Abstract
Phosphorylation on tyrosine, threonine and serine residues represents one of the most important post-translational modifications and is a key regulator of cellular signaling of multiple biological processes that require a strict control by protein kinases and protein phosphatases. Abnormal protein phosphorylation has been associated with several human diseases including Alzheimer disease (AD). One of the characteristic hallmarks of AD is the presence of neurofibrillary tangles, composed of microtubule-associated, abnormally hyperphosphorylated tau protein. However, several others proteins showed altered phosphorylation levels in AD suggesting that deregulated phosphorylation may contribute to AD pathogenesis. Phosphoproteomics has recently gained attention as a valuable approach to analyze protein phosphorylation, both in a quantitave and a qualitative way. We used the fluorescent phosphospecific Pro-Q Diamond dye to identify proteins that showed alterations in their overall phosphorylation in the hippocampus of AD vs. control (CTR) subjects. Significant changes were found for 17 proteins involved in crucial neuronal process such as energy metabolism or signal transduction. These phosphoproteome data may provide new clues to better understand molecular pathways that are deregulated in the pathogenesis and progression of AD.
Keywords: Alzheimer disease, protein phosphorylation, phosphoproteomics, 2-DE
INTRODUCTION
Post-translational modifications are key modulators of protein structure and function [1]. Reversible protein phosphorylation of serine, threonine, and tyrosine residues is the most common peptide modification and plays a crucial role in controlling various cellular functions in living cells. Phosphorylation is regulated by a highly dynamic network of protein kinases and phosphatases and it is [2–4] estimated that at least one-third of eukaryotic proteins are phosphorylated, however only a subset are modified by any given stimulus [5–7].. Phosphorylation often occurs at multiple residues within a protein and by different protein kinases allowing the protein to adapt to several different functions. Complex cooperation between various classes of kinases and phosphatases triggers the dynamics of phosphorylation cycles [8, 9].
Abnormal phosphorylation has been implicated in many human diseases such as cancer and Alzheimer disease (AD) [5, 10]. Thus, analysis of the entire phosphoproteome and its alterations is gaining interest and presently represents an attractive subject for researchers.
AD is the most common neurodegenerative disease affecting more than 20 million people worldwide and the leading cause of dementia. Most cases of Alzheimer disease are sporadic and idiopathic; however, mutations in three genes, APP, presenilin 1 and presenilin 2 have been identified as being responsible for a small fraction of early-onset Alzheimer disease, that accounts for approximately 5% of all Alzheimer disease cases. With increasing life-span and in the absence of interventions to slow or arrest progression of AD, the number of elderly people at risk of developing dementia is growing rapidly [11]. Large number of studies proposed various hypotheses to explain the pathogenesis or progression of sporadic AD that include the amyloid hypothesis, the oxidative stress hypothesis and APOE genotype among others [12–17]. Although the initiating events are still unknown, it is likely that AD is the consequence of the combination of genetic risk factors with different epigenetic events. The neocortex, entorhinal cortex, and hippocampus brain regions of AD patients are characterized by the presence of two principal histopathological hallmarks, extracellular senile plaques (SPs) and intracellular neurofibrillary tangles (NFTs). In addition, the above mentioned brain regions are dramatically atrophied in AD compared to brains from non-demented subjects [18]. SPs are composed of a core of amyloid β-peptide (Aβ) and exist as either diffuse or neuritic variants. Neuritic plaques have dense fibrillary Aβ cores surrounded by dystrophic axons and dendrites, activated microglia, and reactive astrocytes [19–21]. NFTs consist as paired helical filaments composed of hyperphosphorylated tau protein and occur in the cytoplasm of the soma and in neuritic projections [22–26]. Aberrant phosphorylation of several other proteins such as neurofilaments and microtubule-associated protein 1B have also been found to associate with AD pathogenesis, confirming that altered phosphorylation is a common event during AD progression [10].
