Abstract
PADI4 post-translationally converts peptidylarginine to citrulline. PADI4 can disrupt the apoptotic process via the citrullination of histone H3 in the promoter of p53-target genes. The current study focused on PADI4 expression in various subtypes of oesophageal carcinoma (EC) by immunohistochemistry, western blotting and real time PCR. The study also investigated the effect of bile acid deoxycholate (DCA) on PADI4 expression in Eca-109 cells that originated from EC. Apoptosis and DCA-induced toxicity were analyzed by TUNEL, MTT assay and flow cytometry. Additionally, the present study investigated the correlation between single nucleotide polymorphism (SNP) in PADI4 gene and EC risk in Chinese population using Illumina GoldenGate assay. Compared with paraneoplastic tissues, the transcriptional and translational levels of PADI4 were significantly elevated in oesophageal squamous cell carcinoma (ESCC, n=9) and oesophageal adenocarcinoma (EAC, n=5) tissues. Immunolabeling detected expression of PADI4 in ESCC tissues (98.56%, n=139), EAC samples (87.5%, n=16) and oesophageal small cell undifferentiated carcinoma (91.7%, n=12) but not in normal tissues (0%, n=16). Furthermore, PADI4 levels is positively correlated with the pathological classification of ESCC (p=0.009). PADI4 expression levels were consistent with the number of apoptotic cells in the induced Eca-109 cells. rs10437048 [OR= 0.012831; 95% CI, 0.001746~0.094278; p=1.556×10-12] were significantly associated with decreased risk of EC, whereas rs41265997 [OR=12.7; 95% CI, 0.857077~33.207214; p=3.896×10-8] were significantly associated with increased risk of EC. rs41265997 in exon 3 of PADI4 gene is non-synonymous and converts ACG to ATG resulting in a threonine /methionine conversion at position 274 of the protein. Haplotypes GC that carries the variant alleles for rs2501796 and rs2477134 was significantly associated with increased risk of EC (frequency=0.085, p=0.0256, OR=2.7). The results suggest that PADI4 expression is related to the tumorigenic process of EC and the DCA-induced apoptosis. The PADI4 gene may be a valid EC susceptibility gene.
Keywords: Peptidyl arginine deaminase type 4 (PADI4/PAD4), oesophageal squamous cell carcinoma (ESCC), oesophageal adenocarcinoma (EAC), deoxycholate (DCA), apoptosis, genotyping, susceptibility, haplotype.
Introduction
The peptidylarginine deaminase (PAD) enzyme post-translationally modifies arginine, converting it to citrulline in the presence of Ca2+ in a process known as citrullination. PADI4, one of the five different isoforms of PAD, is associated with rheumatoid arthritis (RA) in some populations 1, 2. By immunohistochemistry, western blot analysis and real time PCR, we detected the significant expression of PADI4 in many tumor tissues from the colon, duodenum, oesophagus, fallopian tube, gall gladder, lung, ovary, parotid, pancreas, prostate, rectum, small intestine, stomach, thyroid and uterus 3, 4. We also observed PADI4 co-localization with cytokeratin (CK) in some tumor tissues, preventing the caspase-mediated degradation of CK via citrullination, a process that may contribute to abnormal tumor cell apoptosis 3. However, PADI4 expression in various subtypes of tumors from the same organ has not been well characterized, and the relationship between PADI4 expression and patients' clinical parameters have never been carefully investigated.
The incidence of oesophageal cancer (EC) is increasing in both developing and developed countries 5. Oesophageal cancer is comprised of oesophageal squamous cell carcinoma (ESCC) and oesophageal adenocarcinoma (EAC), which have distinct etiological and pathological characteristics. In the current study, we investigated the expression of PADI4 in ESCC and EAC cells to determine if its expression level correlated to the histological grades and stages of the tumors. It has previously been reported that bile acid deoxycholate (DCA) promoted the tumourigenesis of EC and induced cell apoptosis 6-9. In this study, we investigated how DCA affected apoptosis and PADI4 expression in cultured EC cells. In addition, we investigated the susceptibility of PADI4 to EC in the Chinese Han population.
Materials and Methods
Patients and EC sample preparation
Samples of EC, which included nine samples of ESCC and five samples of EAC, were obtained during excision surgery in Shandong Tumor Hospital (Jinan, China). Normal tissues located 5 cm away from the corresponding tumors were collected during the surgery as controls. Tumor diagnosis was verified by histological methods according to the World Health Organization (WHO) classification system. Tissue arrays containing 183 oesophageal tissue sections were commercially obtained from Chaoying Bioscience (Shanxi, China). The slides contained ESCC (n=139), EAC (n=16), oesophageal small cell undifferentiated carcinoma (n=12), and normal tissues (n=16). Clinical data, including the age, sex, clinical pathological diagnosis, and origin of every participant, were provided by the manufacturer.
Fresh anticoagulated blood was collected from patients with EC (n = 83, 60 males and 23 females). The patients had a mean age of 57.2 years. The patients were selected from the same population living in the Shandong area of Northern China. The diagnosis of EC was determined by barium X-ray tests, CT examination, oesophageal cytology examination and oesophagoscopical examination. A total of 67 healthy people (female 28) having a mean age of 48.0 years. The blood donors with no evidence of any personal or family history of other serious illness were used as controls. Control individuals were age-matched and from the same geographical area as patients diagnosed with EC. The blood samples were collected in Monovette tubes containing 3.8% sodium citrate.
All participants gave their written consent to participate in the study and to allow their biological samples to be genetically analyzed. Approval for the study was given by the Ethical Committee of Shandong Academy of Medicinal Sciences.
Anti-PADI4 antibody
The PADI4 antibody was prepared by immunizing rabbits using oligo-peptides (FGDSCYPSNDSRQMH) specific to the amino acid sequence of PADI4. The immuno-specificity of the antibody was verified in previous studies 3, 4, 10.
Immunohistochemistry
Tissue samples were fixed in 10% neutral buffered formalin and embedded in paraffin. Tissue sections were deparaffinized and rehydrated using standard procedures. To repair the antigen, the sections were heated at 95 ℃ for 10 min in citrate buffer solution (Sigma, USA) and then incubated with Endogenous Peroxidase Inhibitor (MaixinBio, China) for 30 min at room temperature. After washing with PBS buffer (0.132 M NaCl, 0.0066 M K2HPO4, 0.0015 M KH2PO4 in distilled water, pH 7.6), sections were incubated with the anti-PADI4 antibody overnight at 4℃. The immunoreaction was processed using the UltraSensitive TM S-P kit (Maixin-Bio, China) according to the manufacturer's instructions, and immunoreactive signals were visualized using a DAB substrate that stains the target protein yellow. Cell structures were counterstained with hematoxylin.
