Abstract
Telomere capping conceals chromosome ends from exonucleases and checkpoints, but the full range of capping mechanisms is not well defined. Telomeres have the potential to form G-quadruplex (G4) DNA, although evidence for telomere G4 DNA function in vivo is limited. In budding yeast, capping requires the Cdc13 protein and is lost at nonpermissive temperatures in cdc13-1 mutants. Here, we use several independent G4 DNA–stabilizing treatments to suppress cdc13-1 capping defects. These include overexpression of three different G4 DNA binding proteins, loss of the G4 DNA unwinding helicase Sgs1, or treatment with small molecule G4 DNA ligands. In vitro, we show that protein-bound G4 DNA at a 3′ overhang inhibits 5′→3′ resection of a paired strand by exonuclease I. These findings demonstrate that, at least in the absence of full natural capping, G4 DNA can play a positive role at telomeres in vivo.
Linear chromosome ends are capped with nucleoprotein complexes known as telomeres in order to avoid being recognized and processed as double-strand breaks (DSBs). Loss of capping, caused by telomere shortening or failure of capping protein function, can have detrimental consequences, including chromosome end-to-end fusions, cellular senescence or apoptosis1.
Most eukaryotic telomeres consist of tandem repeats in which one strand has runs of guanine bases and terminates as a 3′ single-stranded (ss) DNA overhang. These overhangs are bound by capping proteins, such as POT1 and CTC1 in mammals or Cdc13 in S. cerevisiae2. Cdc13, together with Stn1 and Ten1, forms the CST complex, which is essential for capping3,4. CST inhibits 5′→3′ exonuclease activity on the recessed cytosine-rich strand, prevents telomere termini from being fully recognized by checkpoint response factors and regulates access of telomerase, a reverse transcriptase that maintains telomere length3,5. Although much is known about the capping proteins that bind telomeres, the nucleoprotein structures adopted by telomeres to achieve a functionally capped state are less understood. In higher eukaryotes, structures known as t-loops, in which the 3′ single-stranded overhang invades duplex telomeric DNA, are thought to contribute to capping1. Because they have G-runs, telomere repeats could potentially also adopt G4 configurations, which might influence capping.
G4 DNA is a family of structures composed of Hoogsteen hydrogen-bonded guanines arranged in planar G-tetrads, which stack upon one another to form higher order quadruplex structures6. The structures may be intramolecular (that is, formed within one strand of DNA) or intermolecular, and are stabilized by centrally positioned monovalent cations such as Na+ and K+. Stable intramolecular G4 DNA requires four runs of at least two guanines with spacers of one or more nucleotides (nt); stability increases with longer guanine runs and/or shorter spacers7,8. Guanine-rich telomere repeat sequences, including the imperfect TG1–3 repeats of S. cerevisiae, can form G4 DNA at physiologic pH and salt concentrations in vitro9. In the protist Stylonychia lemnae, genetic knockdown of the telomere-binding protein TEBPβ, which itself catalyzes formation of G4 DNA in vitro, abolished anti-G4 DNA antibody-based detection of telomeres, demonstrating that G4 DNA forms naturally at these eukaryotic telomeres in vivo10. Very recently, the G4 DNA-binding activity of Est1 was linked to its stimulation of telomerase-mediated extension of telomeres in S. cerevisiae11. These findings indicate G4 DNA may form at telomeres, and they raise questions about the range of functions it might have there.
Here, we test potential roles for G4 DNA in telomere capping, using S. cerevisiae mutants deficient for Cdc13-dependent capping as a model system. Specifically, we use a temperature-sensitive mutant form of Cdc13, encoded by cdc13-1, which loses the ability to block exonucleolytic resection of the 5′ recessed telomere strand at elevated temperatures. This yields long tracts of guanine-rich ssDNA and activates G2/M checkpoint arrest3. Several 5′→3′ exonucleases contribute to ssDNA generation at telomeres. Among these, exonuclease I (Exo1) is the principal nuclease, although two more unidentified and weakly active nucleases contribute to resection—the so-called ExoX, which is stimulated by the Rad24-dependent DNA damage checkpoint clamp, and ExoY12. We reasoned that guanine-rich ssDNA exposed at cdc13-1 telomeres at elevated temperatures might form G4 DNA and that this setting would enable determination of whether telomere capping is rescued or further compromised by manipulations that stabilize or inhibit G4 DNA formation, respectively. Here, we describe several independent tests of the hypothesis that G4 DNA can contribute to telomere capping under these conditions. Our findings support the idea that telomere G4 DNA can play a positive role in telomere capping in vivo.
RESULTS
Overexpression of G4 DNA binding proteins rescues cdc13-1
It was previously reported that overexpression of the G4 DNA binding protein Stm1 rescues growth of cdc13-1 mutants at semi-permissive temperature (SPT)13. Stm1 was originally identified as a G4-binding protein and was later shown to associate in vivo with yeast telomeric and subtelomeric DNA as well as with polyribosomes14,15. Stm1 binds diverse G4 structures, including weak quadruplexes that possess runs of only two guanines (in other words, with only two G-quartets)16. We reproduced the rescue of cdc13-1 temperature-sensitive growth by overexpression of Stm1 (Fig. 1a). The rescue occurred in two distinct strain backgrounds, and we confirmed that Stm1 binds to telomeric G4 DNA in vitro and also promotes parallel G4 DNA conformations of several yeast telomere sequences (Supplementary Fig. 1). Stm1 was reported to interact with Cdc13 in a two-hybrid assay, raising the possibility that it might suppress cdc13-1 in an allele-specific fashion13. However, Stm1 overexpression also rescued the growth of cells with the stn1-154 mutant allele, which encodes a temperature-sensitive version of a different member of the CST capping complex, Stn1 (Fig. 1b). Therefore, Stm1 overexpression provides general rescue of growth in cells with CST-capping defects.
Figure 1.
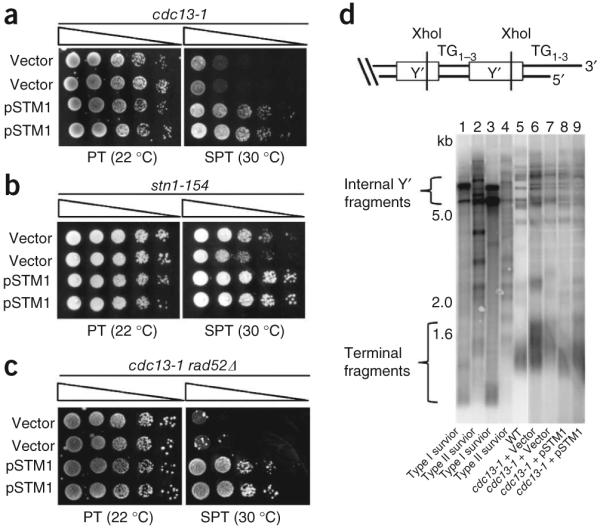
Overexpression of the G4 DNA binding protein Stm1 rescues growth defects caused by telomere uncapping and is independent of RAD52-dependent homologous recombination. (a) Growth of cdc13-1 mutants carrying pSTM1 or vector alone at permissive temperature (PT, 22 °C) or at semipermissive temperature (SPT, 30 °C). (b) pSTM1 overexpression rescues impaired growth caused by telomere uncapping in stn1-154 mutants at SPT. (c) Growth of cdc13-1 rad52Δ mutants carrying pSTM1 or vector. For each assay, serial dilutions of cells were spotted on selective medium and grown for 3 d (b,c) or 4 d (a). (d) Top: map of a typical telomere containing two tandem subtelomeric Y’ elements, separated by interstitial telomere repeats. Bottom: telomere Southern blots of samples grown in liquid culture at SPT for 2 d. Type I and type II survivors of telomerase inactivation are shown for comparison. The different sizes of internal Y’ fragments are due to short and long forms of Y’. Lanes 1–5 and 6–9 are sections from the same Southern blot.
