Abstract
Aims/hypothesis
Rare mutations in the gene (HNF4A) encoding the transcription factor HNF-4A account for ~5% of cases of maturity-onset diabetes of the young (MODY) and more frequent variants in this gene may be involved in multifactorial forms of diabetes. Two low frequency, non-synonymous variants in HNF4A (V255M, minor allele frequency [MAF] ~0.1%, T130I, MAF ~3.0%), known to influence downstream HNF-4A target gene expression, are of interest but previous type 2 diabetes association reports were inconclusive. We aimed to evaluate the contribution of these variants to type 2 diabetes susceptibility through large-scale association analysis.
Methods
We genotyped both variants in at least 5745 cases and 14756 population controls from the UK and Denmark. We also undertook an expanded association-analysis including previously reported and novel genotype data obtained in Danish, Finnish, Canadian and Swedish samples. A meta-analysis incorporating all published association studies of the T130I variant was subsequently carried out in a maximum sample size of 14279 cases and 26835 controls.
Results
We found no association between V255M and type 2 diabetes in either the initial (p=0.28) or expanded analysis (p=0.44). However, T130I demonstrated a modest association with type 2 diabetes in the UK and Danish samples (additive per allele OR 1.17 [1.08-1.28]; p=1.5×10−4), which was strengthened in the meta-analysis (OR 1.20 [1.10-1.30]; p=2.1×10−5).
Conclusions/interpretation
Our data are consistent with T130I as a low frequency variant influencing type 2 diabetes risk, but are not conclusive when judged against stringent standards for genome-wide significance. This study exemplifies the difficulties encountered in association testing of low frequency variants.
Keywords: Type 2 Diabetes, HNF4A, Low frequency variants, T130I, V255M
Introduction
The heritability seen in type 2 diabetes remains largely unexplained, despite substantial progress in identifying genetic variants conferring increased risk of this condition. To date, ~40 such variants have been identified, largely through genome-wide association studies (GWAS) [1-8]. However, the sibling relative risk (λS) of type 2 diabetes conferred by all these variants combined is ~1.15, well below the epidemiological estimate (~3.0) [8, 9]. Of late there has been great interest in the potential role of low frequency (LF) variants in terms of individual susceptibility to complex diseases such as type 2 diabetes. Since GWAS to date have focused on the detection of common variant associations, the contribution to type 2 diabetes risk of variants with a minor allele frequency (MAF) below 5% remains largely unexplored.
A logical place to initiate the search for LF variants influencing multifactorial type 2 diabetes lies in exploring those genes already implicated in diabetes pathogenesis because they contain either rare mutations causal for monogenic forms of diabetes, or common variants associated with multifactorial type 2 diabetes. In genes implicated due to their role in monogenic diabetes, there is evidence that large-effect mutations are compatible with life, and that they result in a phenotype with substantial similarities (and clinical overlap) to type 2 diabetes. It is likely therefore that variants with less dramatic effects on function and/or expression, where they exist, result in less extreme clinical phenotypes including multifactorial type 2 diabetes. Since existing GWAS and linkage study data argue against the possibility of common variants (MAF>5%) of medium- to large-effect size, variants with such effect sizes are also likely to be rare (MAF <0.1%) or of low frequency (MAF 0.1-5%). Low frequency variants have been implicated in the pathogenesis of other complex diseases, such as type 1 diabetes [10], although their contribution to type 2 diabetes predisposition is as yet uncertain. Rare, highly-penetrant mutations in the gene (HNF4A) encoding the transcription factor hepatocyte nuclear factor 4 alpha (HNF-4A) account for approximately 5% of cases of maturity onset diabetes of the young (MODY) [11]. Though HNF4A is expressed in multiple tissues, its expression in the pancreatic beta cells and liver is of particular interest. In pancreatic beta cells, HNF-4A is required for glucose metabolism as well as normal insulin gene expression and secretion [12]. In the liver, HNF4-A is required for hepatic gluconeogenesis [13]. Several studies have shown linkage between multifactorial type 2 diabetes and the region of chr20q where HNF4A is located [14-17]. Previous candidate gene analyses have demonstrated weak evidence of association (p~0.01) between common variants in the P1 and P2 promoters of HNF4A and multifactorial type 2 diabetes [17, 18], but these have not been substantiated in genome-wide association studies to date [1-5, 8]. Since common variants in HNF4A do not explain the findings of linkage studies, it is possible that this region harbours more penetrant low frequency variants that might explain this observation [19].