Immunofluorescence
The tissue sections were processed as described above. After three washes with PBS buffer, the tissue sections were treated with pre-immune serum for 30 min to improve immunoreaction specificity. The slides were incubated with the PADI4 antibody at 4℃ for 12 h and then washed with PBS. TRITC-conjugated 5-goat anti-rabbit IgG (ZSG-BIO, China) antibody was added to the slides, and the slides were incubated for 40 min at room temperature. Immunofluorescence was observed with a Nikon 50i fluorescence microscope (Nikon, Japan). To determine antibody specificity and optimize antibody dilution, a series of control slides were prepared as follows: primary antibodies only, secondary antibodies only, normal rabbit serum and normal goat serum.
A semi-quantitative image analysis and scoring system was applied to evaluate PADI4 immunofluorescent expression. This scoring system assigns the following values, corresponding to the intensity of fluorescent signals: <20 (no expression), 20-30 (low expression), 30-50 (moderate expression) and >50 (strong expression).
The correlation between the expression levels of PADI4 and clinical indices were evaluated using the χ2-test. SPSS 11.5 software (USA) was used for the analyses, and P < 0.05 was considered significant.
Cell culture
Eca-109, an ESCC cell line, was cultured in RPMI 1640 medium (Hyclone Thermo, USA) supplemented with 10% foetal ox blood serum at 37℃ in a humidified atmosphere with 5% CO2. Cells were incubated with increasing concentrations of DCA (0, 10, 25, 50, 75, 100, 150, 200, 250 and 300 μM) for 24 hr.
Western blot analysis
ESCC (n=5) and EAC tissues (n=5) were obtained during excision surgery. Sample tissues (200 μg) were homogenized with RIPA lysis solution (Beyotime) and centrifuged at 16000 g for 5 min at 4 ℃. The supernatant was collected, and protein concentrations were determined using the BCA Protein Assay Kit (Beyotime). Total protein was also extracted from Eca-109 cells by the same method. Ten micrograms of total protein was separated on a 12% SDS-polyacrylamide gel and transferred onto a PVDF membrane. Membranes were incubated with the PADI4 antibody at a dilution of 1:2000 overnight at 4 ℃. Membranes were rinsed with wash solution and incubated with sheep anti-rabbit IgG conjugated to alkaline phosphatase (Jackson). Immunosignals were visualized with the Protein Detector BCIP/NBT Western Blot Kit (Beyotime) following the manufacturer's instructions. The quantification was conducted using ImageQuant 5.2 software. A separate membrane prepared in the same manner was probed with an anti-GAPDH antibody (Santa Cruz, USA) to normalize sample loading.
Real-time PCR
Total RNA was extracted using Tri-blue (Beyotime) from ESCC (n=5) and EAC tissues (n=5) and their corresponding healthy tissue samples. Concentrations of total RNA were determined with a spectrophotometer. Quantitative PCR analysis was performed with a LightCycler 480 thermocycler (Roche). Gene-specific primers were designed using the PADI4 mRNA sequence in Genbank (NM012387) and were as follows: forward 5'-GACTTCAACGGGCTCATTC-3' and reverse 5'-GGACCGATGACTCGTTTGA-3'. The forward primer for human GAPDH was 5'-TGGCACCCAGCACAATGAA-3' and the reverse primer was 5'-CTAAGTCATAGTCCGCCTAGAAGCA-3'. cDNA was prepared with 1 μg of total RNA from each sample using random hexamer priming and reverse transcription with the PrimeScriptTM RT-PCR Kit (TaKaRa). A 387-bp DNA fragment was produced by first strand cDNA synthesis with the primers described above, cloned into the PGEM-T Easy Vector (Promega), and verified by sequence analysis. Taqman real-time PCR was performed in 10 μl reaction mixtures containing 5 μl of SYBR Green (TaKaRa), 1 μl of each primer, 1 μl of cDNA and 2 μl of H2O. The following optimized thermal cycling program was used: 1 cycle of 10 s at 95℃, 45 cycles of 5 s at 60℃ and 10 s at 72℃, and one cycle of 30 s at 65℃. The level of GAPDH mRNA was used as a reference to normalize sample loading. For each tumor sample, two reactions were performed at the same time. One reaction was performed to determine the mRNA level of the target gene, and the second was performed to determine level of GAPDH. The experiment was performed in triplicate. PCR products were confirmed by melting curve analysis. The relative expression of mRNA was calculated using the comparative threshold cycle (Ct) method according to the following formula: Ratio = 2-ΔΔCt = 2-ΔCt (sample), where ΔCt = Ct of target genes - Ct of endogenous control gene (GAPDH). The relative target gene expression was normalized in comparison to GAPDH mRNA. Copy numbers of PADI4 mRNA in the samples were calculated automatically by the data analysis software.
Cell proliferation assay
Eca-109 cells (3×104) were plated in flat-bottomed micro-titer plates and incubated for 24 h at 37℃ in 5% CO2. Cells were incubated either with increasing concentrations of DCA (0-350 μM) for 24 h. Following the treatment, MTT (3-[4,5-dimethylthiazo-2-yl]-2,5-diphenyl tetrazolium bromide) (Amoresco) was added to the culture. After 24 h, absorbance was measured at 490 nm with spectrophotometer. Cell proliferation is expressed as the percentage of cells that remained in cultures treated with DCA relative to their untreated controls.
TUNEL assay
Cell apoptosis was analyzed using the In Situ Cell Death Detection kit (Roche) based on the terminal TUNEL technique. Cells were cultured on chamber glass culture slides in the presence of DCA. To avoid apoptotic cells losing adherence to the slides, the slides were coated with polylysine. After the cells were treated with DCA for the indicated time, the slides were fixed overnight in 100 g/L formaldehyde, treated with proteinase K and H2O2, and labeled with dUTP in a humidified chamber at 37℃ for 1 h. The cells that did not receive TdT enzyme were used as the negative control.
Flow cytometry by annexin V-FITC and PI staining
The cultured cells treated with DCA were collected and pelleted at 1200 U/min. Pellets were washed twice with cold PBS and then resuspended in binding buffer at a concentration of 1×106 cells/mL. One hundred microlitres of the solution (1×105 cells) was transferred to each of two 5 ml culture tubes. Five milliliters of annexin V-FITC (Sigma) and 5 μl of propidium iodide (PI) (Sigma) were added to each 100 μl reaction. The cells were gently vortexed and incubated for 15 min at room temperature in the dark. Then 400 ul of binding buffer was added. Flow cytometry (FCM) was performed with a FACSCalibur (BD Biosciences) within one hour of staining.
