CD25Foxp3GFP+ (TR) cells were sorted from C×43Twt mice and CD4+CD25Foxp3GFP− (conventional) and CD4+CD25+Foxp3GFP− (TR-like) cells were sorted from C×43T−/− mice. IL-2 was added to indicated wells. The plot shows a representative experiment of three. (D) Cytokine production by T cell subsets isolated from C×43Twt and C×43T−/− mice. CD4+CD25Foxp3GFP− (conventional T cells, Tc) were sorted from C×43Twt (wt) and C×43T−/− mice and CD4+CD25+Foxp3GFP+ (TR) or CD4+CD25+Foxp3GFP− (TR-like) cells were sorted from C×43Twt or C×43T−/− mice respectively. Sorted cells were stimulated with plate bound antibodies anti-CD3/anti-CD28 and cytokine concentration was measured by ELISA. Brackets show statistically significant differences in cytokine concentration between cells from C×43Twt (wt) and C×43T−/− (ko) mice. Number above brackets shows p value. Plot shows a representative experiment of three.
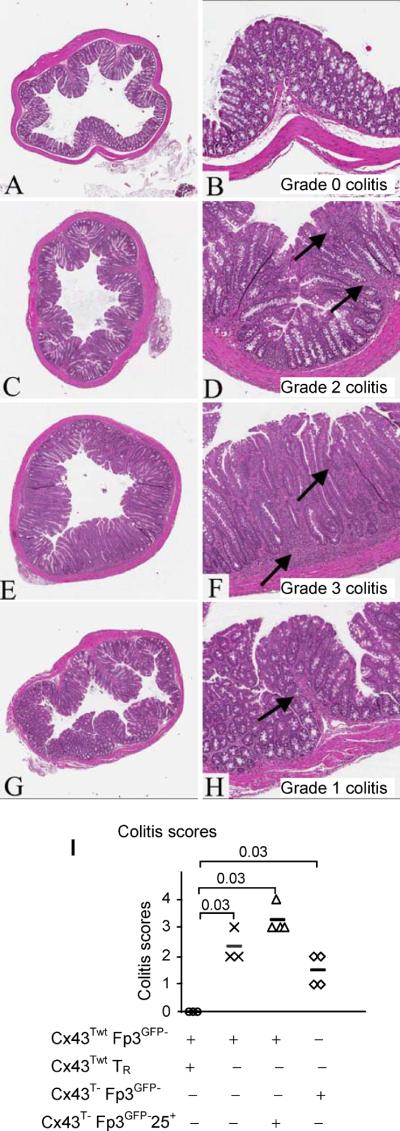
FIGURE 4.
CD4+CD25+Foxp3GFP− T cells from C×43T−/− mice do not protect mice receiving adaptive transfer of effector CD4+Foxp3GFP− T cells isolated from C×43Twt mice from inflammatory bowel disease. Low (×40, left panels) and high (×200, right panels) magnifications are shown. At least three mice in each group were analyzed. Recipient mice were sacrificed after 10 weeks and colon sections were examined for inflammatory bowel disease. Colitis scores are shown on high magnification panels. (A, B) Recipient RAG− mice transferred with CD4+Foxp3GFP− T cells from C×43Twt mice mixed with CD4+Foxp3GFP+ TR cells from C×43Twt mice do not suffer from inflammation of the colon (grade 0 colitis). (C, D) Recipient mice transferred with CD4+Foxp3GFP− T cells from C×43Twt mice show moderate inflammation (grade 2 colitis). High magnification demonstrates moderate inflammation in basal and upper mucosal lamina propria (arrows). (E, F) Recipient RAG− mice transferred with CD4+Foxp3GFP− T cells from C×43Twt mice mixed with CD4+CD25+Foxp3GFP− T cells from C×43T−/− mice show moderate inflammation (grade 3 colitis). High magnification demonstrates severe inflammation in basal and upper mucosal lamina propria (arrows). (G, H) The lymphocyte infiltration in mice receiving CD4+Foxp3GFP− T cells from C×43T−/− mice (grade 1 colitis). High magnification demonstrating mild inflammation in basal lamina propria (arrow). (I) Summary of colitis scores. Colitis scores are shown for individual recipient mice. Recipient mice analyzed in (A, B) are shown by (∘), analyzed in (C, D) are shown by (x), analyzed in (E, F) by (Δ) and mice analyzed in (G, H) are shown by (◊). Transferred cell populations are indicated below the graph. Average values are shown by bars. Statistical differences between data are indicated by brackets and p values are shown above brackets.