RAD52-dependent homologous recombination allows rare cdc13-1 cells to bypass arrest at SPT, giving rise to telomeres that are longer and more heterogeneous in length than those of normal cells, similar to RAD52-dependent survivors of telomerase inactivation17. However, the high frequency of rescue by Stm1 suggests a homologous recombination–independent mechanism. Indeed, rescue was independent of RAD52 (Fig. 1c), and Southern blot analysis of the telomeres of rescued cells confirmed they were more similar to those of wild-type cells than to those of homologous recombination–dependent survivors of telomerase deletion (Fig. 1d).
To test if other G4 DNA–binding proteins rescue cdc13-1 growth at SPT, we overexpressed the Sgs1 RQC domain or the HF1 single-chain antibody (scFv). The Sgs1 helicase binds G4 DNA with high affinity (Kd = 5 nM) through the conserved RQC domain18,19. Whereas Sgs1 unwinds G4 DNA, expression of the RQC domain alone should bind, but not unwind, G4 DNA. Indeed, overexpression of the RQC domain (residues 1002–1212) rescued growth of cdc13-1 sgs1Δ cells at SPT (Fig. 2a). Rescue was not due to interference with endogenous Sgs1, because the cells were sgs1Δ. Moreover, chromatin immunoprecipitation (ChIP) confirmed that the RQC domain was enriched at the telomeres of cdc13-1 mutants (Fig. 2b), which also indicates that any residual Cdc13-1 protein bound at SPT20 does not prevent binding of the RQC domain. Next, we tested the HF1 scFv, which binds a range of G4 DNA structures but not duplex DNA21. As predicted, overexpression of HF1 rescued growth of cdc13-1 cells at SPT (Fig. 2c). Rescue was RAD52-independent, and HF1 bound tightly to G4 DNA formed by two different yeast telomere sequences in vitro (Kd < 75 nM) and promoted G4 DNA formation of a model yeast telomere sequence (Supplementary Fig. 2). As with RQC, we confirmed enrichment of HF1 at cdc13-1 telomeres by ChIP (Fig. 2d); thus, three proteins related only by their G4 DNA–binding capacities can assist with telomere capping when the CST complex is defective.
Figure 2.
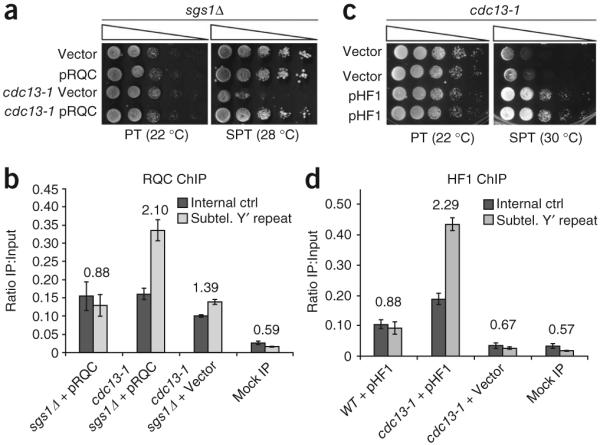
Expression of two additional, distinct G4 DNA binding proteins rescues the cdc13-1 temperature-sensitive phenotype. (a) The G4 DNA-binding Sgs1 RQC domain, overexpressed from a plasmid and under the control of the GAL1 promoter (pRQC), or vector control were transformed into the indicated strain backgrounds and tested in spot assays. (b) A 5× HA-tagged Sgs1 RQC domain is enriched at telomeres of cdc13-1 mutants. The primer sets used were directly adjacent to the telomeres (subtelomeric Y’ repeat) or a control, centromere-proximal primer set (to a portion of SWC4, which has no QFP). The telomeric/centromeric ratio is listed above bars. Error bars are ± 1 s.d., and essentially the same result was obtained in three independent ChIP experiments. Mock IP indicates that cdc13-1 sgs1Δ cells with the RQC-expressing vector were ChIPed without antibody. (c) GAL-induced overexpression of the G4 DNA binding single-chain antibody (scFv), HF1 or empty vector in cdc13-1 mutants. For each assay, serial dilutions of cells were spotted on selective medium containing 2% galactose and grown at the indicated temperatures for 4 d. (d) The 13xMyc-tagged HF1 scFv is enriched at telomeres of cdc13-1 mutants. DNA was amplified as described in (b). Error bars are ± 1 s.d., and essentially the same result was obtained in three independent ChIP experiments.
As an additional test, we treated cdc13-1 cells with G4 DNA–stabilizing compounds. Two structurally related G4 DNA binding bisquinolinium compounds, Bipy-DC6 and Phen-DC6 (recently shown to have activity on G4 DNA sequences in yeast22)23, improved growth of cdc13-1 cells at 30 °C relative to vehicle control, and did so in proportion to the capacity of the drugs to stabilize G4 DNA (Supplementary Fig. 3a,b). Similar results were obtained with a highly specific G4 ligand, N-methyl mesoporphyrin IX (NMM)24,25, whereas a related porphyrin, mesoporphyrin IX, with much less G4 DNA–binding activity26, or the double-stranded DNA–binding agent ethidium bromide, did not improve cell growth (Supplementary Fig. 3c–e); thus, only ligands selective for G4 DNA stimulated growth of cells lacking Cdc13-dependent telomere capping.
Inactivation of a G4 DNA unwinding helicase rescues cdc13-1
Because Sgs1 unwinds G4 DNA efficiently18, we tested if sgs1 deletion would rescue the cdc13-1 temperature-sensitive phenotype. In vivo, evidence for Sgs1 activity at G4 DNA includes the downregulation of gene expression in sgs1Δ mutants preferentially at loci with G-quadruplex-forming potential (QFP)27. Additionally, previous work has showed that Sgs1 overexpression abolishes rescue of the cdc13-1 temperature-sensitive phenotype by Stm1, suggesting the two might oppose one another to unwind or stabilize, respectively, telomeric G4 DNA13. Indeed, the temperature-sensitive phenotype of cdc13-1 rad24Δ rad52Δ mutants was rescued by sgs1 deletion (Fig. 3a). Also, single or combined deletions of rad24, rad52 and/or sgs1 in a CDC13 background did not alter growth (Supplementary Fig. 4a). Note that we were unable to observe rescue when RAD24 was intact (Supplementary Fig. 4b), suggesting that the exonucleolytic activity that is stimulated by Rad24 (the unidentified ‘ExoX’)12 is not inhibited by sgs1 deletion. As expected, Stm1 overexpression also rescued cdc13-1 rad24Δ rad52Δ growth at SPT, and Southern analysis confirmed that rescue by sgs1 deletion was not due to recombination-based survivor formation (Supplementary Fig. 4c–d).