HNF4A has been extensively resequenced not least as part of clinical diagnostic screening for MODY. These resequencing efforts have, inter alia, identified two LF coding non-synonymous variants of particular interest: V255M (c.763G>A p.Val255Met) and T130I (c.389C>T p.Thr130Ile, rs1800961). V255M was first described following resequencing of Danish samples but no evidence of association to type 2 diabetes was seen in analysis of 1434 cases and 4790 controls [20]. T130I, positioned in the DNA binding domain of HNF4A, showed modest (p=0.04) association with type 2 diabetes in the same sample [20], though subsequent efforts at replication failed to confirm this [21]. One arm of a meta-analysis of the association of HNF4A genetic variants to type 2 diabetes [22] also included some previous association studies of T130I (by our estimation including approximately 3500 cases and 3700 controls for this variant), and demonstrated a modest association (p=0.045) [22]. Most recently, and of particular interest, given the relationship between lipids and type 2 diabetes, a significant association between T130I and HDL-cholesterol levels has been demonstrated (p=8×10−10) in a GWAS meta-analysis incorporating 30714 individuals [23].
Both variants have been shown to be functional based on studies of the transcriptional regulation of HNF-4A target genes in a range of cell lines and primary mouse hepatocytes [20, 24-26]. We therefore reasoned that they remain interesting candidates for assessment in larger samples to more clearly establish their likely contribution to type 2 diabetes susceptibility.
Methods
Subjects studied
Three categories of samples were included. Category 1 consisted of samples specifically genotyped for this study. Category 2 was comprised of samples with previously reported genotyping information for these SNPs. Category 3 included samples for which only summary statistics were available from previous published reports.
Category 1 samples were derived from three sources (two UK samples and one Danish sample). UK Sample 1 (“UK1”, n=4124 cases, 5126 controls) included the United Kingdom Type 2 Diabetes Genetics Consortium (UKT2DGC) collection recruited in Tayside, Scotland: these have been previously described [1, 27]. UK Sample 2 (“UK2”) comprised type 2 diabetes cases (n=1853 for V255M; 1193 for T130I) ascertained from a subset of the Diabetes UK Warren 2 repository [28]. The controls for UK2 were taken from the population-based British 1958 Birth Cohort (n=7133), and the United Kingdom Blood Service collection (n=3087) [27].
Danish Sample 1 (“DK1”, n=2646 cases) was also included in category 1 for the study of T130I. DK1 represents samples collected in the Steno Diabetes centre and Danish samples from the Anglo-Danish-Dutch study of Intensive Treatment in People with Screen-Detected Diabetes in Primary Care (ADDITION) [20, 29]. The new samples in DK1 were combined with the previously reported case and control data from DK2 (described below) to generate a combined DK analysis involving 3771 cases and 4727 controls.
Category 2 included samples from Denmark, Sweden, Finland and Canada. Danish Sample 2 (“DK2”, n=1397 cases, 4865 controls) was previously genotyped for T130I and V255M [20]. Two samples from the Finland-United States Investigation of Non Insulin Dependent Diabetes Mellitus Genetics (FUSION) study were included for T130I (FUSION Sample 1, “FS1”, [n=1160 cases, 1173 controls] and FUSION Sample 2, “FS2”, [n=1211 cases, 1264 controls]) [4]. FS1 and FS2 represent the FUSION GWAS and replication samples respectively and have been included in a type 2 diabetes [2] and a lipid GWAS [23] and subsequent follow up of significant findings. Due to DNA availability, the withdrawal of some individuals, and updated type 2 diabetes status of others, the numbers of individuals quoted for FS1 and FS2 vary slightly to those in the reference article. The recruitment criteria for these samples have been previously reported [4]. We also included samples from The Metabolic Syndrome in Men (METSIM) study, (“MS1”, n=801 cases, 3043 controls) recruited in Finland [30]. The T130I genotype data for MS1 were included as part of the lipid GWAS follow up [23] though type 2 diabetes data have not been published. Previously reported genotyping results for T130I from three samples from the Broad institute were also included [21]. These comprised of a Canadian sample (Broad Sample 1, “BR1”, n=127 cases, 127 controls), a combined Swedish, Finnish sample (Broad Sample 2, “BR2”, n=490 cases, 490 controls) and a Swedish sample (Broad sample 3, “BR3”, n=514 cases, 514 controls).
All studies were approved by local ethics committees and were performed in accordance with the principles of the Helsinki Declaration II. Informed consent was obtained from all subjects before participation. Detailed descriptions of category 1 and 2 samples are included in the supplementary information. All subject characteristics are summarised in Table 1.
Table 1.