Genomic DNA isolation and genotyping
Genomic DNA was extracted from whole blood samples with the Omega E-Z 96 Blood DNA kit (Omega, USA) according to the manufacturer's protocol. DNA samples were dissolved in deionized water. The concentration and quality of DNA samples were measured using a spectrophotometer at 260 nm (A260) and 280 nm (A280). An A260/A280 ratio of 1.7-1.9 was considered to be highly pure DNA. Electrophoresis was also used to examine the quality and integrity of DNA.
TagSNPs located in PADI4 were determined by searching the HapMap database. TagSNPs were determined under conditions of a pair-wise r2 ≥ 0.8 and minor allele frequencies (MAF) over 0.05. SNPs that were in exons and untranslated regions (UTR), as well as promoters and introns within 500 bp near exons, were also selected. Candidate SNPs were submitted to Illumina for a design score. The Illumina Assay Design Tool filtered out SNPs that were not suitable for the Illumina platform, such as insertions/deletions, tri- and tetra-allelic SNPs, and SNPs that were not uniquely localized. Finally, 74 SNPs with a design score of “1” were selected; they span 57,000 bases of the chromosome. These SNPs included 4 tag SNPs, 13 coding SNPs, 2 SNPs at the 3'UTR, 55 SNPs in introns and 5 SNPs in the promoter region. These SNP sites, locations and MAF are described in Table 1.
Table 1.
SNP information
| Serial number | SNP ID | Chr position | Locus | Allele | Protein residue |
|---|---|---|---|---|---|
| 1 | rs16825114 | 17633206 | 5' near gene | C/T | |
| 2 | rs11579504 | 17633246 | 5' near gene | A/G | |
| 3 | rs2501795 | 17633499 | 5' near gene | A/G | |
| 4 | rs2501796 | 17633522 | 5' near gene | A/G | |
| 5 | rs2477134 | 17633572 | 5' near gene | A/C | |
| 6 | rs2477133 | 17633892 | 5' near gene | G/T | |
| 7 | rs35381732 | 17634740 | missense | A | His [H] |
| 8 | rs2501799 | 17635072 | intron | A/G | |
| 9 | rs2477132 | 17635141 | intron | G/T | |
| 10 | rs1886301* | 17635411 | intron | C/T | |
| 11 | rs1635598 | 17657321 | intron | A/G | |
| 12 | rs35111705 | 17657650 | intron | -/G | |
| 13 | rs882537 | 17660047 | intron | C/T | |
| 14 | rs34309058 | 17660468 | missense | A | Thr [T] |
| 15 | rs874881 | 17660499 | missense | C | Ala [A] |
| 16 | rs1548323 | 17660697 | intron | A/G | |
| 17 | rs1748034 | 17662541 | intron | G/T | |
| 18 | rs2240340 | 17662639 | intron | A/G | |
| 19 | rs1748033 | 17662662 | synonymous | C | Leu [L] |
| 20 | rs12733102 | 17662705 | missense | C | Thr [T] |
| 21 | rs1621005 | 17662751 | intron | C/G | |
| 22 | rs1748032 | 17662804 | intron | A/G | |
| 23 | rs1748031 | 17662907 | intron | A/T | |
| 24 | rs12089685 | 17664480 | intron | C/T | |
| 25 | rs11588132 | 17664615 | missense | C | Thr [T] |
| 26 | rs1635586 | 17664770 | intron | C/T | |
| 27 | rs1635585 | 17664813 | intron | A/C | |
| 28 | rs1635583 | 17665003 | intron | C/G | |
| 29 | rs2477152 | 17665064 | intron | A/G | |
| 30 | rs11203368* | 17666508 | intron | C/T | |
| 31 | rs35236764 | 17668036 | intron | -/C | |
| 32 | rs1748021 | 17668270 | intron | C/T | |
| 33 | rs16825565 | 17668508 | synonymous | T | Pro [P] |
| 34 | rs41265995 | 17668509 | missense | A | Ser [S] |
| 35 | rs34743635 | 17668570 | missense | T | Leu [L] |
| 36 | rs41265997 | 17668606 | missense | T | Met [M] |
| 37 | rs1748020 | 17668609 | missense | T | Phe [F] |
| 38 | rs1748019 | 17668976 | intron | C/G | |
| 39 | rs1748016 | 17671140 | intron | A/G | |
| 40 | rs1635571 | 17672729 | intron | A/G | |
| 41 | rs2240338 | 17674185 | intron | A/G | |
| 42 | rs2240337 | 17674222 | intron | A/G | |
| 43 | rs6686575 | 17674366 | intron | C/T | |
| 44 | rs2240336 | 17674402 | intron | A/G | |
| 45 | rs2240335 | 17674537 | synonymous | A | Arg [R] |
| 46 | rs1635570 | 17674601 | intron | A/G | |
| 47 | rs41266005 | 17674793 | intron | C/T | |
| 48 | rs12746451 | 17674821 | intron | C/T | |
| 49 | rs766449 | 17674852 | intron | A/G | |
| 50 | rs12731347 | 17674865 | intron | A/G | |
| 51 | rs12731545 | 17675012 | intron | A/G | |
| 52 | rs12754450 | 17675056 | intron | A/C | |
| 53 | rs3094881 | 17680807 | intron | A/G | |
| 54 | rs6683201 | 17681277 | intron | C/T | |
| 55 | rs4920600 | 17681424 | intron | A/G | |
| 56 | rs35348318 | 17681492 | intron | -/T | |
| 57 | rs36097026 | 17681498 | intron | -/T | |
| 58 | rs16825849 | 17682215 | intron | C/G | |
| 59 | rs10437048 | 17682719 | intron | A/C | |
| 60 | rs2301887 | 17682923 | synonymous | T | His [H] |
| 61 | rs1635566 | 17683157 | intron | C/T | |
| 62 | rs34213328 | 17683185 | intron | -/T | |
| 63 | rs1748011 | 17683281 | intron | G/T | |
| 64 | rs1635565 | 17683332 | intron | A/G | |
| 65 | rs1635564* | 17683526 | intron | A/C | |
| 66 | rs34481461 | 17685466 | intron | A/C | |
| 67 | rs6673924 | 17686013 | intron | C/T | |
| 68 | rs6664430 | 17686051 | intron | C/G | |
| 69 | rs1635561* | 17687027 | intron | A/G | |
| 70 | rs34170018 | 17689736 | intron | C/G | |
| 71 | rs2477150 | 17689954 | intron | A/G | |
| 72 | rs34954650 | 17690376 | 3' UTR | C/T | |
| 73 | rs1408423 | 17690877 | 3' near gene | C/T | |
| 74 | rs1408422 | 17690915 | 3' near gene | A/G |
* represents tag SNP
We performed genotyping using a custom-designed Illumina 96-SNP VeraCode microarray (Illumina). Genotyping was completed at the Beijing Institute of Genomics. The genotyping was conducted with the BeadXpress Reader using the Illumina VeraCode GoldenGate Assay kit. 500 ng of sample DNA were used per assay. Genotype clustering and calling were performed using BeadStudio software (Illumina).