Figure 3.
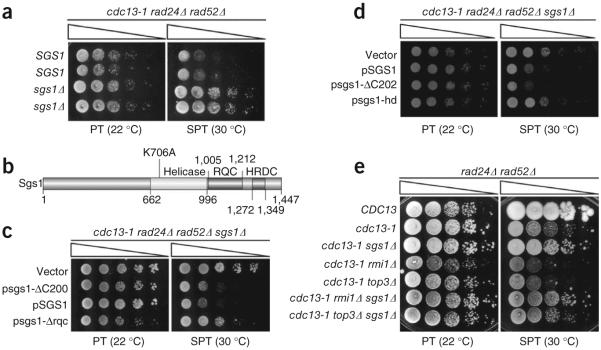
Loss of the SGS1 activities associated with G4 DNA binding and unwinding rescues cdc13-1 temperature sensitivity and is independent of RAD52-dependent homologous recombination. (a) Growth of cdc13-1 rad24Δ rad52Δ, either SGS1 or sgs1Δ, at permissive temperature versus SPT. (b) Map of specific Sgs1 domains mutated or deleted in this study. (c) Deletion of the G-quadruplex binding RQC domain alone is sufficient to rescue the cdc13-1 growth defect. (d) Loss of Sgs1 helicase activity (K706A point mutation, denoted sgs1-hd) is sufficient to rescue cdc13-1 temperature sensitivity. (e) Sgs1 is the sole component of the Sgs1–Top3–Rmi1 complex that, when lost, is sufficient to rescue the cdc13-1 temperature sensitive phenotype. For each assay, serial dilutions of cells were spotted on YPAD (a,e) or selective medium (c,d) and grown at the indicated temperatures for 3 d (a,c,d) or 4 d (e).
Sgs1 affects several cellular processes, including recombination, replication and S-phase checkpoint responses28-32, so we began to map subfunctions of Sgs1 that must be lost to rescue the cdc13-1 temperature-sensitive phenotype. Sgs1 derivatives (see map, Fig. 3b) were expressed from ARS-CEN plasmids driven by the SGS1 promoter (Supplementary Fig. 4e,f). Wild-type pSGS1 caused poor growth in cdc13-1 rad24Δ rad52Δ sgs1Δ quadruple mutants at SPT compared with vector control, as expected, as did constructs containing C-terminal truncation mutants missing the final 200 or 202 residues of Sgs1 (Fig. 3c–d and Supplementary Fig. 4f). These mutants lack S-phase checkpoint functions and also lack the HRDC domain, which binds ssDNA preferentially33-35; thus, rescue by sgs1 deletion is due neither to bypass of the Sgs1-dependent S-phase checkpoint nor to loss of the HRDC domain.
In contrast, mutant alleles specifically lacking only the RQC domain (residues 1005–1212) or possessing a K706A helicase dead point mutation36 did not cause sickness in quadruple mutants (Fig. 3c,d and Supplementary Fig. 4f). Each of these mutant proteins has shown activity in other assays (ref. 34 and Supplementary Fig. 4g), so it is evident they lost specific functions that promote cdc13-1 sickness; thus, the domains associated with binding and unwinding G4 DNA promote poor growth of cdc13-1 mutants at SPT and may therefore promote telomere uncapping in this setting.
Recent data suggest that Sgs1 stimulates 5′→3′ exonucleolytic resection at generic DNA DSBs28,30. Because end resection at telomeres leads to arrest in cdc13-1 mutants grown at SPT3, loss of Sgs-dependent generic end-resection activity might explain how sgs1 deletion rescues cdc13-1 growth. Generic end resection by Sgs1 depends on its cofactors Rmi1 and Top3 (ref. 28), so we tested whether deletion of RMI1 or TOP3 also rescued cdc13-1 rad24Δ rad52Δ growth at SPT. However, neither deletion improved growth (Fig. 3e), although deleting SGS1 in addition to RMI1 or TOP3 did improve growth, confirming that poor growth results from an Sgs1-specific function; thus, the roles of Sgs1 at uncapped telomeres and the Sgs1–Rmi1–Top3 complex at generic DSBs appear to be distinct.
Stm1 overexpression or sgs1 deletion restores capping
Capped telomeres inhibit exonucleolytic resection of the 5′ cytosinerich telomere strand and prevent checkpoint arrest due to accumulation of ssDNA3,37. To determine whether Stm1 overexpression or sgs1 deletion restores capping to cdc13-1 mutants, we assessed subtelomeric ssDNA generation and checkpoint activation at nonpermissive temperature (NPT, 37 °C).
To measure subtelomeric ssDNA generation at NPT, dot blots of native DNA were hybridized to a radiolabeled oligonucleotide capable of binding the guanine-rich strand at the Y’ subtelomeric element ~500 nt in from the telomere repeats, and signals were normalized to denatured loading controls. To assess ssDNA at a cell-cycle point during which exonucleases act upon uncapped telomeres and to normalize for any differences in growth rates among our mutants, we arrested cells in G2/M with nocodazole for 3 h before shifting them to NPT20. In cdc13-1 rad24Δ rad52Δ mutants, substantial amounts of subtelomeric ssDNA accumulated by 2–3 h after shift to NPT (Fig. 4a,b; and Supplementary Fig. 5a,b). In contrast, Stm1 overexpression or sgs1 deletion decreased ssDNA to levels seen in the CDC13 rad24Δ rad52Δ controls. Resection approached that of cdc13-1 exo1Δ double mutants, which is noteworthy because Exo1 is responsible for the majority of telomere resection in cdc13-1 cells at NPT, and when deleted, restores growth to cdc13-1 mutants at SPT12,37.
Figure 4.
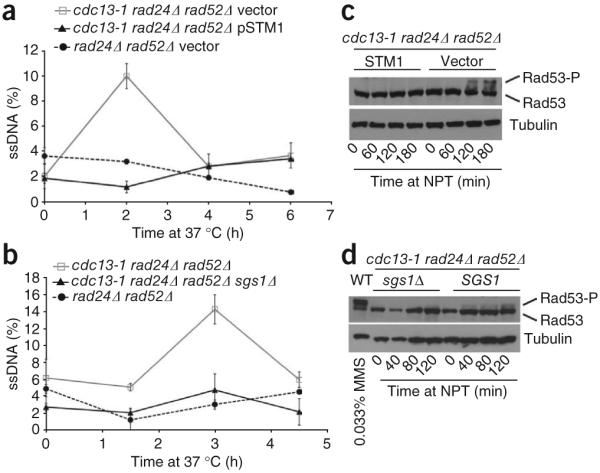
Telomere-proximal single-stranded (ss) DNA accumulation at NPT (37 °C) is attenuated by two G4 DNA-stabilizing mechanisms. (a,b) ssDNA measurements in the context of STM1 overexpression (a) or sgs1 deletion (b) in cdc13-1 rad24Δ rad52Δ backgrounds. Subtelomeric G-strand ssDNA was probed with a complementary end-labeled ssDNA probe against the Y’ element and quantified by normalizing hybridization signals of native to denatured samples. Each sample was spotted in triplicate, and standard errors are shown. Each graph is one representative experiment. The apparent difference in the timing of ssDNA accumulation between the experiments reflects the time points examined rather than true experimental variability. (c,d) Attenuation of Rad53 phosphorylation by Stm1 overexpression or sgs1 deletion in cdc13-1 rad24Δ rad52Δ cells incubated at NPT (37 °C). Treatment of wild-type cells with 0.033% methyl methanesulfonate (MMS) provides a positive control for Rad53 phosphorylation (4d, lane 1).