Study Subject Characteristics
| Sample name | Short form | Sample name in previous publicationsA |
Subjects | % Male | Mean AgeB (years) ± SD |
Mean BMI (kg/m2) ± SD |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | |||
| UK Sample 1 | UK1 | UKT2DGC Sample | 5126 | 4124 | 51 | 55 | 60 ± 13 | 58 ± 12 | 27 ± 5 | 31 ± 6 |
| UK Sample 2 | UK2 | Diabetes UK Warren 2 repository, British 1958 birth cohort, UK blood service collection of common controls |
10220 | 1853 | 50 | 61 | 42 ± 7 | 52 ± 7 | 27 ± 6C | 32 ± 7 |
| Danish Sample 1 | DK1 | Inter99, SDC1, SDC2 |
- | 2646 | - | 59 | - | 57 ± 10 | - | 31 ± 6 |
| Danish Sample 2 | DK2 | SDC2 and ADDITION |
4865 | 1397 | 47 | 60 | 46 ± 9 | 52 ± 10 | 26 ± 4 | 30 ± 5 |
| FUSION sample 1 | FS1 | Stage 1 genotyping Sample for DIAGRAM meta- analysis |
1173 | 1160 | 49 | 56 | 64 ± 7 | 54 ± 9 | 27 ± 4 | 30 ± 5 |
| FUSION Sample 2 | FS2 | Stage 2 genotyping Sample for DIAGRAM meta- analysis |
1264 | 1211 | 61 | 60 | 59 ± 8 | 55 ± 9 | 27 ± 4 | 31 ± 5 |
| METSIM | MS1 | METSIM | 3043 | 801 | 100 | 100 | 59 ± 6 | 58 ± 7 | 26 ± 4 | 30 ± 5 |
| Broad Sample 1 | BR1 | Canada Sample | 127 | 127 | 55 | 55 | 52 ± 8 | 52 ± 8D | 29 ± 4 | 29 ± 5 |
| Broad Sample 2 | BR2 | Scandinavia Sample | 490 | 490 | 52 | 52 | 60 ± 10 | 60 ± 10D | 27 ± 4 | 28 ± 5 |
| Broad Sample 3 | BR3 | Sweden Sample | 514 | 514 | 52 | 52 | 66 ± 12 | 66 ± 12D | 28 ± 4 | 28 ± 4 |
Full details available in Supplementary information
Mean age refers to age at recruitment for controls and age at diagnosis for cases
Based on the British 1958 Birth Cohort (n=7133) and Panel 2 of the United Kingdom Blood Services Collection (n=1643)
Based on age at recruitment (age of diagnosis not available)
For a more complete study of T130I, the results from these category 1 and 2 samples were included in a meta-analysis together with those from all previously published studies (category 3) for which summary statistics were available. For these category 3 samples we had no access to genotype information. These included a Pima Indian sample (“PI1”, n=573 cases, 464 controls) [31] and a Japanese sample (“JP1”, n=423 cases, 354 controls) [24] included in the meta-analysis by Sookoian et al [22] in addition to a Mexican sample (“MX1”, n=100 cases, 75 controls) [32].
Genotyping and Quality Control
Genotyping of T130I in the UK samples was carried out using a Taqman assay on the ABI 7900HT platform (Applied Biosystems, Warrington, Cheshire, UK). A KBioscience allele specific PCR (KASPar) assay (KBioscience, Hoddesdon, U.K.) was utilised in the genotyping of V255M in the UK samples and T130I in DK1. Quality of the genotyping was assured by (a) assessing the genotyping pass rate (>96% globally), (b) evaluating the estimated error rate based on completed duplicate pairs (UK samples: 0.00% for V255M, n=314 duplicate pairs and 0.18% for T130I, 268 duplicate pairs; DK1: 0.20% based on 521 duplicate samples), and (c) assessing for departure (p<0.05) from Hardy Weinberg Equilibrium (none detected). Genotyping methods and quality control measures for DK2 [20], Broad samples [21], FUSION samples [2, 23] and MS1 [23] have been previously reported.