While the development and suppressor function of TR cells are severely compromised in Cx43T−/− mice, proliferation of conventional CD4+ T cells, stimulated in vitro, was not affected (Fig. 3C). These cells were able to express Th subset specific transcription factors T-bet, GATA3 and RORγt and consistently produced similar quantity of IL-2, lower quantities of IFN-γ and increased quantity of IL-4 (Fig. 3D and data not shown). Cx43 is expressed only in cells stimulated to become Th1 which suggests that Cx43 modulates the outcome of activation of only some of conventional CD4+ T cell subsets. It is likely that the magnitude of this modulation depends on the level of Cx43 expression which is regulated by cytokines e.g. it is greatly augmented by TGF-β. In vivo, effector CD4+Foxp3GFP− cells from Cx43T−/− mice were less effective in inducing inflammatory bowel disease than the respective population from Cx43 - sufficient mice (Fig. 4G, H). The weaker functions of effector cells from Cx43T−/− mice couldresult from observed Th2 shift in the cytokine expression profile or impaired migration to the intestine. Alternatively, as reported earlier Cx43 and gap junctions are important for the initiation of the immune response and T cell activation in vivo (46). In summary, the lack of Cx43 in CD4+ T cells selectively impaired the generation of TR cells, while the development of conventional CD4+ cells was not affected.
Lack of TR development in Cx43T−/− mice is TR-intrinsic
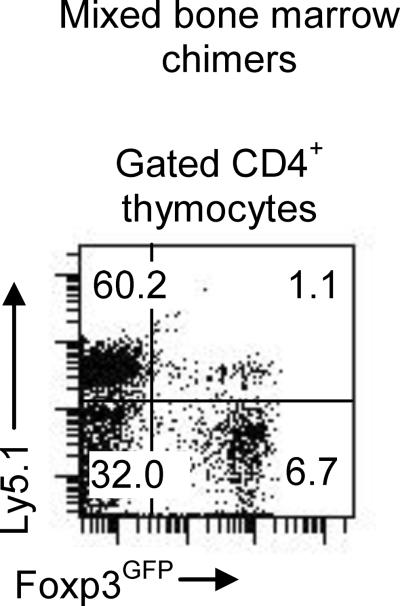
The transgenic expression of cre recombinase may affect Cx43 expression in other cell types besides thymocytes. Alternatively, thymocytes expressing Cx43 could induce Cx43 expression on stromal epithelial cells or other thymocytes. To conclusively establish that a defect of TR cell lineage development in Cx43T−/− mice is T cell intrinsic, we have prepared mixed bone marrow chimeras. Recipient mice were irradiated and reconstituted with wild type bone marrow from Ly5.1−/−Cx43Twt and mutant bone marrow from Ly5.1+/+Cx43T−/− mice. The TR cells developed efficiently only from wild type bone marrow (Fig. 5). A much greater proportion of wild type TR cells (greater than in normal mice) was observed in the single positive thymocytes suggesting that wild type bone marrow compensates for the lack of TR cells originating from the Cx43-deficient cells while TR development from mutant thymocytes remained impaired. This result argues that soluble factors or molecules produced by stromal cells and/or other thymocytes are not able to restore TR cell numbers which argues for the intrinsic developmental defect of Cx43T−/− thymocytes.
FIGURE 5.
Flow cytometry analysis of thymic development of TR cells in mixed bone marrow chimera. RAG−/− recipient mice were reconstituted with the bone marrow from C×43Twt (Ly5.1−) and C×43T−/− (Ly5.1+) mice and chimeras were analyzed 8 weeks after bone marrow transfer. Representative experiment of three with 2–3 mice each is shown.