In the current study we used a proteomic approach to analyze alterations of the AD phosphoproteome compared to control (CTR). Our method takes advantage of a fluorescent phosphosensor, Pro-Q Diamond, capable of sensitive detection of phosphoserine-, phosphothreonine-, and phosphotyrosine-containing proteins [8, 27, 28]. Total protein phosphorylation levels are then normalized to total protein expression within the same gel using multiplexed proteomics technology [29]. By following this approach, we identified several proteins with altered phosphorylation in AD vs. CTR hippocampal samples.
Materials and methods
AD and CTR subjects
Frozen hippocampal samples (n=6 each) from well-characterized subjects with AD and respective age-matched controls (Table 1) were obtained from the University of Kentucky Rapid Autopsy Program of the Alzheimer's Disease Clinical Center (UK ADC) with an average PMI of about 3 h for AD patients and respective control subjects. All the subjects were longitudinally followed and underwent annual neuropsychological testing, and neurological and physical examinations. Control subjects were without history of dementia or other neurological disorders and with intact activities of daily living (ADLs), and they underwent annual mental status testing and semi-annual physical and neurological exams as part of the UK ADC normal volunteer longitudinal aging study. The control subjects showed no significant histopathological alterations and the average Braak score was II (range: I–III) for the age-matched controls. AD patients diagnosis was made according to criteria developed by the National Institute of Neurological and Communicative Disorders and Stroke (NINCDS) and the Alzheimer’s Disease and Related Disorders Association (ADRDA) [30]. All AD patients displayed progressive intellectual decline. The mean Braak score for AD patients was VI (range: V–VI), respectively. The demographic parameters of controls and AD subjects are provided in Table 1.
Table 1.
Demographic information with causes of death, concomitant neuropathologies and known co-morbidities of AD subjects and their respective age-matched controls;
| Age (years) |
Sex | Brain Weight (g) |
PMI (hrs) |
Braak stage |
Cause of Death |
Amyloid Angiopathy |
PD1 | Brain Tumor |
Head Injury |
Co- morbidities |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| CTR 1 | 77 | Male | 1310 | 3.5 | I | No | No | No | No | MI, CABG4 | |
| CTR 2 | 83 | Male | 1275 | 2 | III | MI2 | No | ||||
| CTR 3 | 87 | Male | 1150 | 2 | II | Embolism | No | ||||
| CTR 4 | 72 | Male | 1150 | 3.75 | I | Mild | |||||
| CTR 5 | 85 | Female | 1020 | 2.5 | III | CHF3 | No | No | No | Yes | CHF |
| CTR 6 | 81 | Male | 1410 | 2 | II | No | |||||
| Average | 81 ± 5.5 |
1219 ± 139 |
2.60 ± 0.8 |
Range I-III |
|||||||
| AD 1 | 80 | Female | 1160 | 2.75 | VI | No | No | No | No | Hypertensio n |
|
| AD 2 | 90 | Female | 1050 | 2.6 | VI | No | No | No | No | ||
| AD 3 | 88 | Male | 1230 | 5.75 | V | Mild | No | No | No | Hypertensio n |
|
| AD 4 | 81 | Male | 1260 | 3 | VI | Severe | No | No | No | MI, peptic ulcer |
|
| AD 5 | 81 | Female | 835 | 3 | VI | Moderate | No | No | No | ||
| AD 6 | 92 | Female | 1090 | 2 | VI | Moderate | No | No | No | Cancer | |
| Average | 85 ± 5.3 |
1104 ± 154 |
3.2 ± 1.3 |
Range V-VI |
PD = Parkinson disease;
MI = myocardial infarction;
CHF = congestive heart failure;
CABG = coronary artery bypass
Sample preparation
Hippocampal samples from control and AD subjects were sonicated in Media 1 lysis buffer (pH 7.4) containing 320 mM Sucrose, 1% of 990 mM Tris-HCl (pH=8.8), 0.098 mM MgCl2, 0.076 mM EDTA, proteinase inhibitors leupeptin (0.5 mg/mL), pepstatin (0.7 µg/mL), aprotinin (0.5 mg/mL) and PMSF (40 µg/mL), phosphatase and kinase inhibitor cocktail (Sigma-Aldrich). Homogenates were centrifuged at 14,000g for 10 min to remove debris. Protein concentration in the supernatant was determined by the Pierce BCA method (Pierce, Rockford, IL, USA).