Statistical analysis
No heterogeneity of genotype counts was seen between category 1 cases when assessed by an exact Pearson chi-square test using StatXact (v6.0: Cytel software corps, Cambridge, MA, USA). The same was true of controls. We subsequently carried out a primary association analysis of category 1 samples (UK1, UK2, DK1 and DK2, for T130I, UK1 and UK2 for V255M) (each as a separate stratum) followed by a secondary association analysis including category 1 and category 2 samples (UK1, UK2, DK1, DK2, FS1, FS2, MS1, BR1, BR2, and BR3 for T130I and UK1, UK2 and DK2 for V255M) as separate strata. We used an exact Cochran-Armitage trend test (StatXact v6.0) for all association analyses in this report. The ORs and sample sizes for each stratum from this study were subsequently used in a meta-analysis of T130I incorporating the previously defined category 3 samples [22, 32] utilising an additive model using GWAMA (Genome Wide Association Meta-Analysis) software package (http://www.well.ox.ac.uk/gwama). Though these samples are geographically disparate, a low level of heterogeneity of the T130I association effect sizes was detected using GWAMA (I2= 61%; Q statistic P value= 0.10; quantified by the comparison of the samples in category 1 and 2 to category 3 samples).
Power calculations derived from Quanto [33], using the previously reported OR for T130I [20], indicated 99% power to detect an effect size of 1.3 (for α=0.001) for this variant in our expanded association analysis incorporating UK, Danish, FUSION, METSIM and Broad samples. Power for V255M is less (12% power to detect the same effect size [for α=0.001]) due to the much lower MAF. For this variant, we had 80% power to detect an effect size of 3.0 (for α=0.001).
Results
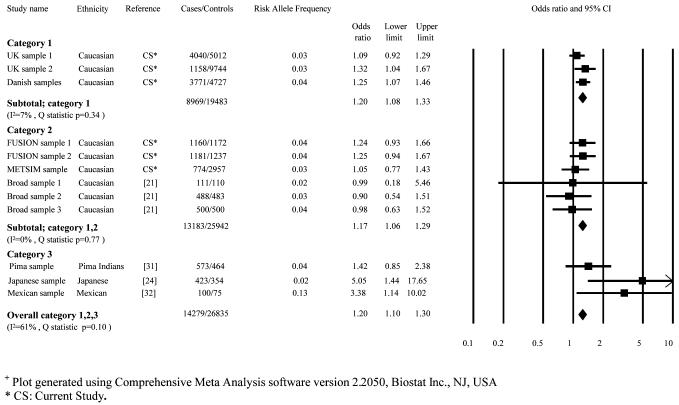
T130I was successfully genotyped in 7645 cases and 14756 controls in category 1. This variant had an MAF of 3.76% in cases and 3.00% in controls. In category 1 samples, a modest association with type 2 diabetes was conferred by the T allele of this variant (additive per allele OR 1.20 [1.08-1.33]; p=5×10−4). The expanded association analysis incorporating the category 1 and 2 samples marginally increased the strength of this association (OR 1.17 [1.08-1.28]; p=1.5×10−4) (Table 2). The meta-analysis (Figure 1) incorporating all available studies of T130I further increased the strength of this association (n=14279 cases, 26835 controls; OR 1.20 [1.10-1.30]; p=2.1×10−5). There was no evidence for heterogeneity in our meta-analysis (I2= 61%; Q statistic P value= 0.10).
Table 2.
Association testing of the T130I variant of HNF4A
| Study sample | N | Cases: Genotype counts (Genotype frequencies) |
Controls: Genotype counts (Genotype frequencies) |
Statistical Analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases/Controls | CC | CT | TT | CC | CT | TT | OR Het (95% CI) |
OR Hom (95% CI) |
PAdd | |
| UK1 | 4040/5012 | 3771 (93.34%) |
266 (6.59%) |
3 (0.07%) |
4711 (94.00%) |
291 (5.81%) |
10 (0.19%) |
1.09 (0.92-1.29) |
1.18 (0.84-1.65) |
0.333 |
| UK2 | 1158A/9744 | 1069 (92.31%) |
89 (7.69%) |
0 (0.00%) |
9175 (94.16%) |
563 (5.78%) |
6 (0.06%) |
1.32 (1.04-1.66) |
1.74 (1.08-2.77) |
0.020 |
| DK1 | 2447/0 | 2240 (91.54%) |
200 (8.17%) |
7 (0.29%) |
- | - | - | - | - | - |
| DK2 | 1324/4727 | 1216 (91.84%) |
106 (8.01%) |
2 (0.15%) |
4412 (93.33%) |
305 (6.45%) |
10 (0.21%) |
1.21 (0.97-1.51) |
1.47 (0.93-2.29) |
0.090 |
| DK1 and DK2 | 3771/4727 | 3456 (91.65%) |
306 (8.11%) |
9 (0.24%) |
4412 (93.33%) |
305 (6.45%) |
