Abstract
The mammalian fatty acid desaturase 2 (FADS2) gene codes for catalytic activity considered to be the rate limited step in long chain polyunsaturated fatty acid (LCPUFA) synthesis. FADS2 catalyzes 6-desaturation in at least five substrates and 8-desaturation in at least two substrates. However, the molecular mechanisms that regulate FADS2-mediated desaturation remain ill-defined. We report here characterization of an alternative transcript (AT1) of primate FADS2 and compare its expression to that of the classical transcript in 12 tissues of a 12 week old neonate baboon, and in human SK-N-SH neuroblastoma (NB) cells. RT-PCR analysis indicates relatively greater abundance of classical transcript than AT1 in all tissues. However, AT1 expression is highly variable, showing greater expression in liver, retina, occipital lobe, hippocampus, spleen, and ovary, than in other tissues, whereas classical transcript displayed little variability. These data suggest that FADS2 AT1 is a candidate for regulation of LCPUFA synthesis.
Keywords: Fatty acid desaturases, Alternative transcript, Long chain polyunsaturated fatty acids, Baboon
Introduction
Fatty acid desaturases are enzymes that catalyze the introduction of cis double bonds at specific positions in a fatty acid chain. FADS1 and FADS2 are genes coding key desaturase enzymes for the biosynthesis of long chain polyunsaturated fatty acids (LCPUFA), and are referred as ‘front end’ desaturases [1,2]. Fatty acid desaturase 2 (FADS2) codes for a protein that catalyzes the first and rate limiting step in the biosynthesis of LCPUFA from 18 carbon PUFA. In mammals, the FADS2 gene product catalyzes 6-desaturation in at least five substrates, 18:2n-6, 18:3n-3, 24:5n-3, 24:4n-6, and 16:0 [3,4]. Recently, we demonstrated that the primate FADS2 gene product also catalyzes 8-desaturation, using substrates 20:2n-6 and 20:3n-3, which were previously thought to be dead end products [5].
We recently reported the first finding of alternate transcripts (AT) of the FADS2 and FADS1 paralog FADS3 elsewhere [6]. In this study, we report a transcript variant of FADS2 in fetal baboon brain generated by alternative splicing, as well as evidence that it is widely expressed in baboon neonate tissues and in human SK-N-SH neuroblastoma (NB) cells.
Materials and methods
Fetal baboon brain tissue was obtained from banked tissue at the Southwest Foundation for Biomedical Research (San Antonio, Tx). Neonate baboon tissues originated from a 12 week old baboon fed an infant formula with no LCPUFA from a previous study [7]. All tissues were maintained at −80°C since necropsy until RNA isolation.
RNA isolation and cDNA synthesis
Total RNA was extracted from baboon tissue homogenates and SK-N-SH neuroblastoma (NB) cells using the RNeasy Mini kit (Qiagen, Valencia, CA).The yield of total RNA was assessed by 260 nm UV absorption.The quality of RNA was analyzed by 260/280 nm ratios of thesamples and by agarose gel electrophoresis to verify RNA integrity.One microgram total RNA was reverse transcribed into first-strand cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). The resulting cDNA was stored at −20°C until use.
PCR conditions and identification of alternative transcript
We have earlier [5] reported the protein coding region of baboon FADS2 (GenBank accession number EU780003) using primers FADS2 forward: ATGGGGAAGGGAGGGAACCAGGGCGA and FADS2 reverse: TCATTTGTGAAGGTAGGCGTCCAGCCA. The above primers were used to perform polymerase chain reaction (30 μl) with baboon fetal brain tissue cDNA as template, 1 μM of each primer, 0.25 mM each of dNTPs, 1.5 mM MgCl2, 5X PCR buffer, and high-fidelity Taq polymerase (Roche Diagnostics Ltd) in an Eppendorf gradient thermal cycler. Cycling conditions were: initial denaturation at 95°C for 5 min followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 72°C for 45 s and extension at 72°C for 1 min, with a final extension at 72°C for 5 min. Two prominent bands were obtained when separated by electrophoresison a 2% agarose gel. The two bands were gel purified, cloned into pGEM T-Easy vector (Promega, USA) and sequenced using T7 forward and SP6 reverse universal primers at the Cornell University life sciences core laboratories.