2D electrophoresis
For the first-dimension electrophoresis, approximately 200 µg of sample solution were applied to 110-mm pH 3–10 IPG® ReadyStrip (Bio-Rad, Hercules CA). The strips were then actively rehydrated in the protean isoelectric focusing (IEF) cell (Bio-Rad) at 50 V for 18 h. The isoelectric focusing was performed in increasing voltages as follows; 300 V for 1 h, then linear gradient to 8000 V for 5 h and finally 20 000 V/h. Strips were then stored at −80 °C until the second dimension electrophoresis was to be performed. For the second dimension, the IPG® Strips, were thawed and equilibrated for 10 min in 50 mM Tris–HCl (pH 6.8) containing 6 M urea, 1% (w/v) sodium dodecyl sulfate (SDS), 30% (v/v) glycerol, and 0.5% dithiothreitol, and then re-equilibrated for 15 min in the same buffer containing 4.5% iodacetamide instead of dithiothreitol. Linear gradient precast criterion Tris–HCl gels (8–16%) (Bio-Rad) were used to perform second dimension electrophoresis. Precision Protein™ Standards (Bio-Rad, CA) were run along with the samples at 200 V for 65 min.
Gel Staining
For the detection of phosphoproteins, the gels were stained using a Pro-Q Diamond phosphoprotein gel stain kit (Molecular Probes) according to the manufacturer’s protocol. Briefly, 2-DE gels were fixed with 50% methanol and 10% acetic acid (100 mL per gel) for 30 min two times. Gels were then washed three times with ultrapure water (100 mL per gel) for 30 min for each washing. Gels were incubated in Pro-Q Diamond phosphoprotein stain solution (100 mL per gel) for 90 min, and destained by washing three times in 50 mM sodium acetate, pH 4.0, 20% acetonitrile (100 mL per gel) for 30 min. Gels were then washed again three times with ultrapure water (100 mL per gel) for 30 min for each washing. After the fluorescence scanning the gels were incubated in fixing solution (7% acetic acid, 10% methanol) for 20 min and stained overnight at room temperature with 50 mL SYPRO Ruby gel stain (Bio-Rad). The SYPRO ruby gel stain was then removed and gels stored in deionized water.
Image analysis
Pro-Q Diamond images were acquired using a Typhoon transilluminator (GE Healthcare) with excitation/emission wavelength of 555/580 nm. SYPRO ruby-stained gel images were obtained using a STORM phosphoimager (GE Healthcare). All the images were saved in TIFF format. Gel imaging was software-aided using PD-Quest (Bio-Rad) imaging software. Briefly, a master gel was selected followed by normalization of all gels (Control and AD) according to the total spot density. Gel-to-gel analysis was then initiated in two parts. First, manual matching of common spots that could be visualized among the differential 2D gels was performed. After obtaining a significant number of spots the automated matching of all spots was then initiated. Automated matching is based on user-defined parameters for spot detection. These parameters are based on the faintest spot, the largest spot, and the largest spot cluster that occur in the master gel and are defined by the user. Based on these parameters the software defines spot centers for the gel. This process generates a large pool of data, approximately 400 spots. Only proteins showing computer-determined significant differential levels between the two groups being analyzed were considered for identification. To determine significant differential levels of proteins, analysis sets were created using the analysis set manger software incorporated into the PD-Quest software. The numbers of pixels that occur in a protein spot were computed by the software corresponding to an increase/decrease in protein level. The gel image analysis was conducted first on phosphostained gels and then on Sypro Ruby-stained expression gels. The two analyses were compared by software to normalize phosphorylation value to expression value for each spot matched. A quantitative analysis set was created that recognized matched spots with differences in phosphorylation intensity (normalized to expression intensity) that occur in each spot and a statistical analysis set was created that used a Student’s t-test at 95% confidence to identify spots with p-values of p < 0.05. Spots with p < 0.05 were considered significant. A Boolean analysis set was created that identified overlapping spots from the aforementioned quantitative and statistical sets. These spots were selected for subsequent mass spectrometric analysis.