10 (0.21%) |
1.26 (1.07-1.47) |
1.57 (1.15-2.16) |
0.004 |
| Category 1 | 7645/14756 | 1.20 (1.08-1.33) |
1.44 (1.17-1.77) |
5×10−4 | ||||||
| (UK1, UK2, DK1, DK2) | ||||||||||
| FS1 | 1160/1172 | 1057 (91.12%) |
98 (8.44%) |
5 (0.43%) |
1087 (92.75%) |
82 (7.00%) |
3 (0.26%) |
1.24 (0.93-1.66) |
1.54 (0.86-2.77) |
0.134 |
| FS2 | 1181/1237 | 1069 (90.52%) |
109 (9.23%) |
3 (0.25%) |
1142 (92.32%) |
93 (7.52%) |
2 (0.16%) |
1.25 (0.94-1.67) |
1.57 (0.89-2.78) |
0.122 |
| MS1 | 774/2957 | 720 (93.02%) |
52 (6.72%) |
2 (0.26%) |
2760 (93.34%) |
191 (6.46%) |
6 (0.20%) |
1.05 (0.77-1.43) |
1.11 (0.59-2.03) |
0.758 |
| BR1 | 111/110 | 107 (96.40%) |
4 (3.60%) |
0 (0.00%) |
106 (96.36%) |
4 (3.64%) |
0 (0.00%) |
0.99 (0.18-5.46) |
- | 1.000 |
| BR2 | 488/483 | 460 (94.26%) |
26 (5.33%) |
2 (0.41%) |
451 (93.37%) |
31 (6.42%) |
1 (0.21%) |
0.90 (0.54-1.51) |
0.81 (0.29-2.29) |
0.711 |
| BR3 | 500/500 | 461 (92.20%) |
36 (7.20%) |
3 (0.60%) |
459 (91.80%) |
39 (7.80%) |
2 (0.40%) |
0.98 (0.63-1.52) |
0.96 (0.39-2.31) |
1.000 |
| Category 1 and 2 (UK1, UK2, DK1, DK2, FS1, FS2, MS1, BR1, BR2, BR3) |
13183/25942 | 1.17 (1.08-1.28) |
1.38 (1.17-1.63) |
1.5×10−4 | ||||||
| Category 1, 2 and 3 (UK1, UK2, DK1, DK2, FS1, FS2, MS1, BR1, BR2, BR3, PI1, JP1, MX1) |
14279/26835 | 1.20 (1.10-1.30) |
2.1×10−5 | |||||||
CC: Common Homozygote; CT: Heterozygote; TT: Rare Homozygote
N: Total number successfully genotyped only
OR Het: Odds Ratio for Heterozygote carriers compared to CC; OR Hom: Odds Ratio for rare allele homozygotes compared to CC
PAdd: P value for additive model
A: Full sample set not available in Oxford at the time of genotyping (n=1193 cases available for genotyping)
Figure 1.
Meta-analysis of the association studies of the T130I variant to type 2 diabetes+
+ Plot generated using Comprehensive Meta Analysis software version 2.2050, Biostat Inc., NJ, USA
* CS: Current Study.
The V255M variant was successfully genotyped in 5745 cases and 15044 controls in the UK study. The MAF for V255M was far lower than for T130I (cases 0.08%, controls 0.10%), and no type 2 diabetes association was observed in either the UK sample (p=0.28) or the larger association analysis incorporating Danish genotyping data (p=0.40) (Table 3).
Table 3.
Association testing of the V255M variant of HNF4A
| Study sample | N | Cases Genotype counts (Genotype frequencies) |
Controls Genotype counts (Genotype frequencies) |
Statistical Analysis | |||||
|---|---|---|---|---|---|---|---|---|---|
| Cases/Controls | GG | GA | AA | GG | GA | AA | OR Het (95% CI) |
PAdd | |
| UK1 | 3949/4925 | 3942 (99.82%) |
7 (0.18%) |
0 (0.00%) |
4911 (99.72%) |
14 (0.28%) |
0 (0.00%) |
0.62 (0.21-1.65) |
0.38 |
| UK2 | 1796/10119 | 1794 (99.89%) |
2 (0.11%) |
0 (0.00%) |
10103 (99.84%) |
16 (0.16%) |
0 (0.00%) |
0.76 (0.08-2.99) |
0.76 |
| Category 1 (UK1, UK2) |
5745/15044 | 0.64 (0.26-1.44) |
0.28 | ||||||
| DK 2 | 1360/4791 | 1357 (99.78%) |
3 (0.22%) |
0 (0.00%) |
4781 (99.79%) |
10 (0.21%) |
0 (0.00%) |
1.05 (0.19-4.11) |
1.00 |
| Category 1 and 2 (UK1, UK2, DK2) |
7105/19835 | 0.73 (0.34-1.47) |
0.44 | ||||||
GG: Common Homozygote; GA: Heterozygote; AA: Rare Homozygote
N: Total number successfully genotyped only
OR Het: Odds Ratio for Heterozygote carriers compared to GG
PAdd: P value for additive model
We did not find any evidence for linkage disequilibrium (r2<0.005) between either T130I or V255M and the common variants in the promoter region of HNF4A that had previously shown a weak association to type 2 diabetes susceptibility.