Mammalian cell culture and sample preparation
SK-N-SH neuroblastoma (NB) cells (ATCC, USA) were grown in a humidified environment at 37°C with 5% CO2 using DMEM/F-12 with 10% FBS, 2mM L-alanyl-glutamine, and 15 mM HEPES for undifferentiated cells and routine passaging. NB cell differentiation was carried out in serum free DMEM/F-12 with 1X N-2 supplement (Invitrogen, USA) containing 100 mg/L human transferrin, 5 mg/L recombinant full chain insulin, 0.0063 mg/L progesterone, 16.1 mg/L putrescine, and .0052 mg/L sodium selenite [8,9]. Cells were seeded in parallel and grown to 70% confluence in regular growth media with 10% FBS. One flask of undifferentiated cells was then harvested for RNA extraction, while the remaining cells were switched to the chemically defined N-2 supplemented media for an additional six days in order to halt growth and induce differentiation before harvesting. RNA was extracted using the QIAshredder and RNeasy Mini kit (Qiagen, Valencia, CA).
Expression of FADS2 and AT1 in baboon tissues and human cells
To analyze the expression of FADS2 CS, a forward primer was designed within the exonic region that is deleted in the alternative transcript. To amplify FADS2 AT1, the forward primer bridged the deleted parts of exons.
FADS2 CS Primer sequences:
Forward: AGGCCCAAGCTGGATGGCTGCAA
Reverse: AGTTGGCAGAGGCACCCTTTAAG
FADS2 AT1 Primer sequences:
Forward: AGAAGCATAACCTGTCTGTCTACA
Reverse: ATGATTCCACCAGTTGGCAGAG
The tissue distribution of each transcript was measured by RT-PCR using cDNA from 12 normal tissues from a baboon neonate (12 weeks old), and in undifferentiated (embryonic stage) and differentiated SK-N-SH neuroblastoma cells. PCR amplification reactions were performed using 1 μM of each primer, 0.25 mM each of dNTPs, 1.5 mM MgCl2 and AmpliTaq Gold (ABI, Foster City, CA) in a final volume of 30 μl. Cycling conditions were: initial denaturation at 95°C for 5 min followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 67°C for 45 s and extension at 72°C for 1 min, with a final extension at 72°C for 5 min. PCR products were separated on 2% agarose gels, and the PureLink Gel Extraction Kit (Invitrogen) was used to isolate the PCR bands. The amplicons were sequenced to confirm identity.
Results and Discussion
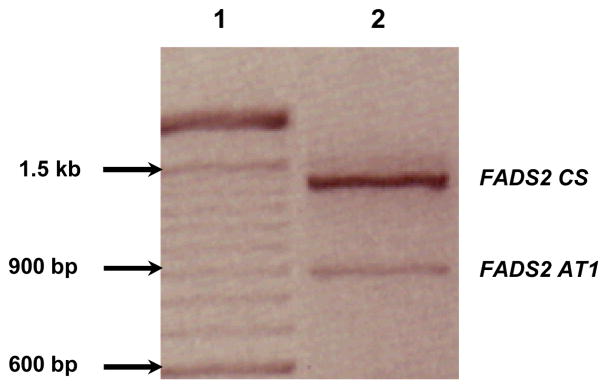
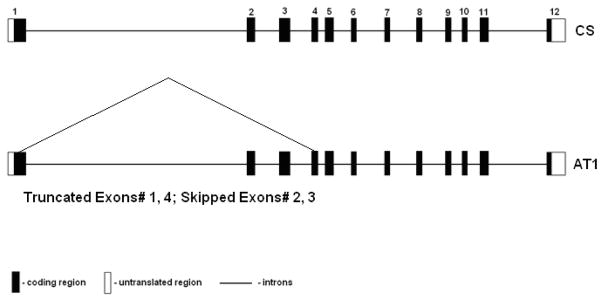
Previously, we cloned and sequenced the entire protein coding region of baboon FADS2, GenBank accession number EU780003, using baboon neonate liver cDNA. However, in addition to the expected size PCR band we noticed another prominent amplified product using baboon fetal brain cDNA (Figure 1). Product sequencing resulted in the identification of an 873 bp FADS2 alternative transcript (AT1) with a C/T substitution at nucleotide position 327 (GenBank Accession# FJ901343). The splice variant has truncated exons 1 and 4 and skipped exons 2 and 3 (Figure 2). The putative coding region 0.9 kb for AT1 resulting from an in-frame loss of 154 aa was identified using ORF finder <http://www.ncbi.nlm.nih.gov/projects/gorf/>. Two of the conserved motifs characteristic of front end desaturases (“HPGG” Cytochrome b5 motif and the first histidine repeat “HDYGH”) are lost during the splicing event.