To assess the effect of Stm1 overexpression or sgs1 deletion on checkpoint activation, cell extracts were immunoblotted for Rad53. Uncapped telomeres activate a Rad53-dependent checkpoint pathway, as evidenced by phosphorylation and decreased mobility of Rad53 on SDS-PAGE. Stm1 overexpression or sgs1 deletion attenuated accumulation of phosphorylated Rad53 (Fig. 4c,d and Supplementary Fig. 5c–e). We therefore conclude that Stm1 and sgs1 deletion support telomere capping in cdc13-1 mutants, as evidenced by inhibited telomere resection and attenuated Rad53 phosphorylation.
Decreased telomere QFP attenuates cdc13-1 rescue by sgs1Δ
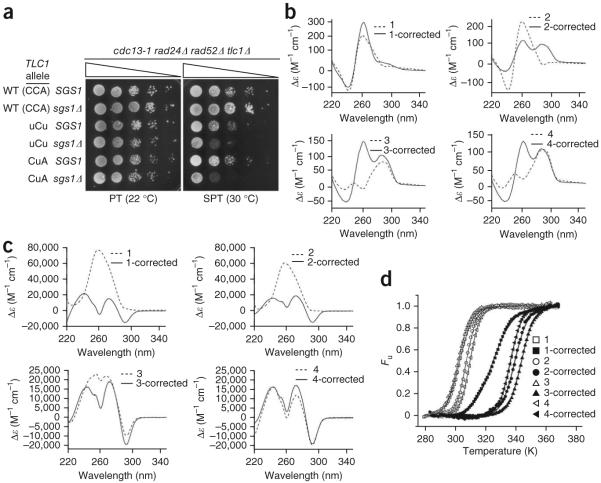
To determine if telomeric QFP (G-quadruplex forming potential) is important for rescue, it was reduced using mutant alleles of the TLC1 telomerase RNA template38. Mutations disrupting the CCC portion of the template (3′-CACACACCCACACCAC-5′) should decrease QFP by coding for fewer GGG triplets, which contribute more to highly stable G-quadruplexes than do GG doublets39. We compared a plasmid-based wild-type allele of TLC1 (pTLC1-CCA) to reduced QFP mutants previously shown to support nearly normal levels of growth: ptlc1-uCu and ptlc1-CuA38. The plasmids were transformed into cdc13-1 rad24Δ rad52Δ tlc1Δ strains, either SGS1+ or sgs1Δ, 45–50 generations after telomerase deletion, and individual colonies were then restreaked six times to allow for equilibration of telomere lengths. The absence of Rad52 ensured telomere maintenance by telomerase and not homologous recombination. All strains grew normally at permissive temperature, but the rescue at SPT afforded by sgs1 deletion was lost in strains carrying the mutant templates (Fig. 5a). Because the correspondence between a mutant TLC1 template and the encoded telomere sequence is not always straightforward, we cloned and sequenced examples of the chromosome (Chr.) I-L telomere38,40. The tlc1-uCu and tlc1-CuA mutants each encoded telomere sequences having the expected alterations and with apparently reduced QFP (see examples, Table 1). We used circular dichroism and UV-visible thermal difference spectroscopy (TDS)41,42 to test four different examples of mutant sequences (two examples each from cells expressing tlc1-uCu and tlc1-CuA) for the ability to form stable G4 DNA in comparison with their wild-type equivalents. TDS spectra were obtained by subtracting the molar extinction coefficients at low temperature (DNA folded) from those at high temperature (DNA unfolded). Various secondary DNA structures have specific TDS signatures that allow their identification (for example, G4 DNA structures, both parallel and antiparallel, yield positive peaks at 242 and 272 nm and a negative peak at 295 nm)42. Some of the mutant telomere sequences weakly formed G4 DNA or other secondary structures (Fig. 5b,c), whereas each of the wild-type equivalents formed stable G4 DNA (Fig. 5b,c; CD peaks at 263 nm and/or 295 nm43, and TDS peaks at 242, 273 and 295 nm41,42) with melting temperatures 21–42 degrees higher than those for the mutant sequences (Table 1 and Fig. 5d). Among the mutant sequences, oligonucleotide 4 showed the strongest propensity to form G4 DNA (based on TDS) and consistent with this, also had the highest Tm of the mutant sequences. Although not all telomere ends were examined, the mutant tlc1 templates should affect all telomeres. Moreover, because the loss of a single telomere is sufficient to elicit growth arrest44, the presence of reduced-QFP sequences even in a small subset of telomere ends should be sufficient to effect arrest due to loss of G4-based capping. Altogether, these data support the idea that rescue of the cdc13-1 temperature-sensitive phenotype by sgs1 deletion depends on the capacity of telomeres to form G4 DNA.
Figure 5.
Diminished rescue of cdc13-1 growth at SPT by sgs1 deletion in cells with mutant telomerase RNA templates that decrease QFP at telomeres. (a) cdc13-1 rad24Δ rad52Δ tlc1Δ cells, either SGS1 or sgs1Δ, and with plasmid-borne wild-type TLC1 or the uCu or CuA mutant tlc1 alleles, were spotted on SC-HIS medium and grown for 4 d. The sequence of the TLC1 template, including the mutated region (bold), is as follows: 3′-CACACACCCACACCAC-5′. (b) CD spectra of representative tlc1 mutant telomere sequences and their corrected counterparts (see Table 1). (c) Thermal difference spectra (TDS) of mutant versus corrected sequences, shown as the difference between molar extinction coefficients. G4 DNA yields a negative peak at 295 nm and positive peaks at 273 and 242 nm. All four corrected sequences showed evidence of G4 DNA formation by means of CD (parallel quadruplex for 1-corr and mixed parallel and antiparallel for 2, 3 and 4-corr) and TDS. (d) Thermal denaturation of oligonucleotides was followed by CD absorbance at 263 nm (for oligonucleotides 1, 1-corrected (corr), 2 and 2-corr) or 290 nm (for oligonucleotides 3, 3-corr, 4 and 4-corr).