Discussion
When genes undergo monogenic screening, variants discovered tend to be put into two categories: they are either considered to be causal for monogenic diabetes or neutral “polymorphisms”. The latter group have been assumed to have no role in disease susceptibility. With the large sample sizes now available, it is possible to go back to some of these coding variants that are clearly not causal for monogenic diabetes and re-examine whether they could nevertheless be influencing susceptibility to common forms of diabetes. Previous functional and association studies had highlighted two coding LF variants within HNF4A as interesting candidates in this respect and we have carried out the largest association analysis for the V255M and T130I variants of HNF4A to better understand their role in type 2 diabetes pathogenesis.
We found no association between the V255M variant of HNF4A and type 2 diabetes risk in the UK samples or in our larger analysis. It is worth emphasising that the low MAF of this variant means that our power to detect association was limited to large effect sizes only.
In contrast, evidence of association between the T130I variant of HNF4A and type 2 diabetes risk was found in our analysis of category 1 and 2 samples. The evidence for association was increased when we added category 3 samples reaching a P of 2.1×10−5. To determine whether there was any additional evidence for association available from recent large-scale GWA meta-analyses for type 2 diabetes, we examined data from the recently published DIAGRAM+ meta-analysis [8], after excluding samples already in our meta-analysis. T130I (rs1800961) is represented on Illumina arrays but is neither present on nor can it be reliably-imputed into genome-wide genotypes obtained on early Affymetrix platforms, limiting the data available from DIAGRAM+ to 3590 cases and 32326 controls. In these samples, there was a directionally consistent but non-significant association with T130I (P=0.13) such that the combined analyses (17869 cases and 59197 controls) reached a P of 1.0×10−5.
This association however, fails by some margin, to reach widely-accepted thresholds for genome-wide significance, which in the context of LF variants should be even more stringent than those required for common variants (perhaps around α=5×10−9) given the larger number of independent tests that are possible once lower-frequency variants are considered. We estimate that to achieve this level of significance for T130I (using the effect size [an OR of 1.20] observed in our expanded meta-analysis) would require almost 100,000 samples (in fact, 48697 cases and 48697 controls). Recent evidence (achieving such levels of genome wide significance) that T130I is associated with altered HDL-cholesterol levels raises the prior odds that the type 2 diabetes association we observe here is genuine, as does the strong biological candidacy of this gene given its proven causal role in monogenic forms of diabetes. The previously reported association with lipid levels also raises an interesting question as to whether or not the type 2 diabetes association is mediated by a direct influence of the variant (or a causal variant with which it is in linkage disequilibrium) on beta cell function, or a primary effect on lipid physiology. The latter question could in principle be answered by a suitably-scaled Mendelian randomisation experiment. The broad transcriptional effects of HNF-4A would be consistent with pleiotropic effects of the variant on multiple systems.
As efforts are increasingly aimed at understanding the full allelic spectrum of variants involved in multifactorial disease pathogenesis, large-scale genotyping will be required to clarify the role played by LF variants. As our results exemplify, the numbers required for association testing of such variants are substantial when the effect size is modest. It is clear that large collaborative efforts will be needed to maximise the samples available for any such studies.
Supplementary Material
Acknowledgements
We acknowledge use of genotype data from the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. We acknowledge Diabetes UK for funding the collection of the Warren 2 resource and would also like to thank Professors Graham Hitman, Barts and The London School of Medicine and Dentistry, UK, and Mark Walker, Newcastle University, UK. We thank the staff and senior management of the UK Blood Services responsible for the UK Blood Services Collection. The UK Type 2 Diabetes Genetics Consortium collection was supported by the Wellcome Trust (Biomedical Collections Grant GR072960). BJM is a Diabetes UK Clinical Training Fellow. ALG is a Medical Research Council (MRC) New Investigator (Grant Reference 81696). This work was part funded in Oxford by the Oxford NIHR Biomedical Research Centre Program.