Figure 1.
FADS2 CS - Classical splicing, FADS2 AT1 is alternative splicing. The products were separated on 2% agarose gel, visualized with ethidium bromide and a negative image of photograph is shown. Lane 1: 100 base pair molecular weight marker and Lane 2: PCR products amplified by RT-PCR
Figure 2.
FADS2 CS - Classical splicing, FADS2 AT1 is the novel alternative transcript. Missing spans within AT1 are shown. Numbers 1 to 12 are exons, CS- Classical Splicing, AT1- alternative transcript
Expression profiling
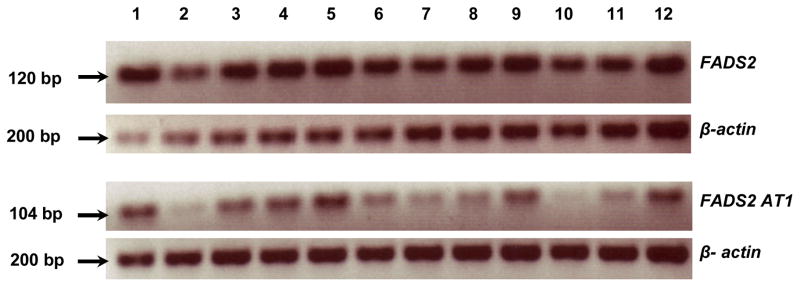
To investigate if AT1 expression is limited to the fetus, we studied the expression of FADS2 classically spliced (FADS2 CS) and AT1 in 12 neonate baboon tissues and in human SK-N-SH neuroblastoma (NB) cells (figure 3 and figure 4). FADS2 CS expression was greater than AT1 in all tissues examined, and displayed relatively little variability, with noticeably lower expression only in kidney. AT1 expression was highly variable, showing greater expression in liver, retina, occipital lobe, hippocampus, spleen, and ovary, than in other tissues. FADS2 CS and AT1 were also expressed in undifferentiated and differentiated NB cells with no obvious difference in expression levels.
Figure 3.
FADS2 and AT1 expression in 12 neonate baboon tissues (lane 1: liver, lane 2: kidney, lane 3: retina, lane 4: occipital lobe, lane 5: hippocampus, lane 6: heart, lane 7: skeletal muscle, lane 8: lung, lane 9: spleen, lane 10: thymus, lane 11: pancreas, lane 12: ovary)
Figure 4.
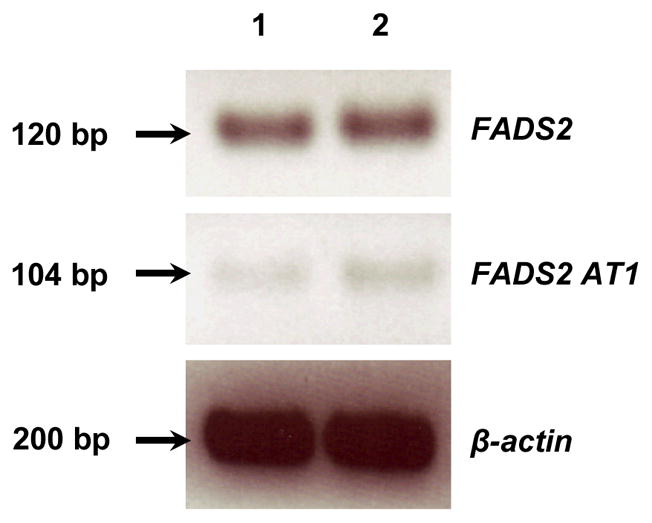
FADS2 and AT1 expression in human SK-N-SH neuroblastoma (NB) cells (lane 1: undifferentiated NB cells, lane 2: differentiated NB cells)
Putative subcellular localization
FADS2 CS and FADS2 AT1 putative subcellular localization prediction was achieved using the web-based tool, Proteome Analyst Specialized Subcellular Localization Server (PA-SUB) [10]. The software takes amino acid sequence information in FASTA format and provides an estimate of the probability that the protein location within a cell. Both FADS2 CS and FADS2 AT1 were found to be localized to endoplasmic reticulum (ER) with a probability score of 1.000 and to be localized to mitochondria with probability scores of 0.562 and 0.999, respectively. FADS2 CS is known to code for ER proteins. However, the above results suggest a mitochondrial role, as well as an ER role, for FADS2 AT1.