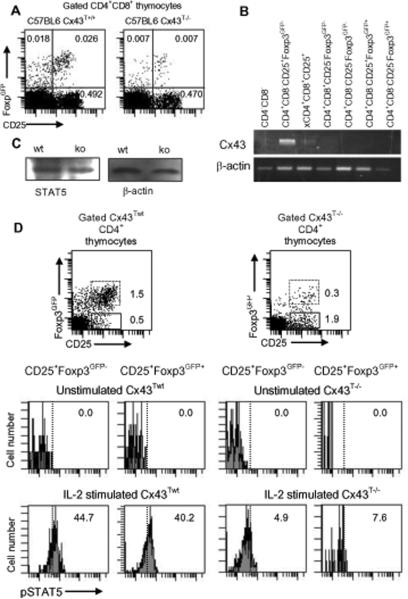
Cx43 is expressed in TR precursors at the double and single positive stage
To further determine the stage of thymocytes when Cx43 is first expressed, we have sorted double negative, double positive and single positive thymocytes and investigated the presence of Cx43 transcripts by RT-PCR. Flow cytometry analysis of double positive thymocytes shows that Cx43T−/− mice have much lower proportion of Foxp3GFP+ T cells, but similar proportion of CD4+CD8+CD25+ cells as the wild type mice (Fig. 6A). RT-PCR analysis detected low level of Cx43 expression in CD4+CD8+CD25+ cells. Much higher levels of Cx43 were detected in single positive CD4+CD25+Foxp3GFP− thymocytes but not in CD4+CD25−Foxp3GFP− conventional thymocytes or CD25−Foxp3GFP+ or CD25+Foxp3GFP+ TR thymocytes expressing Foxp3 (Fig. 6B). Altogether, the gene expression study showed that Cx43 is detected in double and single positive subsets expressing CD25, but is downregulated in thymocytes expressing Foxp3. In summary, gene expression studies place Cx43 expression at the stage of thymocyte development just before the onset of Foxp3 expression, when the critical decision of the TR lineage commitment is made. Our studies are consistent with the recent report that the single positive thymocyte subset CD4+CD25+Foxp3GFP− represents immediate precursors of TR thymocytes (47).
FIGURE 6.
Flow cytometry of CD25 and Foxp3 an C×43Twt and C×43T−/− mice and C×43 gene expression analysis in thymocyte subsets. (A) Analysis of CD25 and Foxp3GFP expression in CD4+CD8+ thymocytes from C×43Twt and C×43T−/− mice. (B) RT-PCR analysis of C×43 expression in populations of thymocytes sorted from C×43Twt mice. (X) denotes that this cell population contained cells that were Foxp3GFP+ or Foxp3GFP−. Experiment was repeated two times. (C) CD4+ T cells from C×43Twt and C×43T−/− mice have the same level of STAT5. Western blot analysis of sorted CD4+ cells (2×105 cells/sample) from C×43Twt and C×43T−/− mice. Samples were normalized for mouse β-actin. (D) STAT5 phosphorylation in single positive thymocytes from C×43Twt (left upper dot plot and histograms) and C×43T−/− (right upper dot plot and histograms) mice. STAT5 phosphorylation of CD4+CD25+Foxp3GFP− (dot plots, continuous line) and CD4+CD25+Foxp3GFP+ (dot plots, dotted line) thymocytes was detected by flow cytometry. Thymocytes were stained with antibodies specific for CD4, CD8, CD25 and pSTAT5 (antibody pY694, BD). Percentage of pSTAT5+ cells is shown in gated CD4+CD25+Foxp3GFP− cells (left histograms) and CD4+CD25+Foxp3GFP+ cells (right histograms). Cells not incubated with IL-2 (Unstimulated) served as negative controls. Experiment was done 3 times with cells from 1–2 mice of each kind.
When sorted, CD4+CD8+CD25+ double positive thymocytes included rare CD25+Foxp3GFP+ cells. Since single positive Foxp3GFP+ thymocytes do not express Cx43, we did not formally determine if Cx43 was still downregulated in double positive TR thymocytes, following Foxp3 expression, or later at the stage of single positive CD4+Foxp3GFP+ thymocytes. However, regardless of the exact time of onset of Foxp3 expression, all single positive Foxp3GFP+ thymocytes downregulate Cx43. Altogether, experimental data shows that Cx43 expression is tightly controlled, and that Cx43 transcripts are detected in thymocyte subsets defined as immediate precursors of Foxp3+ TR cells.
To further examine possible outcome of Cx43 deficiency on the thymic TR precursors, we analyzed their sensitivity to IL-2 by analyzing phosphorylation of STAT5. IL-2 signaling which depends on STAT5 phosphorylation regulates both thymic generation of TR cells and their peripheral homeostasis and function (48). We have found that IL-2 results in suboptimal phosphorylation of STAT5 in CD4+CD25+Foxp3− thymocytes, which are immediate precursors of CD4+CD25+Foxp3+ TR thymocytes (Fig. 6C, D). Expression of CD25 is similar in wild type and Cx43T−/− mice and is considered to be the result of high affinity interaction between the MHC/peptide complexes and TCRs on TR precursors. Our result suggests that at least one mechanism how Cx43 regulates TR thymocyte generation is by increasing the sensitivity of TR precursors to IL-2 signaling.