The Danish studies were supported by the Danish Agency for Science Technology and Innovation (grant no. 271-06-0539). The ADDITION study was initiated by: Knut Borch-Johnsen, Steno Diabetes Center, Gentofte, Denmark (PI), Torsten Lauritzen, University of Aarhus, Denmark (PI) and Annelli Sandbæk, University of Aarhus, Denmark. The study was supported by the National Health Services in the counties of Copenhagen, Aarhus, Ringkøbing, Ribe and South Jutland, together with the Danish Research Foundation for General Practice, Danish Centre for Evaluation and Health Technology Assessment, the diabetes fund of the National Board of Health, the Danish Medical Research Council, the Aarhus University Research Foundation and the Novo Nordisk Foundation. The study received unrestricted grants from Novo Nordisk, Novo Nordisk Scandinavia, Astra Denmark, Pfizer Denmark, GlaxoSmithKline Pharma Denmark, Servier Denmark and HemoCue Denmark.
We acknowledge use of the Finland-United States Investigation of NIDDM Genetics (FUSION) genotyping data. Support for this study was provided by US National Institutes of Health grants (DK062370, DK072193, HL084729, HG002651 and U54 DA021519) as well as National Human Genome Research Institute intramural project number 1 Z01 HG000024. The METSIM Study was supported by the Academy of Finland and the EVO grant (no. 5263). The Broad study would like to acknowledge the many clinical researchers involved with these sample collections. The Scandinavian collections are part of the Botnia Study, and are principally supported by the Sigrid Juselius Foundation, the Academy of Finland, the Finnish Diabetes Research Foundation, the Folkhalsan Research Foundation, EC (BM4-CT95-0662, GIFT), the Swedish Medical Research Council, the JDF Wallenberg Foundation, and the Novo Nordisk Foundation. We thank other members of the DIAGRAM consortium (see Voight et al [8], Nature Genetics, 2010 for a full listing) for sharing data. Funding sources for the DIAGRAM consortium is included in reference [8]. D.A. is a Burroughs Wellcome Fund Clinical Scholar in Translational Research, which supported this work.
Abbreviations used
- MAF
Minor Allele Frequency
- GWAS
Genome Wide Association Study
- OR Het
Odds Ratio for Heterozygote carriers
- OR Hom
Odds Ratio for Homozygote carriers
Footnotes
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
References
- [1].Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- [4].Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature genetics. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature genetics. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Saxena R, Hivert MF, Langenberg C, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nature genetics. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kong A, Steinthorsdottir V, Masson G, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature genetics. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lango H, Palmer CN, Morris AD, et al. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes. 2008 doi: 10.2337/db08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ellard S, Bellanne-Chantelot C, Hattersley AT. Best practice guidelines for the molecular genetic diagnosis of maturity-onset diabetes of the young. Diabetologia. 2008;51:546–553. doi: 10.1007/s00125-008-0942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bartoov-Shifman R, Hertz R, Wang H, Wollheim CB, Bar-Tana J, Walker MD. Activation of the insulin gene promoter through a direct effect of hepatocyte nuclear factor 4 alpha. J Biol Chem. 2002;277:25914–25919. doi: 10.1074/jbc.M201582200. [DOI] [PubMed] [Google Scholar]
- [13].Rhee J, Inoue Y, Yoon JC, et al. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Weedon MN, Owen KR, Shields B, et al. Common variants of the hepatocyte nuclear factor-4alpha P2 promoter are associated with type 2 diabetes in the U.K. population. Diabetes. 2004;53:3002–3006. doi: 10.2337/diabetes.53.11.3002. [DOI] [PubMed] [Google Scholar]
- [15].Ji L, Malecki M, Warram JH, Yang Y, Rich SS, Krolewski AS. New susceptibility locus for NIDDM is localized to human chromosome 20q. Diabetes. 1997;46:876–881. doi: 10.2337/diab.46.5.876. [DOI] [PubMed] [Google Scholar]
- [16].Zouali H, Hani EH, Philippi A, et al. A susceptibility locus for early-onset non-insulin dependent (type 2) diabetes mellitus maps to chromosome 20q, proximal to the phosphoenolpyruvate carboxykinase gene. Hum Mol Genet. 1997;6:1401–1408. doi: 10.1093/hmg/6.9.1401. [DOI] [PubMed] [Google Scholar]