Statistical analysis
After genotyping, SNP markers were evaluated for significant deviation from the Hardy-Weinberg equilibrium (HWE). SNPs were analyzed for association by comparing the MAF between cases and controls. Associations of SNPs with EC were evaluated using odds ratios (OR) with 95% confidence intervals (CI). Fisher's exact test was used for comparison between categorical variables. P values less than 0.05 were considered statistically significant. A test of deviation from HWE was performed for each SNP in both case and control population using software Plink v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) 11. Association tests for allele and genotype frequencies for each SNP were also performed using this software. Multiple-test correction was performed by Plink v1.07 including Genomic-control correction, Bonferroni single-step correction, Holm step-down correction and Sidak single-step correction. Linkage disequlibrium (LD), coefficient (D′ and r2) and haplotype were estimated by software Haploview 4.2 (http://www.broad.mit.edu/mpg/haploview/) 12.
Results
Immunodetection of PADI4 in oesophageal tumor tissues
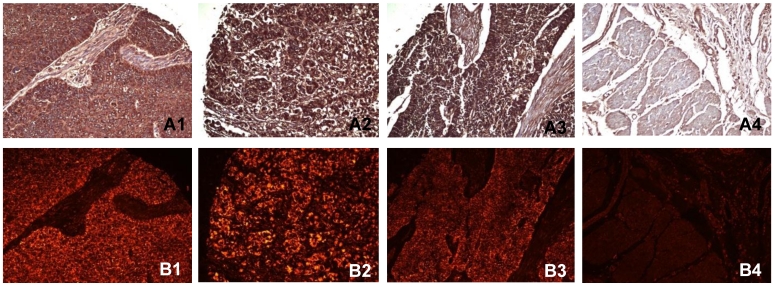
Immunohistochemistry and immunofluorescent labeling were performed to determine the expression of PADI4 in ESCC and EAC tissues. The enzyme was detected in 98.6% (137/139) of ESCC tumors, 87.5% (14/16) of EAC tumors and 91.7% (11/12) of small cell undifferentiated carcinomas. We failed to detect the enzyme in any normal tissues (0/16 cases). In most cases, PADI4 was located in the cytoplasm of tumor cells. However, for small cell undifferentiated carcinoma, PADI4 was present in both the nucleus and cytoplasm of the tumor cells. The enzyme was not significantly detected in normal oesophageal tissues except in a few mesenchymal cells. No signal was observed in negative controls that were incubated with either the primary or secondary antibody alone. The results of these immunohistochemical studies are shown in Figure 1.
Figure 1.
Detecting PADI4 in various oesophageal tumor subtypes using immunohistochemistry (A) and immunofluorescent labelling (B). 1, ESCC tissues; 2, EAC tissues; 3, oesophageal small cell undifferentiated carcinoma tissues; 4, normal tissues. (SP × 400)
The immunofluorescent signals of PADI4 were semi-quantified using SimplePCI software. Within ESCC tissues (n=139), the PADI4 level was positively correlated with a pathological classification (p=0.009). PADI4 expression was increased in ESCC tissues that had higher pathological tumor grades. However, PADI4 levels did not correlate with clinical tumor stage, gender, or age (p>0.05). These results are presented in Table 2. These correlations were not detectable in EAC tumors or small cell undifferentiated carcinomas.
Table 2.
The correlation of PADI4 levels with clinical indexes
| - | ± | + | ++ | P | ||
|---|---|---|---|---|---|---|
| Pathological grade | ||||||
| Ⅰ | 2 | 14 | 34 | 4 | 0.009 | |
| Ⅱ | 0 | 20 | 24 | 22 | ||
| Ⅲ | 0 | 5 | 6 | 8 | ||
| Gender | ||||||
| Female | 0 | 0 | 9 | 22 | 13 | 0.253 |
| Male | 1 | 2 | 31 | 44 | 18 | |
| Age | ||||||
| <50 | 1 | 0 | 0 | 3 | 1 | 0.768 |
| 50~60 | 2 | 0 | 16 | 25 | 11 | |
| >60 | 3 | 2 | 24 | 38 | 19 | |
| Clinical stage | ||||||
| 1 | 0 | 0 | 1 | 0 | 0.853 | |
| 2 | 1 | 17 | 24 | 9 | ||
| 3 | 1 | 23 | 41 | 22 |
* -: negative; ±:weak positive; +: positive; ++: strong positive
Quantifying PADI4 expression levels by western blot analysis and real time PCR
PADI4 protein levels were quantified by western blot analysis. The 67 kDa protein was detected in ESCC (n=5), EAC (n=5) and corresponding non-malignant tissues. The expression level of PADI4 was significantly higher in ESCC and EAC than in their corresponding paraneoplastic tissues (p=0.0029 and 0.0048, respectively). These results are shown in Figure 2A, B and C.
Figure 2.
Detecting PADI4 expression with western blotting. A. Western blotting shows expression of PADI4 in ESCC tumors (lanes 1-5) and paraneoplastic tissues (lanes 6-10). B. Western blotting detected expression of PADI4 in EAC tumors (lanes 11-15) and paraneoplastic tissues (lanes 16-20). C. PADI4 expression is significantly increased in ESCC and EAC tissues as compared to that in paraneoplastic tissues. (p=0.0029 and 0.0048, respectively). Lanes 1-2, PADI4 protein levels in ESCC and corresponding paraneoplastic tissues; Lanes 3-4, PADI4 protein levels in EAC and corresponding paraneoplastic tissues.
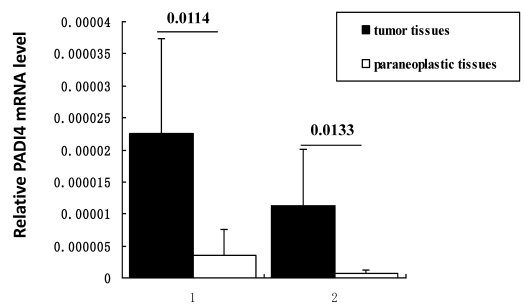
Transcription of PADI4 was quantified by real-time PCR. All samples of ESCC and EAC (n=5 for each subtype) tissues exhibited a higher level of PADI4 mRNA than the corresponding non-malignant tissues (p=0.0114 and 0.0133, respectively). These results are shown in Figure 3.