Table 1. Mutant and corrected telomere sequences analyzed for G4 structure.
| Clone | Template mutation |
Sequence |
Tm, K (assuming ΔCp = 0) |
ΔTm (corrected – mutant) |
|---|---|---|---|---|
| 1 | uCu | GAG AGT GGT GTG TGT GAG AGT GGT GAG AGT GG | 302.6 ± 0.3 | 22.9 |
| 1-corrected | uCu | GGG TGT GGT GTG TGT GGG TGT GGT GGG TGT GG | 325.5 ± 0.8 | |
| 2 | uCu | GAG AGT GGT GAG AGT GTG AGA GTG GGT G | 307.2 ± 0.3 | 32.1 |
| 2-corrected | uCu | GGG TGT GGT GGG TGT GTG GGT GTG GGT G | 339.3 ± 0.3 | |
| 3 | CuA | GGA TGT GGA TGT GTG GAT GTG GAT GTG GA | 301.4± 0.8 | 42.0 |
| 3-corrected | CuA | GGG TGT GGG TGT GTG GGT GTG GGT GTG GG | 343.4 ± 0.4 | |
| 4 | CuA | GGA TGT GGT GTG TGG ATG TGG ATG TGG A | 309.0 ± 0.9 | 27.2 |
| 4-corrected | CuA | GGG TGT GGT GTG TGG GTG TGG GTG TGG G | 336.2 ± 0.3 |
The mutated bases are shown in bold font. Mutant sequences were obtained by cloning and sequencing the Chr. I-L telomere and the corrected sequences were generated by replacing the mutant bases with those that would have been encoded by the wild-type TLC1 template. Tm, melting temperature of each structure obtained from CD analyses. Cp, specific heat capacity at constant pressure.
Protection of a model telomere from Exo1 by G4 DNA
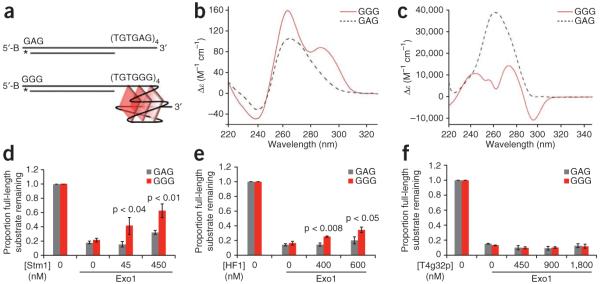
To test if G4 DNA and binding proteins can directly protect telomeres from exonucleolytic attack, we constructed a simplified in vitro system comprising model telomere substrates and purified Exo1 and DNA binding proteins (Stm1, HF1, T4 gene 32 protein). The model substrates had 24 nt 3′ overhangs that could (GGG = (TGTGGG)4) or could not (GAG = (TGTGAG)4) form G4 DNA structures at the ends of identical 51-base-pair (bp) non-telomere repeat duplex DNA (Fig. 6a). The 3′ terminal base at the end of the duplex opposite the overhang was radiolabeled, and the cognate 5′ base was biotinylated to prevent exonucleolytic attack at the non-telomeric end. Even though yeast telomere repeats are irregular, we note that the (TGTGGG)4 sequence exists naturally within these repeats, and other versions of the yeast telomere repeat can also form G4 DNA9 (and see Fig. 5). Exo1 was selected because it is the principal exonuclease that resects telomere ends in cdc13-1 mutants at NPT12,37. DNA substrates were incubated beforehand with 100 mM KCl at 30 °C to promote G4 DNA formation, as well as with DNA binding proteins where indicated. G4 DNA formation under these conditions was verified by CD and TDS spectroscopy (Fig. 6b,c). The GGG substrate formed a mixture of parallel and antiparallel G-quadruplexes42,43. In contrast, although the GAG control substrate yielded a CD signal similar to a parallel quadruplex (with positive and negative peaks at 263 and 240 nm, respectively), its TDS signature was clearly not that of G4 DNA but rather possibly that of GA homoduplex DNA, which has a positive peak at around 258 nm42 (Supplementary Fig. 6).
Figure 6.
G4 DNA-binding proteins and G4 DNA-forming sequences cooperate to inhibit end-resection by Exo1 in vitro. (a) Synthetic substrates having 3′ overhangs that form (bottom) or do not form (top) G-quadruplex DNA. Each contains 51 bp of identical duplex DNA. The 5′ end of each top strand was biotinylated (B) to inhibit resection from the incorrect end, and the terminal base of each bottom strand was labeled with α-32P-dCTP. We illustrate a parallel intramolecular quadruplex (bottom), although this may not be the actual or only structure formed by this overhang. (b) Analysis of overhang substrates by CD shows a mixture of parallel and anti-parallel G4 DNA (GGG overhang) and a possible GA homoduplex (GAG overhang) (see Supplementary Fig. 6) under the exact conditions used for the Exo1 assay. (c) Thermal difference spectra of GGG and GAG yield a G-quadruplex-specific signal for only the GGG overhang. (d,e) Both Stm1 (d) and HF1 (e) cooperate with G4 DNA overhang to partially rescue Exo1 5′→3′ resection. Quantification of the fraction of remaining full-length sequence; shown are the averages of two (d) or three (e) independent experiments with standard errors and P-values from unpaired two-tailed Student’s t-tests. (f) Quantification of protection by T4 gene 32 protein (T4g32p). An average of two experiments is shown. P > 0.7 for all concentrations of T4g32p.
Following G4 DNA formation, substrates were incubated with purified Exo1-FLAG and resection was assessed by the disappearance of the labeled bottom strand using denaturing PAGE and autoradiography (note that the FLAG tag did not inhibit Exo1 in vivo, Supplementary Fig. 7). The GGG and GAG substrates alone were degraded similarly, indicating that either G4 DNA alone is not sufficient to confer protection, or that the particular type of G4 DNA tested is not stable enough under these conditions to do so. However, both Stm1 and HF1, which each stabilize G4 DNA at the GGG overhang (Supplementary Figs. 1j,k and 2c,d), conferred dose-dependent and significant levels of protection of the GGG substrate versus the GAG overhang control (Fig. 6d,e and Supplementary Fig. 8a,b). Rescue was specific to these G4 DNA binding proteins, as the single-stranded DNA binding T4 gene 32 protein protected neither substrate, although it bound to both (Fig. 6f and Supplementary Fig. 8c,d); thus, mere binding of proteins to the overhang does not confer protection. Additionally, two control proteins, BSA and lysozyme, provided no protection (Supplementary Fig. 8e,f). Na+ and K+ can promote different G4 DNA conformations, so we tested protection by Stm1 in the presence of 100 mM Na+, rather than K+, and found that protection was lost (data not shown). CD analysis revealed substantially lower G4 DNA signal in the presence of Na+ versus K+ (Supplementary Fig. 8g–j), and we hypothesize that either K+-specific G4 DNA conformations or highly stable G4 DNA are required for protection. Additionally, because both Stm1 and HF1 promote stabilization of parallel G4 DNA structures (Supplementary Figs. 1 and 2), we hypothesize that end protection might be specific to parallel G4 DNA conformations bound by G4-binding factors. We emphasize that although G4 DNA forms on the guanine-rich strand, it enabled Stm1 and HF1 to protect the cytosine-rich strand, and we discuss possible mechanisms below. Altogether, these data suggest that G4 DNA in conjunction with G4-binding proteins is sufficient to protect the telomere cytosine-rich strand from degradation by Exo1 (Fig. 7).
Figure 7.