Functional characterization studies remain to be implemented to understand the physiological significance of the co-expression of AT1 along with FADS2. Although FADS2 AT1 is missing apparently conserved domains, in the flowering plant Anemone leveillei, a desaturase has been described without the cytochrome b5 domain catalyzed synthesis of non-methylene-interrupted (non-conjugated) PUFA. This observation shows that the four common domains are not required for production of functional protein [11]. Qualitatively, expression levels appear to follow the tissue concentrations of 22 carbon LCPUFA. Confirmation of this observation would suggest a role in the regulation or biosynthesis of LCPUFA from precursors.
Acknowledgments
This work was supported by NIH grant GM071534 and by a Cornell University Center for Vertebrate Genomics seed grant. The authors thank Yun-sang Huh for technical assistance.
References
- 1.Marquardt A, Stohr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66(2):175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura MT, Nara TY. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Ann Rev Nutr. 2004;24:345–376. doi: 10.1146/annurev.nutr.24.121803.063211. [DOI] [PubMed] [Google Scholar]
- 3.D’Andrea S, Guillou H, Jan S, Catheline D, Thibault JN, Bouriel M, Rioux V, Legrand P. The same rat Delta6-desaturase not only acts on 18- but also on 24-carbon fatty acids in very-long-chain polyunsaturated fatty acid biosynthesis. Biochem J. 2002;364(Pt 1):49–55. doi: 10.1042/bj3640049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillou H, D’Andrea S, Rioux V, Jan S, Legrand P. The surprising diversity of Delta6-desaturase substrates. Biochem Soc Trans. 2004;32(Pt 1):86–87. doi: 10.1042/bst0320086. [DOI] [PubMed] [Google Scholar]
- 5.Park WJ, Kothapalli KS, Lawrence P, Tyburczy C, Brenna JT. An alternate pathway to long-chain polyunsaturates: the FADS2 gene product Delta8-desaturates 20:2n-6 and 20:3n-3. J Lipid Res. 2009;50(6):1195–1202. doi: 10.1194/jlr.M800630-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park WJ, Kothapalli KS, Reardon HT, Kim LY, Brenna JT. Novel fatty acid desaturase 3 (FADS3) transcripts generated by alternative splicing. Gene. 2009 doi: 10.1016/j.gene.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh AT, Anthony JC, Diersen-Schade DA, Rumsey SC, Lawrence P, Li C, Nathanielsz PW, Brenna JT. The influence of moderate and high dietary long chain polyunsaturated fatty acids (LCPUFA) on baboon neonate tissue fatty acids. Pediatr Res. 2007;61(5 Pt 1):537–545. doi: 10.1203/pdr.0b013e318045bec9. [DOI] [PubMed] [Google Scholar]
- 8.Bottenstein JE. Environmental Influences on Cells in Culture. In: Boulton A, Baker G, Walz W, editors. Neuromethods. Vol. 23. The Humane Press Inc; 1992. pp. 63–85. [Google Scholar]
- 9.Dong JM, Lim L. Selective up-regulation of alpha 1-chimaerin mRNA in SK-N-SH neuroblastoma cells by K+/−induced depolarisation. Eur J Biochem. 1996;236(3):820–826. doi: 10.1111/j.1432-1033.1996.00820.x. [DOI] [PubMed] [Google Scholar]
- 10.Lu Z, Szafron D, Greiner R, Lu P, Wishart DS, Poulin B, Anvik J, Macdonell C, Eisner R. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics. 2004;20(4):547–556. doi: 10.1093/bioinformatics/btg447. [DOI] [PubMed] [Google Scholar]
- 11.Sayanova O, Haslam R, Venegas Caleron M, Napier JA. Cloning and characterization of unusual fatty acid desaturases from Anemone leveillei: identification of an acyl-coenzyme A C20 Delta5-desaturase responsible for the synthesis of sciadonic acid. Plant Physiol. 2007;144(1):455–467. doi: 10.1104/pp.107.098202. [DOI] [PMC free article] [PubMed] [Google Scholar]