Figure 3.
Detecting PADI4 expression levels using real time PCR. Real time PCR measurement of PADI4 mRNA levels in ESCC (lane 1, n=5) and EAC (lane 2, n=5) samples. This expression was normalized to the expression of GAPDH. These results indicate that PADI4 transcription is significantly increased in ESCC and EAC tissues compared to that in the oesophageal paraneoplastic tissues. (p=0.0114 and 0.0133, respectively).
Effect of DCA on cell proliferation and PADI4 expression in Eca-109 cells
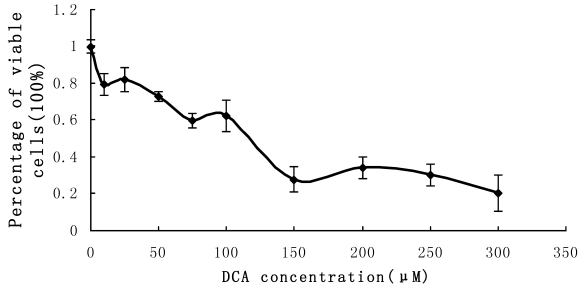
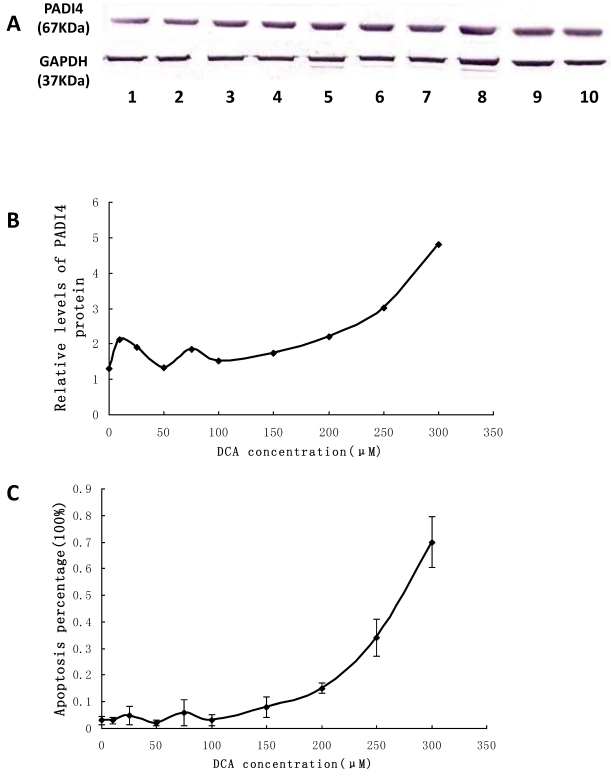
Cultured Eca-109 cells were treated with DCA at different concentrations as described above. The percent of viable cells were assessed using the MTT assay. The results indicated that cell proliferation was suppressed in the presence of increasing DCA concentrations following 24 hrs of treatment. Cell growth was suppressed by 80% after treatment with 300 μM DCA for 48 h. The MTT results fluctuated when the concentration of DCA was between the range of 0 μM-100 μM. These results are shown in Figure 4.
Figure 4.
MTT measurement of cell proliferation. Cultured Eca-109 cells were incubated with 0-350 µM DCA. Cell proliferation was suppressed by increasing concentrations of DCA after 24 h of treatment. The error bars represent the standard deviation from 3 replicate measurements.
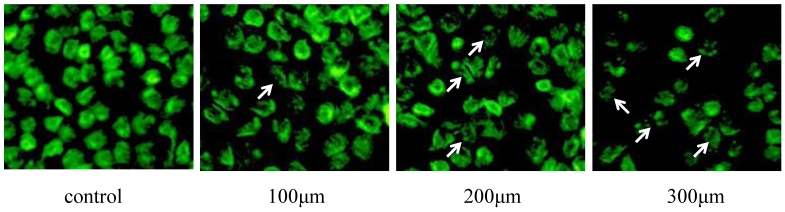
TUNEL assay was used to analyze apoptosis of Eca-109 cells in the presence of DCA. These results revealed that DCA-induced apoptosis in Eca-109 cells occurred in a dose-dependent manner. Morphology indicative of apoptosis was observed after 24 h of DCA treatment at 100, 200 and 300 μM concentrations. These morphologic changes included chromatin condensation, membrane blebbing and shrinkage, and an increase in apoptotic bodies. Compared to the controls, the numbers of apoptotic cells were increased after the addition of increased DCA concentrations. These results are shown in Figure 5.
Figure 5.
TUNEL detection of Eca-109 cell apoptosis after DCA treatment. Cells were stained with acridine orange and then imaged with a fluorescent microscope. Apoptotic cells are indicated with arrows.
The expression level of PADI4 in Eca-109 cells was examined by western blot analysis. The protein levels were significantly increased in the cultured cells treated with 100 μM to 300 μM DCA. The expression level of PADI4 fluctuated when the concentration of DCA was in the range of 0 μM-100 μM. These results are shown in Figure 6A and B.
Figure 6.
Detecting PADI4 expression and apoptosis of Eca-109 cells after DCA treatment. A. The expression level of PADI4 in the cultured cells was analyzed using western blotting. B. The result was normalized to the expression of GAPDH. C. Apoptosis of cultured Eca-109 cells was measured with FCM. Eca-109 cells were treated without DCA (1) or with DCA at 10 μM (2), 25 μM (3), 50 μM (4), 75 μM (5), 100 μM (6), 150 μM (7), 200 μM (8), 250 μM (9) and 300 μM (10). The error bars represent the standard deviation from 3 replicate measurements.
Eca-109 cells treated with 0-300 μM DCA were stained with annexin V and PI to assess apoptosis by FCM. Both “early” apoptotic cells and “late” apoptotic cells were annexin V-FITC-positive and PI-positive. The percentage of apoptotic cells was significantly increased after 100 μM to 300 μM treatment of DCA. When the expression of PADI4 was increased, there was also an increase in the percentage of apoptotic cells. Furthermore, the percentage of apoptotic cells significantly corresponded to the expression levels of PADI4, and fluctuated when the concentration of DCA was in the range of 0 μM-100 μM. These results are shown in Figure 6C.