In gel trypsin digestion
Protein spots in AD gels statistically different than controls were digested in-gel by modified trypsin using protocols previously described and modified by Thongboonkerd et al. [31]. Spots were taken from individual gels and not pooled for mass spectrometric analysis. The amount of protein from one gel-spot is sufficient for identification. Briefly, spots of interest were excised using a clean blade and placed in Eppendorf tubes, which were then washed with 20 µL of 0.1 M ammonium bicarbonate (NH4HCO3) at room temperature for 15 min. 30 µL of 100% acetonitrile was then added to the gel pieces and incubated at room temperature for 15 min. This solvent mixture was then removed and gel pieces dried in a flow hood for 15 min. The protein spots were then incubated with 20 µL of 20 mM DTT in 0.1 M NH4HCO3 at 56 °C for 45 min. The DTT solution was removed and replaced with 20 µL of 55 mM iodoacetamide in 0.1 M NH4HCO3. The solution was then incubated at room temperature for 30 min. The iodoacetamide was removed and replaced with 0.2 mL of 50 mM NH4HCO3 and incubated at room temperature for 15 min. Acetonitrile (200 µL) was added. After 15 min incubation, the solvent was removed, and the gel spots were dried in a flow hood for 30 min. The gel pieces were rehydrated with 20 ng/µL-modified trypsin (Promega, Madison, WI) in 50 mM NH4HCO3 with the minimal volume sufficient to cover the gel pieces. The gel pieces were incubated overnight at 37 °C in a shaking incubator.
Mass spectrometry
The resulting tryptic peptides were analyzed with an automated nanospray Nanomate Orbitrap XL MS platform. The Orbitrap MS was operated in a data-dependent mode whereby the 8 most intense parent ions measured in the FT at 60,000 resolution were selected for ion trap fragmentation with the following conditions: injection time 50 ms, 35% collision energy. MS/MS spectra were measured in the FT at 7500 resolution, and dynamic exclusion was set for 120 seconds. Each sample was acquired for a total of ∼2.5 minutes. MS/MS spectra were searched against the ipi_Human Database using SEQUEST with the following criteria: Xcorr > 1.5, 2.0, 2.5, 3.0 for +1, +2, +3, and +4 charge states, respectively, and P-value (protein and peptide) < 0.01. IPI accession numbers were cross-correlated with SwissProt accession numbers for final protein identification.
Statistical analysis
All statistical analysis was performed using a two-tailed Student’s t-test. P < 0.05 was considered significantly different from control.
RESULTS
Proteomics analysis
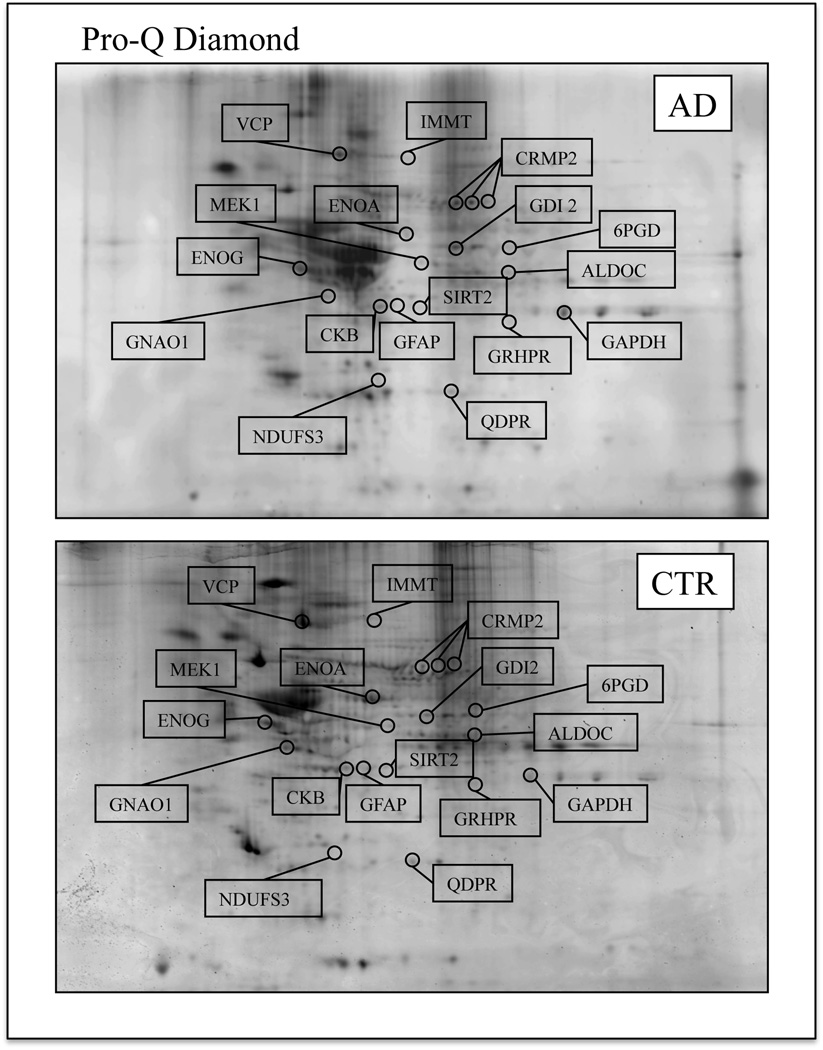