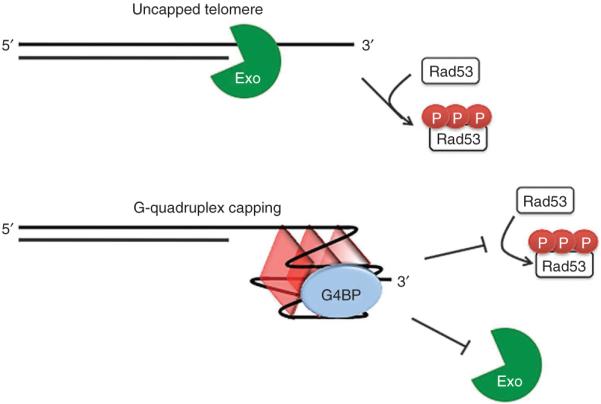
Model of telomere capping in which G4 DNA cooperates with G-quadruplex binding proteins (G4BP) to inhibit exonucleolytic degradation and checkpoint activation caused by loss of Cdc13 function.
DISCUSSION
We have shown that three different types of G4 DNA stabilizing treatments each enable bypass of telomere capping defects in yeast deficient for Cdc13. These include overexpression of several unique G4 DNA binding proteins, treatment with small-molecule G4 DNA ligands or inactivation of a G4 DNA unwinding helicase. Rescue was independent of homologous recombination and was associated with diminished resection of the cytosine-rich strand and a blunted check-point response. These data suggest G4 DNA can contribute to a rudimentary telomere cap, at least when natural capping is perturbed.
The growth defects in cdc13-1 mutants can be suppressed by the individual deletion of a large number of genes (37 strong and 201 weak suppressors45,46). Assessing the possible involvement of these genes in G4 DNA processing should be informative. For example, deletion of the Pif1 DNA helicase rescues cdc13-1 growth46 and unwinds G4 DNA in vitro and inhibits hyperrecombination associated with G4 DNA forming sequences in vivo47. In addition, the cgi121Δ mutant, which lacks a component of the KEOPS complex and which rescues cdc13-1 growth46, is also hypersensitive to growth inhibition by NMM27, consistent with a role for Cgi121 in modulating G4 DNA formation or function.
G4 DNA might facilitate telomere capping by inhibiting 5′→3′ exonucleolytic resection and by blunting checkpoint responses (Fig. 7). Exo1, the primary exonuclease active at uncapped yeast telomeres12,37, appears to be particularly affected by G4 DNA stabilizing treatments because we observed near wild-type levels of resection following Stm1 overexpression or sgs1 deletion, because rescue occurred in the absence of Rad24-mediated ExoX activity, and because Stm1 and HF1 cooperated selectively with a G-quadruplex-forming 3′ overhang to block resection by Exo1 in vitro. The formation of G4 DNA alone was unable to block resection in vitro, consistent with another recent study48; thus, G4 DNA might occlude access of exonucleases to the cytosine-rich strand, in part by providing a binding site for other factors. The contribution of G4 DNA to blunted checkpoint responses may be due not only to diminished end-resection but perhaps also to reduced binding of the exposed guanine-rich strand to checkpoint response factors such as RPA, which binds preferentially to the single-stranded conformation49. Consistent with these ideas, checkpoint responses in human cell nuclear extracts initiated by duplexes with 3′ overhangs were blocked when the overhang formed G4 DNA50. We thus propose that G4 DNA formation might contribute to the ‘anticheckpoint’ function of yeast telomere repeats that is observed when they are placed adjacent to DSBs51.
Given the cooperative role of the Sgs1 cofactors Rmi1 and Top3 in stimulating exonucleolytic resection at generic DSBs28, it is note-worthy that Sgs1 promoted the uncapping of telomeres in cdc13-1 mutants, whereas Top3 and Rmi1 did not. The function of Sgs1 at telomeres in cdc13-1 mutants is thus different from its function at generic DNA ends, consistent with a G4-related role. This interpretation is supported by our finding that rescue by sgs1 deletion was suppressed by mutations in the TLC1 template that encode telomere repeats with reduced QFP. Previous observations also support the idea that resection of telomeres involves different combinations of enzymatic activities than resection of other DNA ends. In particular, Sgs1 stimulates resection at DSBs by the Dna2 exonuclease but not by Exo128,30, whereas at native telomeres it stimulates resection by both exonucleases to generate the 3′ overhang52.
Our findings demonstrate a capping role for G4 DNA under conditions in which natural capping is impaired, but the extent to which G4 DNA is important for telomere capping under natural conditions is still unclear. One possibility is that telomeric G4 DNA may reflect a vestigial role for G-quadruplexes in telomere capping. In this case, G4 DNA-mediated capping might only be revealed in the absence of the primary mechanisms of telomere capping. However, G4 DNA-mediated capping might also exist in a natural setting as a complement to other mechanisms. The high degree of conservation of telomeric QFP, as well as the existence of proteins that interact with G4 DNA and have telomere roles—such as RecQ family helicases18, S. cerevisiae Rap1 (ref. 53) and Pif1 (ref. 47), and mammalian TRF2 (ref. 54)—suggest that this feature may be an important component of normal telomere function.
In mammalian cells, G-quadruplexes have been generally thought to interfere with telomere function. For example, telomeres were recently shown to be fragile sites, prone to breakage during DNA replication55. Telomere fragility is suppressed by the BLM and RTEL helicases, which are known or thought, respectively, to unwind G4 DNA; thus, G-quadruplexes may contribute to fragility18,56. Further, in human cancer cell lines, high concentrations of G4 DNA small-molecule ligands can displace capping proteins, such as TRF2 and POT1, from mammalian telomeres and activate checkpoint responses57,58. These findings have led to suggestions that G4 DNA negatively impacts telomeres, but this begs the question of why telomere repeats have not evolved toward sequences that do not form troublesome secondary structures. Our findings indicate that telomere G4 DNA can sometimes be of net benefit, a view supported by the recent demonstration that yeast Est1 may stimulate telomerase-mediated telomere extension by means of G4 DNA formation11, and consistent with the extension of telomeres in a G4-conformation by Tetrahymena thermophila telomerase59. Perhaps optimal capping requires the natural protein-based cap, which can be disrupted by high concentrations of G-quadruplex ligands, but when the natural cap is disrupted in some other way (for example, as in cdc13-1 mutants), G4 DNA may be of net benefit. Alternatively, G4 DNA might play some positive role under natural conditions, but when cells are treated with small molecule G4-ligands, this interferes with normal telomere G4 DNA functions (for example, by displacing G4-bound factors), or causes telomere G4 DNA to persist when it might be detrimental (for example, during DNA replication). Further investigation of both the positive and negative roles of G4 DNA at telomeres will likely provide new insight into telomere biology and offer new approaches to target telomeres for therapeutic benefit.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Johnson and Yatsunyk labs for helpful discussions and comments on the manuscript; N. Maizels, M. Fry, K. Runge, V. Zakian, J.-L. Mergny, S. Berger, R. Marmorstein and P. Adams for discussions; S. Murakami (Kanazawa University), E. Blackburn (University of California, San Francisco), M. Charbonneau (École Normale Supérieure), S. Brill (Rutgers University), S. Gasser (Friedrich Miescher Institute) and M. Van Dyke (MD Anderson Cancer Center) for providing strains and plasmids, and D. Durocher (University of Toronto) for the Rad53 antibody. This work was supported by US National Institutes of Health grants R01 AG021521 (F.B.J.), P01 AG031862 (F.B.J.), T32 GM008216-22 (J.S.S.) and T32 GM07229 (J.S.S.), a Camille and Henry Dreyfus Faculty Startup Award (L.A.Y.), Research Corporation Award no. 7843 (L.A.Y.), a Deutscher Akademischer Austausch Dienst Fellowship (R.K.) and by a Cancer Research UK program grant (S.B.).