Cx43 could be directly involved in signaling complexes regulating Foxp3 expression
Signaling functions mediated by Cx43 depend on the intercellular communication through gap-junctions, both on transporting molecules via hemichannels (without communicating with other cells) and on channel-independent mechanisms. We have observed differences between Cx43-sufficient and -deficient CD4+ T cells in our in vitro tests when cellular interactions probably do not involve gap junction formation. Gap-junction independent signaling is mediated by the C-terminal cytoplasmic domain which interacts with a number of proteins regulating cell motility, intracellular protein trafficking and components of multiple signaling pathways (26, 49). To investigate the role of Cx43 in regulating Foxp3 expression, we took advantage of EL-4 cell line which was recently used as a cellular model of TR cells to identify regulatory regions of the Foxp3 gene (16). Low level of Foxp3 is spontaneously expressed in EL-4 cells, which is greatly upregulated in cells stimulated through the TCR and further enhanced in the presence of TGF-β.
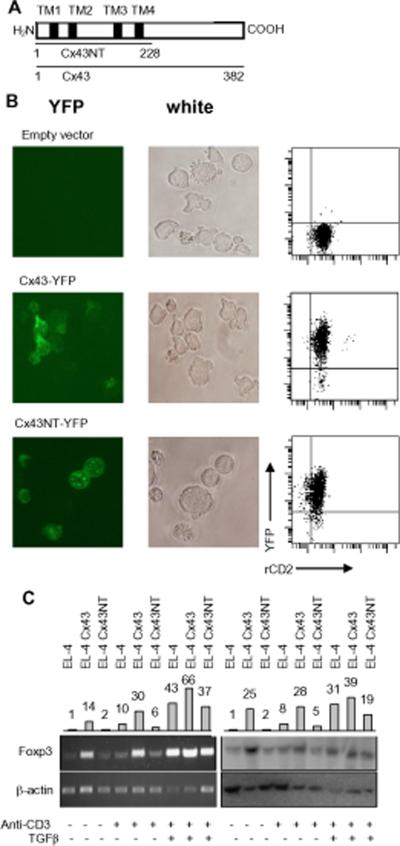
Cx43 was overexpressed in EL-4 cells (which constitutively express low level of Cx43) using retroviral vectors expressing rat CD2 reporter molecule and encoding wild-type Cx43 or Cx43 mutant lacking C-terminal cytoplasmic domain both tagged with YFP (Fig. 7A). Cx43 construct without C-terminal cytoplasmic tail is able to form functional gap junctions, but lacks adaptor sites for signaling molecules (50). EL-4 cells transduced with control vector or constructs encoding wild-type or mutant Cx43 and expressing similar levels of rat CD2 were sorted, and the level of Foxp3 expression was examined by RT-PCR and Western blotting (Fig. 7B, C). EL-4 cells overexpressing wild-type Cx43 but not C-terminal deficient mutant produced increased level of Foxp3 transcript and protein. Stimulation of EL-4 transfectants through the TCR in the presence of TGF-β further increased Foxp3 expression (Fig. 7C). This result suggests that one hypothesis of how Cx43 may augment Foxp3 expression is to contribute to the assembly of a signaling complex around its C-terminal domain. This mode of Cx43 activity is consistent with the reported participation of Cx43 in the immunological synapse where signaling molecules involved in T cell activation assemble and interact (34). We may speculate that Cx43 is a component of the synapse whose role is to increase and sustain Foxp3 expression. This function of Cx43 could be important in activated, conventional CD4+ and TR cells, in particular cells exposed to TGF-β what greatly enhances Cx43 expression. In summary, Cx43 may regulate Foxp3 expression in activated CD4+ T cells independently of its membrane channel forming function.
FIGURE 7.
Analysis of C×43 expression in EL-4 Cells. (A) Mouse C×43 protein showing transmembrane domains (TM1–4), whole length protein (C×43 1–382) and protein truncated at the carboxy end (C×43NT 1–228). (B) Expression of full length wild type and N-terminal membrane domain of C×43 in EL-4 lymphoma. Right column shows imaging for YFP-C×43 fusion protein, middle column shows EL-4 cells in normal light, right column shows flow cytometry analysis of EL-4 cells. EL-4 cells were transduced with empty pMSCV vector expressing rat CD2 (upper panels) or constructs encoding full length C×43-YFP fusion protein (middle panels) or N-terminal part of C×43 fused with YFP (lower panels). Transduced cells (rat CD2+) were analyzed on Nikon Eclipse TE2000-S fluorescence microscope. Both wild type and truncated C×43 incorporate into cell membrane. Green spots on microscope images are C×43 complexes in the plasma membrane. (C) Full length C×43 (EL-4 C×43), but not truncated C×43 (EL-4 C×43NT), enhances Foxp3 expression in unstimulated EL-4 lymphoma cell line. Foxp3 expression is further increased in cells stimulated through the TCR and in cells stimulated in the presence of TGF-β. Left images show RT-PCR reactions with Foxp3-specific primers and right panels show Western blot with Foxp3-specific antibody. Bars above images show relative quantity of Foxp3 transcript and protein standardized for β-actin with expression of Foxp3 in EL-4 cells arbitrarily set as one.