Single SNP and haplotype analysis
We genotyped 74 SNPs within the PADI4 gene. All SNPs yielded usable genotyping data and the study sample success rate was 98.1%. Differences in the allele and genotype frequencies between cases and controls were compared. Overall, two SNPs (rs10437048 and rs41265997) were significantly associated with a risk of EC. rs10437048 [OR= 0.012831; 95% CI, 0.001746~0.094278; p=1.556×10-12] were significantly associated with decreased risk of EC, whereas rs41265997 [OR=12.7; 95% CI, 0.857077~33.207214; p=3.896×10-8] were significantly associated with increased risk of EC. Another SNP rs2501796 was associated with EC before multi-test correction (p=0.02). However, the association was not detected following Bonfferni correction. Table 3contains the allele frequencies, genotype frequencies and the unadjusted and adjusted p-values after the statistical analysis of these SNPs. All SNPs retained in analyses were in Hardy-Weinberg equilibrium (P>0.05) in the overall samples. Other SNPs located in PADI4 gene were not significantly different in allelic or genotypic frequencies between the EC patients and controls. In addition, age and sex were matched between case and control (p>0.05). We also performed multivariate logistic regression analysis using software Plink v1.07 to test the correlations between the SNP and ESCC. The p values after logistic regression is also shown in Table 3.
Table 3.
Allele and genotype frequencies in a case control cohort of patients with EC
| dbSNP identity | Genotype | No. of patients with EC (%) | No. of controls (%) | P value | P value after multivariate logistic regression | p value after multiple testing correction (Bonferroni method) | odds Ratio (%95 CI) | |
|---|---|---|---|---|---|---|---|---|
| rs10437048 | allele | C | 58(0.3766) | 1(0.0077) | 2.255×10-14 | 4.182×10-6 | 1.55610-12 | 0.012831 [0.001746~0.094278] |
| A | 96(0.7234) | 129(0.9923) | ||||||
| genotype | CC | 6(0.0779) | 0(0) | 2×10-14 | 4.182×10-6 | 1.556×10-12 | - | |
| CA | 46(0.5974) | 1(0.0154) | ||||||
| AA | 25(0.3247) | 64(0.9846) | ||||||
| rs41265997 | allele | A | 46(0.3333) | 5(0.0378) | 5.646×10-10 | 1.745×10-9 | 3.89×10-8 | 12.7 [4.857077~33.207214] |
| G | 92(0.6667) | 127(0.9622) | ||||||
| genotype | AA | 0(0) | 0(0) | 1.15×10-12 | 1.745×10-9 | 3.89×10-8 | - | |
| AG | 46(0.6667) | 5(0.0757) | ||||||
| GG | 23(0.3333) | 61(0.9243) |
67 controls & 83 cases observed
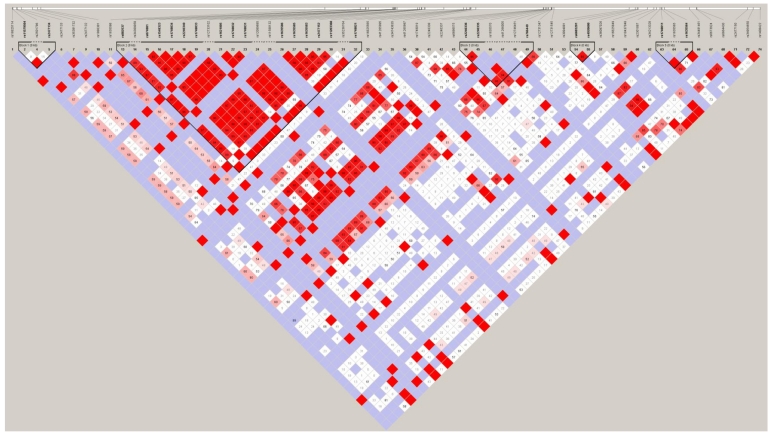
Linkage disequilibrium (LD) analysis was performed within the 74 SNPs genotyped. Pair-wise D′ values between all SNPs were calculated to determine the extent of LD. LD analysis defined five blocks in the PADI4 region. These results are shown in Figure 7. The second block is the largest and most extensive, at 8.2 kb in length. Both rs10437048 and rs41265997 were not localized in any block.
Figure 7.
LD plot of the genotyped 74 SNPs in the PADI4 gene. Numbers in squares are D' values. Red areas represent higher levels of LD. Blue areas represent LD comparisons with a low estimation confidence. Dark triangles represent haplotype blocks.
Haplotypes analysis was performed using Haploview 4.2. Twelve different haplotypes in these blocks were identified. Haplotype AA, AC and GC were detected in block 1 (frequencies as 0.796, 0.12 and 0.085, respectively). Haplotype GGGCGGCAAGCCAAG, ACAAAAGGTAAGGGA and ACAAAAGGTAAGGGG were in block 2 (frequencies as 0.541, 0.358 and 0.022, respectively). Haplotype AAG and GCA were in block 3. Haplotype GG and AA were in block 4 (frequencies as 0.596 and 0.366, respectively). Haplotype AC and CA (frequencies as 0.743 and 0.247, respectively) were in block 5. We omitted the haplotypes whose frequency under 0.05. In haplotype analysis using logistic model, we assigned the most frequent haplotype as reference (OR=1), then compare other haplotypes with this reference haplotype. The GC haplotypes carrying the variant allele rs2501796 and rs2477134 in the first block were significantly associated with the increased risk of EC (frequency=0.085, p=0.0256, OR=2.7). Other haplotypes located in PADI4 gene were not significantly associated with the risk of EC (p>0.05).
Discussion
We previously demonstrated the over-expression and absence of PADI4 in a variety of malignant and normal tissues, respectively 4. The present study focused on PADI4 expression in a cohort of oesophageal tumors. In comparison to normal oesophageal tissues, immunohistochemistry showed the increased expression of PADI4 in ESCC tumors, EAC tumors and small cell undifferentiated carcinomas. Real time PCR and western blot analysis confirmed the significantly higher PADI4 expression in ESCC and EAC tissues compared to that in paraneoplastic tissues. These results agree with our previous findings. Furthermore, we found that PADI4 expression directly correlated to the pathological tumor grade of ESCC tissues; increased PADI4 correlated to more severe cases of the disease. We recently reported that the expression level of PADI4 is related to Pathological Tumour-Node-Metastasis (pTNM) in serious cystadenocarcinoma. The tumors at T1 and T2 stages of pTMN had strong expression of PADI4, whereas the tumors at T3 and T4 stages exhibited weak or moderate PADI4 staining 10. Studies from other groups have also reported that PADI4 plays a role in tumourigenesis. Cuthbert et al. and Wang et al. found that the citrullination of PADI4 prevented arginine methylation of histone H3 and H4 during the transcriptional activation of oestrogen-responsive genes 13, 14. Yao et al. and Li et al. reported that PADI4 repressed expression of p53-mediating genes, including OKL38, p21, CIP1 and WAF1, by deaminasing the methylated arginine sites in histone H3, which consequently interrupted the apoptotic process and the cell cycle 15, 16. We also demonstrated the citrullination of cytokeratin, antithrombin and fibronectin, proteins known to be involved in abnormal apoptosis, high coagulation, disordered cell proliferation and aberrant differentiation 3, 4.