Appendix
ONLINE METHODS
Yeast strains and plasmids
Unless indicated otherwise, strains were derived from the PSY316 background; see Supplementary Table 1 for complete list. Cells were cultured in YPAD or synthetic complete (SC) medium; temperature sensitive strains were propagated at room temperature (22 °C), whereas other strains were grown at 30 °C. pTLC1 template mutant, pSGS1 mutant and the YEpTSTM1 plasmids were kindly provided by L. Blackburn, S. Brill and S. Murakami, respectively. The psgs1-rqcΔ plasmid was generated from WT pSGS1 using two-round PCR to precisely delete amino acids 1005–1212. The Sgs1 RQC and HF1 overexpression constructs were generated by PCR amplification of the desired coding region with primers possessing an N-terminal SV40 NLS, followed by Gateway (Invitrogen) recombination-based cloning into a GAL1-driven ARS-CEN vector.
Spot assays
Spot assays were done using cultures growing in log phase. Beginning with 105 cell per spot, tenfold serial dilutions were spotted onto YPAD, or the relevant selective SC medium, and grown at 22 °C or at 28–30 °C for the indicated number of days.
Single-stranded DNA quantification
Log phase cells growing at 22 °C were arrested at G2/M by treatment with 20 μg ml−1 nocodazole for 3 h, diluted 1:3 with medium prewarmed to 55 °C and shifted to NPT (37 °C) with continued nocodazole arrest. Cells were collected at the indicated time points and DNA was isolated. Detection and quantification of single-stranded DNA was done as follows. Undigested native DNA or DNA denatured in 0.2 M NaOH at 65 °C was diluted to a final volume of 300 μl with 10× SCP buffer (1 M NaCl, 0.3 M Na2HPO4, 0.02 M EDTA, pH 6.8, with HCl) and then spotted and drawn through a Hybond-XL membrane (GE Healthcare) by application of a vacuum, followed by three washes with tetramethylammonium chloride (TMAC) wash buffer (3 M TMAC, 50 mM Tris-HCl, 0.1 mM EDTA, pH 8.0). DNA was then cross-linked to the membrane using a 254-nm Stratalinker (12 μJ × 100), and the membrane was prehybridized at 55 °C in buffer containing 6× SSC, 5× Denhardt’s solution, 20 mM sodium phosphate, pH 7.0, and 25 μg ml−1 boiled salmon sperm DNA. Blots were washed four times for 5 min each with TMAC hybridization buffer (3M TMAC, 5× Denhardt’s solution, 50 mM Tris-HCl, pH 8.0, 10 μg ml−1 salmon sperm DNA, 0.1 mM EDTA, pH 8.0, 0.6% (w/v) lithium dodecylsulfate). An oligonucleotide probe specific for Y’ subtelomeric DNA was end-labeled with γ-32P-ATP using T4 polynucleotide kinase and hybridized overnight to the membrane in TMAC hybridization buffer at 65 °C. Membranes were washed in TMAC wash buffer, then imaged and quantified with a Molecular Dynamics Phosphorimager and ImageQuant analysis software.
In vitro exonuclease assays
Partial duplex telomeric substrates were formed using 35 pmol each of HPLC-purified top (GGG: 5′-BIO/GTACTTATTTCGGAATGAGCTTCATAAGATAATGGTCAGATCAATATGAGA(TGTGGG)4-3′; GAG: 5′-BIO/GTACTTATTTCGGAATGAGCTTCATAAGATAATGGTCAGATCAATATGAGA(TGTGAG)4-3′), where “BIO” indicates biotinylation of the 5′ end, and bottom oligonucleotide (5′-TCTCATATTGATCTGACCATTATCTTATGAAGCTCATTCCGAAATAAGTA-3′) diluted in 15 μl TEM buffer (10 mM Tris 8.0, 1 mM EDTA, 10 mM MgCl2) denatured at 94 °C for 5 min and annealed by shifting to 79 °C and decreasing temperature at 1 °C min−1 to 55 °C, followed by 55 °C for 3 h. Duplex substrates were then end-labeled by mixing 7.5 μl duplex, 2 μl 10× Klenow reaction buffer, 9 μl H2O, 1 μl (5 U) Klenow polymerase (3′→5′ exo−) and 0.5 μl α-32P-dCTP and incubating for 30 min at 30 °C. Unincorporated dCTP was removed by gel filtration, and samples were diluted to a final volume of 80 μl. To promote G-quadruplex formation, 220 fmol (1 μl) of duplex substrate was incubated for 20 min at 30 °C in a final volume of 15 μl of Exo1 buffer (100 mM KCl, 25mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1 mM DTT, 4% (v/v) glycerol and 100 μg ml−1 BSA) containing the indicated amounts of protein. Exonuclease digestion was done by addition of 3 pmol of Exo1 in 10 μl of Exo1 buffer and incubating at room temperature (21 °C) for 20 min. Reactions were stopped by addition of 5 μl 80% (v/v) formamide, 0.1% (w/v) bromophenol blue, 0.1% (w/v) xylene cyanol, boiled for 8 min and separated on 8 M urea, 8% (w/v) acrylamide gels run in 0.5× TBE. Phosphorimager images of gels were quantified using ImageJ analysis software.
Immunoblots
Proteins were isolated using mechanical disruption of cells in the presence of 20% TCA. For Rad53 immunoblots, log phase cultures grown at room temperature (21 °C) were diluted 1:3 with medium prewarmed to 55 °C at the start of the 37 °C time course, and samples were collected at the specified time intervals. Proteins were run on 4%–12% (w/v) SDS-PAGE gradient gels and transferred to PVDF or nitrocellulose membrane. Blots were probed with the following antibodies: 1:1,000 anti-Rad53 (generously provided by D. Durocher), 1:150 anti-Rad53 (yC-19) (Santa Cruz sc-6749), 1:200 anti-HA-tag (Y-11) (Santa Cruz sc805), 1:1,000 anti–α-tubulin [YL1/2] (Abcam ab6160) or 1:1,000 anti-actin (I-19) (Santa Cruz sc-1616), followed by 1:5,000 anti-goat–HRP secondary (Santa Cruz sc2020), 1:2,500 anti-rat–HRP (Jackson ImmunoResearch, 112-035-167) or 1:1,000 anti-rabbit secondary (GE Healthcare, NA9340) antibody.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was done as described60. Lysate was sonicated with a Bioruptor (30 s on/30 s off for 20 min), and immunoprecipitations were done on 0.4 mg protein with an HA tag antibody (Abcam cat. no. ab9110) or a Myc tag antibody (Abcam cat. no. ab9132). Six washes were done (2 × FA-Lysis buffer, 1 × FA-Lysis buffer/500 mM NaCl. Quantitative PCR was done using the Sybr Green Jumpstart Taq ReadyMix (Sigma) on a Roche LightCycler 480.