Discussion
We discovered that Cx43 supports generation of TR cells and is necessary for the suppressor function of TR cells. Thus, Cx43 joins a growing number of molecules like CD28, cytokines and cytokine receptors CD25 (IL-2Rα), CD122 (IL-2Rβ), CD132 (the common γ-chain), TGF-β, signaling molecules STAT5, CARMA-1, c-rel, that are selectively involved in the development of TR cells (6, 10, 12, 14, 15, 51, 52). In contrast to previously defined molecules that function downstream of the TCR complex, or accessory molecules or cytokine receptors, Cx43 may represent a new autonomous molecular pathway important for TR lineage commitment. This pathway may depend on intercellular communication through the gap junctions, which is a characteristic function of Cx43. Thus, it is likely that Cx43 regulates unique aspects of TR development which are independent of TCR specificity of the developing thymocyte and require exchange of signaling molecules between cells. This possibility is supported by a report that Cx43 forms gap junctions between thymocytes and stromal epithelial cells and by our data demonstrating that Cx43 expression is tightly controlled in thymocytes and coincides with thymocyte stages immediately preceding Foxp3 expression (30). This raises a possibility that Cx43 is a marker of early TR precursors found in populations of double or single positive thymocytes (47, 53). Since not all Foxp3+ thymocytes express CD25 and only a fraction of CD4+CD25+ thymocytes eventually becomes TR cells, it is tempting to speculate that expression of Cx43 is a more specific marker of TR lineage commitment than CD25 (47).
Signaling circuits downstream of Cx43 are currently not known but they may involve STAT5. STAT5 phosphorylation is induced by IL-2 but also by signaling through the TCR, IL-7 and thymic stromal lymphopoietin (TSL) (54, 55). In particular, signaling through IL-7 and TSL, which likely occur at the double positive stage, may be important for the induction of TR precursors. We have detected decreased phosphorylation of STAT5 in Cx43-deficient thymocytes exposed to IL-2 in vitro. Considering a well known role of IL-2 in the development of TR thymocytes it becomes possible that Cx43 facilitates TR lineage decision by temporarily increasing the sensitivity to IL-2, and perhaps TCR, signaling in thymocytes which would become TR precursors. This function of Cx43 concludes once Foxp3 is expressed, limiting the number of developing TR cells. This scenario is consistent with observed downregulation of Cx43 expression in TR thymocytes once they initiate Foxp3 expression. Deficient IL-2 signaling could also explain lower Foxp3 expression observed in the TR cells still remaining in Cx43T−/− mice.
Despite a greatly reduced proportion of TR cells, Cx43T−/− mice have an expanded population of thymic and peripheral CD4+CD25+Foxp3− T cells. These cells do not inhibit in vitro proliferation of effector CD4+ T cells but have some characteristics of TR phenotype. CD4+CD25+Foxp3− cells express elevated levels of GITR and Helios, two molecules found on Foxp3+ TR cells, do not efficiently proliferate when stimulated through the TCR in vitro, and do not produce inflammatory cytokines. Considering the phenotype and functional properties of CD4+CD25+ T cells, we hypothesize that this population resembles Foxp3-deficient TR-like cells that develop in the absence of Foxp3 expression. Such cells retained most functional properties of TR cells and were earlier described in scurfy mice and in mice where Foxp3 gene was deleted (21–23). However, as earlier described, Foxp3-deficient TR cells transcribed Foxp3 gene while CD4+CD25+ thymocytes in C×43T−/− mice produce, at most, only very little Foxp3 transcripts. This places CD4+CD25+ T cells at the stage of TR cell precursors which depend on C×43 for Foxp3 expression. In the absence of C×43, TR precursors develop as CD4+CD25+Foxp3− cells. This shows that not only Foxp3-deficient TR cells with transcriptionally active Foxp3 locus are able to advance to mature thymocytes and leave the thymus, but also TR precursors that do not express Foxp3 are able to progress along the TR developmental path. Thus, TR cell development consists of Foxp3-independent molecular events that produce TR lineage cells with many properties of functional TR cells but without suppressor function, and Foxp3-dependent differentiation steps that confer and regulate TR cell suppressor function. We propose that the later steps are enhanced by C×43. It is commonly accepted that TR development and Foxp3 expression occur in the developing thymocytes due to instructive signal delivered through the TCR which recognizes selecting MHC/peptide ligands with increased affinity (2, 3, 56). Our data show that TCR dependent signals are modulated by cellular interactions that are not TCR-specific. We would like to suggest that C×43 is one of the molecules that act before or in concert with, signals from the TCR to induce Foxp3 expression.