Studies have shown that duodenal reflux contributes to the development of EC 17-19. Bile acids are major constituents of the gastroesophageal refluxate and have an important role in malignant development in the oesophagus 20, 21. Previous studies have demonstrated that bile acids cause mucosal injury, induce cyclooxygenase-2 (COX-2) expression, and promote tumourigenesis and apoptosis 22, 23. Bile acids, in particular DCA, inhibit cell proliferation and are potent inducers of apoptosis in several cell types, including hepatocytes and colonic cells 24, 25. Zhang R et al. reported that DCA could inhibit growth and induce apoptosis of Eca-109 cells 26. However, the exact molecular mechanism of how DCA contributes to this process has not been elucidated. In the present study, we found that DCA significantly stimulated the expression of PADI4 and promoted Eca-109 apoptosis. Furthermore, the percentage of apoptotic cells significantly correlated with PADI4 expression levels. This result not only confirmed other previous studies, but it also demonstrated an important role of PADI4 in DCA-induced apoptosis. Liu GY et al. demonstrated that PADI4 over-expression induced haematopoietic cell apoptosis and suggested that PADI4 could induce apoptosis mainly through arrest of the cell cycle through a mitochondria-mediated pathway 27. Tanikawa C et al. recently reported that DNA damage induced the citrullination of various proteins in a p53/PADI4-dependent manner 28.
As described above, PADI4 is increased in malignant tumors, and the level of this expression is directly related to tumor development in ESCC and serious cystadenocarcinoma. However, over-expression of PADI4 enhanced apoptotic cell death of cultured HL-60, Jurkat (human acute leukaemic T cells), and Eca-109 cells 27. These observations remain controversial, and the reason for them is yet unclear. Most of these previous studies were conducted with cultured cells under non-physiologic conditions, and as such, these results probably do not reflect the physiologic function of PADI4. Citrullinated proteins not only comprise histones and cytokeratin, but also extracellular matrix components such as antithrombin, fibrin and fibronectin 3, 4. These extracellular matrices are also involved in abnormal apoptosis, increased coagulation, disordered cell proliferation and cell differentiation 29. Thus, it is possible that in vitro experiments show different results from in vivo studies. Further study is needed to understand the exact pathogenic mechanism of PADI4 in tumourigenesis.
In this study, 74 SNPs located in the PADI4 gene were genotyped within the Han Chinese population. The results indicated that two SNPs at rs104437048 and rs41265997 were significantly associated with EC risk in the population, indicating that PADI4 may be a valid EC susceptibility gene. The SNP at rs41265997 is located in exon 3 of PADI4 gene and is non-synonymous. The mutation converts ACG to ATG resulting in a Threonine /Methionine conversion at position 274 of the PADI4 protein. The amino acid substitution in a region of the protein could significantly affect the protein secondary structure or function.
After haplotype analysis, a significant correlation to EC was observed when haplotype frequencies of the patients were compared with that of the health. GC haplotype carrying the variant alleles for rs2501796 and rs2477134 in block 1 was significantly elevated in the EC patients. Block 1 is located in the promoter of PADI4 gene. It is possible that genetic variation mediates the gene expression by alternating the gene's regulatory mechanism. Dong et al. found that oestrogen regulates the expression of PADI4 through both the classical and nonclassical pathways 30. In the classical way, oestrogen binds to the oestrogen receptor to form a complex and then regulates PADI4 expression via the oestrogen response element on the promoter. In the non-classical pathway, the oestrogen-oestrogen receptor complex acts on the transcription factors AP-1, NF-Y, and SP-1, which bind to the PADI4 promoter to specifically regulate PADI4 expression. Stacey SN et al. analyzed 304,083 SNPs and observed significant association signals from the loci at 1p36 and 1q421p36. The most significant signals were from rs7538876 on 1p36 and rs801114 on 1q42 31. The candidate genes in the 1p36 locus contain PADI4; their previous finding corresponds exactly to our results. Suzuki et al. used a case-control linkage disequilibrium study to show that PADI4 is a susceptible locus for RA. They found that rs2240340/padi4-94 in intron 3 was significantly associated with RA. They also found that susceptible mRNA had more stable versus non-susceptible mRNA in vitro 32. In our study, rs2240340 did not show a strong association with EC. It is possible that PADI4 has different genetic effects on these diseases by the utilization of different pathogenic pathways.
In summary, we demonstrated the increased expression of PADI4 in different types of oesophageal cancer. The expression level of PADI4 was positively related to the pathological classification of ESCC. We also demonstrated that apoptosis corresponded to the expression of PADI4 in cultured EC cells treated with DCA. Furthermore, we found that PADI4 may be a valid EC susceptibility gene by single SNP and haplotype analyses.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (NTFC) (30972720), the National Basic Research Program of China (2010CB529105), the Provincial Natural Science Foundation of Shandong (ZR2010CM1032, 2009ZHZX1A1004) and the Shandong Taishan Scholarship.
Abbreviations
- PADI4/PAD4
Peptidylarginine deiminase type 4
- EC
oesophageal carcinoma
- ESCC
oesophageal squamous cell carcinoma
- EAC
oesophageal adenocarcinoma
- DCA
bile acid deoxycholate
- SNP
single nucleotide polymorphism.