- [17].Love-Gregory LD, Wasson J, Ma J, et al. A common polymorphism in the upstream promoter region of the hepatocyte nuclear factor-4 alpha gene on chromosome 20q is associated with type 2 diabetes and appears to contribute to the evidence for linkage in an ashkenazi jewish population. Diabetes. 2004;53:1134–1140. doi: 10.2337/diabetes.53.4.1134. [DOI] [PubMed] [Google Scholar]
- [18].Silander K, Mohlke KL, Scott LJ, et al. Genetic variation near the hepatocyte nuclear factor-4 alpha gene predicts susceptibility to type 2 diabetes. Diabetes. 2004;53:1141–1149. doi: 10.2337/diabetes.53.4.1141. [DOI] [PubMed] [Google Scholar]
- [19].Lillioja S, Wilton A. Agreement among type 2 diabetes linkage studies but a poor correlation with results from genome-wide association studies. Diabetologia. 2009;52:1061–1074. doi: 10.1007/s00125-009-1324-9. [DOI] [PubMed] [Google Scholar]
- [20].Ek J, Rose CS, Jensen DP, et al. The functional Thr130Ile and Val255Met polymorphisms of the hepatocyte nuclear factor-4alpha (HNF4A): gene associations with type 2 diabetes or altered beta-cell function among Danes. J Clin Endocrinol Metab. 2005;90:3054–3059. doi: 10.1210/jc.2004-2159. [DOI] [PubMed] [Google Scholar]
- [21].Winckler W, Graham RR, de Bakker PI, et al. Association testing of variants in the hepatocyte nuclear factor 4alpha gene with risk of type 2 diabetes in 7,883 people. Diabetes. 2005;54:886–892. doi: 10.2337/diabetes.54.3.886. [DOI] [PubMed] [Google Scholar]
- [22].Sookoian S, Gemma C, Pirola CJ. Influence of hepatocyte nuclear factor 4alpha (HNF4alpha) gene variants on the risk of type 2 diabetes: a meta-analysis in 49,577 individuals. Molecular genetics and metabolism. 2010;99:80–89. doi: 10.1016/j.ymgme.2009.08.004. [DOI] [PubMed] [Google Scholar]
- [23].Kathiresan S, Willer CJ, Peloso GM, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nature genetics. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhu Q, Yamagata K, Miura A, et al. T130I mutation in HNF-4alpha gene is a loss-of-function mutation in hepatocytes and is associated with late-onset Type 2 diabetes mellitus in Japanese subjects. Diabetologia. 2003;46:567–573. doi: 10.1007/s00125-003-1067-y. [DOI] [PubMed] [Google Scholar]
- [25].Moller AM, Urhammer SA, Dalgaard LT, et al. Studies of the genetic variability of the coding region of the hepatocyte nuclear factor-4alpha in Caucasians with maturity onset NIDDM. Diabetologia. 1997;40:980–983. doi: 10.1007/s001250050778. [DOI] [PubMed] [Google Scholar]
- [26].Navas MA, Munoz-Elias EJ, Kim J, Shih D, Stoffel M. Functional characterization of the MODY1 gene mutations HNF4(R127W), HNF4(V255M), and HNF4(E276Q) Diabetes. 1999;48:1459–1465. doi: 10.2337/diabetes.48.7.1459. [DOI] [PubMed] [Google Scholar]
- [27].Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wiltshire S, Hattersley AT, Hitman GA, et al. A genomewide scan for loci predisposing to type 2 diabetes in a U.K. population (the Diabetes UK Warren 2 Repository): analysis of 573 pedigrees provides independent replication of a susceptibility locus on chromosome 1q. Am J Hum Genet. 2001;69:553–569. doi: 10.1086/323249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lauritzen T, Griffin S, Borch-Johnsen K, Wareham NJ, Wolffenbuttel BH, Rutten G. The ADDITION study: proposed trial of the cost-effectiveness of an intensive multifactorial intervention on morbidity and mortality among people with Type 2 diabetes detected by screening. Int J Obes Relat Metab Disord. 2000;24(Suppl 3):S6–11. doi: 10.1038/sj.ijo.0801420. [DOI] [PubMed] [Google Scholar]
- [30].Stancakova A, Javorsky M, Kuulasmaa T, Haffner SM, Kuusisto J, Laakso M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009;58:1212–1221. doi: 10.2337/db08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Muller YL, Infante AM, Hanson RL, et al. Variants in hepatocyte nuclear factor 4alpha are modestly associated with type 2 diabetes in Pima Indians. Diabetes. 2005;54:3035–3039. doi: 10.2337/diabetes.54.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Menjivar M, Granados-Silvestre MA, Montufar-Robles I, et al. High frequency of T130I mutation of HNF4A gene in Mexican patients with early-onset type 2 diabetes. Clinical genetics. 2008;73:185–187. doi: 10.1111/j.1399-0004.2007.00928.x. [DOI] [PubMed] [Google Scholar]
- [33].Gauderman W, Morrison J. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006. http://hydra.usc.edu/gxe.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.