The proportion of CD25+Foxp3GFP+ TR cells in C×43T−/− mice varied between 5–15% of the original number present in the lymph nodes of a wild type mice. The reason for the persistence of some TR cells in C×43T−/− mice is currently unknown. At least some of these cells are present due to incomplete excision of the C×43 gene (data not shown). Alternatively, CD25+Foxp3GFP+ cells may represent a TR subset that does not require C×43 for development which is consistent with our observation that more TR cells persists in older C×43T−/− mice. Finally, since both cre recombinase, controlled by the CD4 promoter, and C×43 are expressed at the same or very close developmental stage, it is also possible that, in some double positive thymocytes, expression of the cre recombinase does not occur before C×43 expression. Once these cells pass the critical stage when C×43 is required for TR lineage commitment they progress to become Foxp3+ TR thymocytes which do not depend on C×43 for further development. This last possibility is consistent with higher proportion of Foxp3GFP+ TR cells in the lymph nodes of eight week old C×43T−/− mice than in younger mice which may be due to higher proportion of selected TR cells in older mice and expansion of the remaining TR cells in peripheral lymph nodes.
Despite severe deficiency of the TR cells, C×43T−/−R mice do not suffer from manifest autoimmunity. Instead, the number of CD4+ T cells with activated phenotype in the peripheral lymph nodes gradually increases, in an age-dependent manner. The lack of acute autoimmune disease in these animals could have several causes. First, sufficient CD25+Foxp3GFP+ TR cells remain in C×43T−/− mice to control autoreactive cells. It has been shown that elimination of as much as 80–90% of TR cells does not result in rampant autoimmunity (57, 58). Second, conventional CD4+ T cells in C×43T−/− mice may have altered effector functions and are less prone to autoimmunity. This interpretation is supported by the data showing that C×43-deficient conventional T cells produce less IFN-γ and by the analysis of mixed bone marrow chimeras demonstrating that C×43-sufficient conventional CD4+ T cells have higher proportion of activated CD44hiCD62L− cells than C×43-deficient cells (data not shown). Finally, one can not exclude the possibility that the TCR repertoire in C×43 mice have lower frequency of autoreactive T cells.
In summary, functional analyses of T cell subsets in C×43T−/− mice supports the view that C×43 supports TR lineage commitment and differentiation but also participates in regulating functions of mature TR and effector cells. Reported and our own data suggest that the suppressor function of TR cells is regulated by C×43 directly, by a gap junction dependent mechanism, or indirectly which may include enhancement of Foxp3 expression (36). Moreover, both Foxp3 and C×43 expression in mature T cells depend on TCR stimulation and are greatly augmented by TGF-β. In conclusion, we speculate that C×43 is an important component of the molecular circuitry, which includes TGF-β, and regulates peripheral tolerance.
Acknowledgements
We thank L. Ignatowicz and members of his laboratory for reading the manuscript and helpful discussions. The authors declare no conflict of interests.
This work was supported by NIH grants R01 CA107349-01A1 to P.K.
Footnotes
The GeneChip expression data were deposited in the GEO database http://www.ncbi.nlm.nih.gov/geo/. The accession numbers are GSE11775 and GSE28130.
Disclosures The authors have no conflicting financial interests.