References
- 1.Cornélis F, Fauré S, Martinez M. et al. New susceptibility locus for rheumatoid arthritis suggested by a genome-wide linkage study. Proc Natl Acad Sci USA. 1998;95(18):10746–10750. doi: 10.1073/pnas.95.18.10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiozawa S, Hayashi S, Tsukamoto Y. et al. Identification of the gene loci that predispose to rheumatoid arthritis. Int Immunol. 1998;10(12):1891–1895. doi: 10.1093/intimm/10.12.1891. [DOI] [PubMed] [Google Scholar]
- 3.Chang X, Han J. Expression of Peptidylarginine Deiminase Type 4 (PAD4) in Various Tumors. Mol Carcinog. 2006;45(3):183–196. doi: 10.1002/mc.20169. [DOI] [PubMed] [Google Scholar]
- 4.Chang X, Han J, Pang L. et al. PADI4 has increased expression in blood and tissues of malignant tumors. BMC cancer. 2009;9:40. doi: 10.1186/1471-2407-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pisani P, Parkin DM, Bray F. et al. Estimates of the worldwide mortality from 25 cancers in 1990. Int J Cancer. 1999;83(1):18–29. doi: 10.1002/(sici)1097-0215(19990924)83:1<18::aid-ijc5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 6.Martinez JD, Stratagoules ED, LaRue JM, et al.Different bile acids exhibit distinct biological effects. the tumor promoter deoxycholic acid induces apoptosis and the chemopreventive agent ursodeoxycholic acid inhibits cell proliferation. Nutr Cancer. 1998;31(2):111–118. doi: 10.1080/01635589809514689. [DOI] [PubMed] [Google Scholar]
- 7.Debruyne PR, Bruyneel EA, Li X. et al. The role of bile acids in carcinogenesis. Mutat Res. 2001;480-481:359–369. doi: 10.1016/s0027-5107(01)00195-6. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein H, Payne CM, Bernstein C. et al. Activation of the promoters of genes associated with DNA damage, oxidative stress, ER stress and protein malfolding by the bile salt, deoxycholate. Toxicol Lett. 1999;108(1):37–46. doi: 10.1016/s0378-4274(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 9.Qiao D, Stratagouleas ED, Martinez JD. Activation and role of mitogen-activated protein kinases in deoxycholic acid-induced apoptosis. Carcinogenesis. 2001;22(1):35–41. doi: 10.1093/carcin/22.1.35. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Chang X, Yuan G, Zhao Y, Wang P. Expression of Peptidylarginine deiminase type 4 in Ovarian Tumors. International Journal of Biological Sciences. 2010;6(5):454–464. doi: 10.7150/ijbs.6.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuthbert GL, Daujat S, Snowden AW. et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118(5):545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Wysocka J, Sayegh J. et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306(5694):279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 15.Yao H, Li P, Venters BJ. et al. Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J Biol Chem. 2008;283(29):20060–20068. doi: 10.1074/jbc.M802940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li P, Yao H, Zhang Z. et al. Regulation of p53 target gene expression by peptidylarginine deiminase. Mol Cell Biol. 2008;28(15):4745–4758. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein SR, Yang GY, Curtis SK. et al. Development of esophageal metaplasia and adenocarcinoma in a rat surgical model without the use of a carcinogen. Carcinogenesis. 1997;18(11):2265–2270. doi: 10.1093/carcin/18.11.2265. [DOI] [PubMed] [Google Scholar]
- 18.Miwa K, Sahara H, Segawa M. et al. Reflux of duodenal or gastro-duodenal contents induces esophageal carcinoma in rats. Int J Cancer. 1996;67(2):269–274. doi: 10.1002/(SICI)1097-0215(19960717)67:2<269::AID-IJC19>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Pera M, Brito MJ, Poulsom R. et al. Duodenal-content reflux esophagitis induces the development of glandular metaplasia and adenosquamous carcinoma in rats. Carcinogenesis. 2000;21(8):1587–1591. [PubMed] [Google Scholar]
- 20.Kauer WK, Peter JH, DeMeester TR. et al. Mixed reflux of gastric and duodenal juices is more harmful to the esophagus than gastric juice alone. The need for surgical therapy re-emphasized. Ann Surg. 1995;222(4):525–531. doi: 10.1097/00000658-199522240-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillen P, Keeling P, Byrne PJ. et al. Implication of duodenogastric reflux in the pathogenesis of Barrett's oesophagus. Br J Surg. 1988;75(6):540–543. doi: 10.1002/bjs.1800750612. [DOI] [PubMed] [Google Scholar]
- 22.Zhang F, Altorki NK, Wu YC. et al. Duodenal reflux induces cyclooxygenase-2 in the esophageal mucosa of rats: evidence for involvement of bile acids. Gastroenterology. 2001;121(6):1391–1399. doi: 10.1053/gast.2001.29781. [DOI] [PubMed] [Google Scholar]
- 23.Looby E, Abdel-Latif MM, Athié-Morales V. et al. Deoxycholate induces COX-2 expression via Erk1/2-, p38-MAPK and AP-1-dependent mechanisms in esophageal cancer cells. BMC Cancer. 2009;9:190. doi: 10.1186/1471-2407-9-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rust C, Karnitz LM, Paya CV. et al. The bile acid taurochenodeoxycholate activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J Biol Chem. 2000;275(26):20210–20216. doi: 10.1074/jbc.M909992199. [DOI] [PubMed] [Google Scholar]
- 25.Washo-Stultz D, Crowley-Weber CL, Dvorakova K. et al. Role of mitochondrial complexes I and II, reactive oxygen species and arachidonic acid metabolism in deoxycholate-induced apoptosis. Cancer Lett. 2002;177(2):129–144. doi: 10.1016/s0304-3835(01)00786-8. [DOI] [PubMed] [Google Scholar]
- 26.Zhang R, Gong J, Wang H. et al. Bile salts inhibit growth and induce apoptosis of human esophageal cancer cell line. World J Gastroenterol. 2005;11(33):5109–5116. doi: 10.3748/wjg.v11.i33.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu GY, Liao YF, Chang WH. et al. Over-expression of peptidylarginine deiminase IV features in apoptosis of haematopoietic cells. Apoptosis. 2006;11(2):183–196. doi: 10.1007/s10495-006-3715-4. [DOI] [PubMed] [Google Scholar]
- 28.Tanikawa C, Ueda K, Nakagawa H. et al. Regulation of protein Citrullination through p53/PADI4 network in DNA damage response. Cancer Res. 2009;69(22):8761–8769. doi: 10.1158/0008-5472.CAN-09-2280. [DOI] [PubMed] [Google Scholar]
- 29.Chang X, Fang K. PADI4 and tumourigenesis. Cancer Cell Int. 2010;10:7. doi: 10.1186/1475-2867-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong S, Zhang Z, Takahara H. Estrogen-enhanced peptidylarginine deiminase type IV gene (PADI4) expression in MCF-7 cells is mediated by estrogen receptor-alpha-promoted transfactors activator protein-1, nuclear factor-Y, and Sp1. Mol Endocrinol. 2007;21(7):1617–1629. doi: 10.1210/me.2006-0550. [DOI] [PubMed] [Google Scholar]
- 31.Stacey SN, Gudbjartsson DF, Sulem P. et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nature Genetics. 2008;40(11):1313–1318. doi: 10.1038/ng.234. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki A, Yamada R, Chang X. et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genetics. 2003;34(4):395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]