Circular dichroism
CD spectra (Figs. 5 and 6) were collected using an AVIV 410DS spectrometer equipped with a Peltier heating unit with the following settings: path length, 0.2–1 cm; windows, 220 nm–350 nm; temperature, 25 °C; averaging time, 1 s; and total of 5 scans. To determine molar ellipticity, UV-visible data were collected using a Cary300 Varian UV-visible spectrophotometer. Data were analyzed using Origin analysis software. For Fig. 5, oligonucleotides were prepared by heating 25 mM oligonucleotide in 10 mM K2HPO4 (pH 7), 100 mM KCl to 100 °C for 3 min, transferring to 75 °C and slowly cooling to 30 °C overnight, followed by dilution in the same buffer to 5 mM for CD studies.
Additional and more highly detailed methods are described in Supplementary Methods.
- 60.Kozak ML, et al. Inactivation of the Sas2 histone acetyltransferase delays senescence driven by telomere dysfunction. EMBO J. 2010;29:158–170. doi: 10.1038/emboj.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit. Rev. Biochem. Mol. Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 6.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Risitano A, Fox KR. Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- 9.Venczel EA, Sen D. Parallel and antiparallel G-DNA structures from a complex telomeric sequence. Biochemistry. 1993;32:6220–6228. doi: 10.1021/bi00075a015. [DOI] [PubMed] [Google Scholar]
- 10.Paeschke K, Simonson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 11.Zhang ML, et al. Yeast telomerase subunit Est1p has guanine quadruplex-promoting activity that is required for telomere elongation. Nat. Struct. Mol. Biol. 2010;17:202–209. doi: 10.1038/nsmb.1760. [DOI] [PubMed] [Google Scholar]
- 12.Zubko MK, Guillard S, Lydall D. Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13–1 mutants. Genetics. 2004;168:103–115. doi: 10.1534/genetics.104.027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi N, Murakami S. STM1, a gene which encodes a guanine quadruplex binding protein, interacts with CDC13 in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2002;267:806–813. doi: 10.1007/s00438-002-0712-3. [DOI] [PubMed] [Google Scholar]
- 14.Frantz JD, Gilbert W. A yeast gene product, G4p2, with a specific affinity for quadruplex nucleic acids. J. Biol. Chem. 1995;270:9413–9419. doi: 10.1074/jbc.270.16.9413. [DOI] [PubMed] [Google Scholar]
- 15.Van Dyke MW, Nelson LD, Weilbaecher RG, Mehta DV. Stm1p, a G4 quadruplex and purine motif triplex nucleic acid-binding protein, interacts with ribosomes and subtelomeric Y’ DNA in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:24323–24333. doi: 10.1074/jbc.M401981200. [DOI] [PubMed] [Google Scholar]
- 16.Nelson LD, Musso M, Van Dyke MW. The yeast STM1 gene encodes a purine motif triple helical DNA-binding protein. J. Biol. Chem. 2000;275:5573–5581. doi: 10.1074/jbc.275.8.5573. [DOI] [PubMed] [Google Scholar]
- 17.Zubko MK, Lydall D. Linear chromosome maintenance in the absence of essential telomere-capping proteins. Nat. Cell Biol. 2006;8:734–740. doi: 10.1038/ncb1428. [DOI] [PubMed] [Google Scholar]
- 18.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber MD, Duquette ML, Shiels JC, Maizels NA. Conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 2006;358:1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 20.Vodenicharov MD, Wellinger RJ. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell. 2006;24:127–137. doi: 10.1016/j.molcel.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 21.Fernando H, et al. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009;37:6716–6722. doi: 10.1093/nar/gkp740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piazza A, et al. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 2010;38:4337–4348. doi: 10.1093/nar/gkq136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Cian A, DeLemos E, Mergny J-L, Teulade-Fichou M-P, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 24.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 25.Smith JS, Johnson FB. Isolation of G-quadruplex DNA using NMM-sepharose affinity chromatography. Methods Mol. Biol. 2010;608:207–221. doi: 10.1007/978-1-59745-363-9_13. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Geyer R, Sen D. Recognition of anionic porphyrins by DNA aptamers. Biochemistry. 1996;35:6911–6922. doi: 10.1021/bi960038h. [DOI] [PubMed] [Google Scholar]
- 27.Hershman SG, et al. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Z, Chung W-H, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom’s and Werner’s Syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frei C, Gasser S. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 32.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein DA, Keck JL. Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure. 2005;13:1173–1182. doi: 10.1016/j.str.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 34.Mullen JR, Kaliraman V, Brill SJ. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000;154:1101–1114. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, et al. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure. 1999;7:1557–1566. doi: 10.1016/s0969-2126(00)88346-x. [DOI] [PubMed] [Google Scholar]
- 36.Lu J, et al. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 37.Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin J, Smith DL, Blackburn EH. Mutant telomere sequences lead to impaired chromosome separation and a unique checkpoint response. Mol. Biol. Cell. 2004;15:1623–1634. doi: 10.1091/mbc.E03-10-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Förstemann K, Hoss M, Lingner J. Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res. 2000;28:2690–2694. doi: 10.1093/nar/28.14.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saccà B, Lacroix L, Mergny J-L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mergny J-L, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 45.Addinall SG, et al. A genomewide suppressor and enhancer analysis of cdc13–1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics. 2008;180:2251–2266. doi: 10.1534/genetics.108.092577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Downey M, et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell. 2006;124:1155–1168. doi: 10.1016/j.cell.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 47.Ribeyre C, et al. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallur AC, Maizels N. Distinct activities of exonuclease 1 and flap endonuclease 1 at telomeric G4 DNA. PLoS ONE. 2010;5:e8908. doi: 10.1371/journal.pone.0008908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salas TR, et al. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai Y-C, Qi H, Liu LF. Protection of DNA ends by telomeric 3′ G-tail sequences. J. Biol. Chem. 2007;282:18786–18792. doi: 10.1074/jbc.M702171200. [DOI] [PubMed] [Google Scholar]
- 51.Michelson RJ, Rosenstein S, Weinert T. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 2005;19:2546–2559. doi: 10.1101/gad.1293805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol. Cell. 2009;35:70–81. doi: 10.1016/j.molcel.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 53.Giraldo R, Rhodes D. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 1994;13:2411–2420. doi: 10.1002/j.1460-2075.1994.tb06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedroso IM, Hayward W, Fletcher TM. The effect of the TRF2 N-terminal and TRFH regions on telomeric G-quadruplex structures. Nucleic Acids Res. 2009;37:1541–1554. doi: 10.1093/nar/gkn1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding H, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 57.Gomez D, et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 58.Phatak P, et al. Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Br. J. Cancer. 2007;96:1223–1233. doi: 10.1038/sj.bjc.6603691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oganesian L, Graham ME, Robinson PJ, Bryan TM. Telomerase recognizes G-quadruplex and linear DNA as distinct substrates. Biochemistry. 2007;46:11279–11290. doi: 10.1021/bi700993q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.