References
- 1.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 2.Jordan MS, Boesteanu A, Reed AJ, Petrone AL, Holenbeck AE, Lerman MA, Naji A, Caton AJ. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat. Immunol. 2001;2:301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 3.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat. Immunol. 2002;3:756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 4.van Santen HM, Benoist C, Mathis D. Number of T reg cells that differentiate does not increase upon encounter of agonist ligand on thymic epithelial cells. J. Exp. Med. 2004;200:1221–1230. doi: 10.1084/jem.20041022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pennington DJ, Silva-Santos B, Silberzahn T, Escorcio-Correia M, Woodward MJ, Roberts SJ, Smith AL, Dyson PJ, Hayday AC. Early events in the thymus affect the balance of effector and regulatory T cells. Nature. 2006;444:1073–1077. doi: 10.1038/nature06051. [DOI] [PubMed] [Google Scholar]
- 6.Tai X, Cowan M, Feigenbaum L, Singer A. CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat. Immunol. 2005;6:152–162. doi: 10.1038/ni1160. [DOI] [PubMed] [Google Scholar]
- 7.Tang Q, Henriksen KJ, Boden EK, Tooley AJ, Ye J, Subudhi SK, Zheng XX, Strom TB, Bluestone JA. Cutting edge: CD28 controls peripheral homeostasis of CD4+CD25+ regulatory T cells. J. Immunol. 2003;171:3348–3352. doi: 10.4049/jimmunol.171.7.3348. [DOI] [PubMed] [Google Scholar]
- 8.Lio CW, Dodson LF, Deppong CM, Hsieh CS, Green JM. CD28 facilitates the generation of Foxp3- cytokine responsive regulatory T cell precursors. J. Immunol. 2010;184:6007–6013. doi: 10.4049/jimmunol.1000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malek TR, Bayer AL. Tolerance, not immunity, crucially depends on IL-2. Nat. Rev. Immunol. 2004;4:665–674. doi: 10.1038/nri1435. [DOI] [PubMed] [Google Scholar]
- 10.Ouyang W, Beckett O, Ma Q, Li MO. Transforming growth factor-beta signaling curbs thymic negative selection promoting regulatory T cell development. Immunity. 2010;32:642–653. doi: 10.1016/j.immuni.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J. Exp. Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long M, Park SG, Strickland I, Hayden MS, Ghosh S. Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 2009;31:921–931. doi: 10.1016/j.immuni.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruan Q, Kameswaran V, Tone Y, Li L, Liou HC, Greene MI, Tone M, Chen YH. Development of Foxp3(+) regulatory T cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molinero LL, Yang J, Gajewski T, Abraham C, Farrar MA, Alegre ML. CARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cells. J. Immunol. 2009;182:6736–6743. doi: 10.4049/jimmunol.0900498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 18.Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, Macisaac KD, Levine SS, Fraenkel E, von BH, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 20.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 21.Lin W, Haribhai D, Relland L, Truong N, Carlson M, Williams C, Chatila T. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 22.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 23.Kuczma M, Podolsky R, Garge N, Daniely D, Pacholczyk R, Ignatowicz L, Kraj P. Foxp3-deficient regulatory T cells do not revert into conventional effector CD4+ T cells but constitute a unique cell subset. J. Immunol. 2009;183:3731–3741. doi: 10.4049/jimmunol.0800601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan F, Yu H, Dang EV, Barbi J, Pan X, Grosso JF, Jinasena D, Sharma SM, McCadden EM, Getnet D, Drake CG, Liu JO, Ostrowski MC, Pardoll DM. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–1146. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouyang W, Beckett O, Ma Q, Paik JH, Depinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 26.Dbouk HA, Mroue RM, El-Sabban ME, Talhouk RS. Connexins: a myriad of functions extending beyond assembly of gap junction channels. Cell Commun. Signal. 2009;7:4. doi: 10.1186/1478-811X-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laird DW. Life cycle of connexins in health and disease. Biochem. J. 2006;394:527–543. doi: 10.1042/BJ20051922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herve JC, Bourmeyster N, Sarrouilhe D, Duffy HS. Gap junctional complexes: from partners to functions. Prog. Biophys. Mol. Biol. 2007;94:29–65. doi: 10.1016/j.pbiomolbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Neijssen J, Pang B, Neefjes J. Gap junction-mediated intercellular communication in the immune system. Prog. Biophys. Mol. Biol. 2007;94:207–218. doi: 10.1016/j.pbiomolbio.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Alves LA, Campos de Carvalho AC, Cirne Lima EO, Rocha e Souza CM, Dardenne M, Spray DC, Savino W. Functional gap junctions in thymic epithelial cells are formed by connexin 43. Eur. J. Immunol. 1995;25:431–437. doi: 10.1002/eji.1830250219. [DOI] [PubMed] [Google Scholar]
- 31.Fonseca PC, Nihei OK, Urban-Maldonado M, Abreu S, De Carvalho AC, Spray DC, Savino W, Alves LA. Characterization of connexin 30.3 and 43 in thymocytes. Immunol. Lett. 2004;94:65–75. doi: 10.1016/j.imlet.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen TD, Taffet SM. A model system to study Connexin 43 in the immune system. Mol. Immunol. 2009;46:2938–2946. doi: 10.1016/j.molimm.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 33.Montecino-Rodriguez E, Leathers H, Dorshkind K. Expression of connexin 43 (C×43) is critical for normal hematopoiesis. Blood. 2000;96:917–924. [PubMed] [Google Scholar]
- 34.Oviedo-Orta E, Evans WH. Gap junctions and connexins: potential contributors to the immunological synapse. J. Leukoc. Biol. 2002;72:636–642. [PubMed] [Google Scholar]
- 35.Oviedo-Orta E, Perreau M, Evans WH, Potolicchio I. Control of the proliferation of activated CD4+ T cells by connexins. J. Leukoc. Biol. 2010;88:79–86. doi: 10.1189/jlb.0909613. [DOI] [PubMed] [Google Scholar]
- 36.Bopp T, Becker C, Klein M, Klein-Hessling S, Palmetshofer A, Serfling E, Heib V, Becker M, Kubach J, Schmitt S, Stoll S, Schild H, Staege MS, Stassen M, Jonuleit H, Schmitt E. Cyclic adenosine monophosphate is a key component of regulatory T cell-mediated suppression. J. Exp. Med. 2007;204:1303–1310. doi: 10.1084/jem.20062129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao Y, Day KH, Damon DN, Duling BR. Endothelial cell-specific knockout of connexin 43 causes hypotension and bradycardia in mice. Proc. Natl. Acad. Sci. USA. 2001;98:9989–9994. doi: 10.1073/pnas.171305298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 39.Kuczma M, Pawlikowska I, Kopij M, Podolsky R, Rempala GA, Kraj P. TCR repertoire and Foxp3 expression define functionally distinct subsets of CD4+ regulatory T cells. J. Immunol. 2009;183:3118–3129. doi: 10.4049/jimmunol.0900514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asseman C, Mauze S, Leach MW, Coffman RL, Powrie F. An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J. Exp. Med. 1999;190:995–1004. doi: 10.1084/jem.190.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinsella TM, Nolan GP. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene. Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 42.Good P. Permutation Tests: A Practical Guide to Resampling Methods for Testing Hypotheses. 2nd. ed. Springer; New York: 2000. [Google Scholar]
- 43.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 45.McHugh RS, Whitters MJ, Piccirillo CA, Young DA, Shevach EM, Collins M, Byrne MC. CD4(+)CD25(+) immunoregulatory T cells: gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity. 2002;16:311–323. doi: 10.1016/s1074-7613(02)00280-7. [DOI] [PubMed] [Google Scholar]
- 46.Elgueta R, Tobar JA, Shoji KF, De CJ, Kalergis AM, Bono MR, Rosemblatt M, Saez JC. Gap junctions at the dendritic cell-T cell interface are key elements for antigen-dependent T cell activation. J. Immunol. 2009;183:277–284. doi: 10.4049/jimmunol.0801854. [DOI] [PubMed] [Google Scholar]
- 47.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bayer AL, Yu A, Malek TR. Function of the IL-2R for thymic and peripheral CD4+CD25+ Foxp3+ T regulatory cells. J. Immunol. 2007;178:4062–4071. doi: 10.4049/jimmunol.178.7.4062. [DOI] [PubMed] [Google Scholar]
- 49.Wei CJ, Xu X, Lo CW. Connexins and cell signaling in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:811–838. doi: 10.1146/annurev.cellbio.19.111301.144309. [DOI] [PubMed] [Google Scholar]
- 50.Omori Y, Yamasaki H. Gap junction proteins connexin32 and connexin43 partially acquire growth-suppressive function in HeLa cells by deletion of their C-terminal tails. Carcinogenesis. 1999;20:1913–1918. doi: 10.1093/carcin/20.10.1913. [DOI] [PubMed] [Google Scholar]
- 51.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 52.Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 2002;17:167–178. doi: 10.1016/s1074-7613(02)00367-9. [DOI] [PubMed] [Google Scholar]
- 53.Liston A, Nutsch KM, Farr AG, Lund JM, Rasmussen JP, Koni PA, Rudensky AY. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc. Natl. Acad. Sci. USA. 2008;105:11903–11908. doi: 10.1073/pnas.0801506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isaksen DE, Baumann H, Trobridge PA, Farr AG, Levin SD, Ziegler SF. Requirement for stat5 in thymic stromal lymphopoietin-mediated signal transduction. J. Immunol. 1999;163:5971–5977. [PubMed] [Google Scholar]
- 55.Van Parijs L, Refaeli Y, Lord JD, Nelson BH, Abbas AK, Baltimore D. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity. 1999;11:281–288. doi: 10.1016/s1074-7613(00)80103-x. [DOI] [PubMed] [Google Scholar]
- 56.Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 57.Feuerer M, Shen Y, Littman DR, Benoist C, Mathis D. How punctual ablation of regulatory T cells unleashes an autoimmune lesion within the pancreatic islets. Immunity. 2009;31:654–664. doi: 10.1016/j.immuni.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lahl K, Loddenkemper C, Drouin C, Freyer J, Arnason J, Eberl G, Hamann A, Wagner H, Huehn J, Sparwasser T. Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 2007;204:57–63. doi: 10.1084/jem.20061852. [DOI] [PMC free article] [PubMed] [Google Scholar]