In this study we used multiplexed proteomics technology that couples the classic proteomics approach with Pro-Q diamond fluorescent staining allowing the parallel determination of protein and phosphorylation levels within a single experiment. The Pro-Q Diamond phosphoprotein dye technology is reliable for the fluorescent detection of phosphoserine-, phosphothreonine-, and phosphotyrosine-containing proteins directly in sodium dodecyl sulfate (SDS)-polyacrylamide gels and two-dimensional (2-D) gels. The staining is rapid, easy to perform, readily reversible, and fully compatible with modern microchemical analysis procedures, such as MS/MS. The specificity and sensitivity of Pro-Q Diamond phosphoprotein stain has been previously confirmed by several authors [32]. Fluorescence signal intensity correlates with the total number of phosphorylated residues on the protein. Through combination of Pro-Q Diamond phosphoprotein stain with SYPRO Ruby protein gel stain, multiplexed proteomics technology permits quantitative, fluorescence detection of proteins in 2-D gels permitting the determination of altered post-translational modification patterns normalized for protein expression level changes within a single 2-D gel experiment [28, 29]. Our samples had an average PMI of about 3h, a short time and an advantage in this study since previous studies showed that the level of protein phosphorylation might be reduced by the activities of phosphatases during extended postmortem intervals. Our analysis reveals a total of 17 proteins differentially phosphorylated in AD hippocampus compared with age-matched controls (Table 2; Figures 1, 2, 3). Nine of these show increased phosphorylation in AD: gamma enolase (ENOG) and alpha enolase (ENOA) (29.6- and 2.5-fold), guanine nucleotide-binding protein G (o) subunit alpha (GNAO1) (16.4-fold), fructose-bisphosphate aldolase C (ALDOC) (4.1-fold), dihydropyrimidine related protein 2 (CRMP2) (2.5-, 7.1-, 5.6-fold), 6-phosphogluconate dehydrogenase, decarboxylating (6PGD) (10.1-fold), glial fibrillary acidic protein (GFAP) (1.3-fold), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1.4-fold) and mitogen activated protein kinase kinase 1 (MEK 1) (2.1-fold). Multiple spots for CRMP2 were found suggesting that differential post-translational modification of this particular protein affects the isoelectric point (pI) of CRMP2. In contrast to the above results, eight of the identified phosphoproteins showed decreased phosphorylation in AD hippocampus: NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial (NDUFS3) (0.12-fold), creatine kinase B-type (CKB) (0.25-fold), NAD-dependent deacetylase sirtuin-2 (SIRT2) (0.63-fold), Rab GDP dissociation inhibitor beta (GDI2) (0.61-fold), glyoxylate reductase/hydroxypyruvate reductase (GRHPR) (0.12-fold), mitochondrial inner membrane protein (IMMT) (0.2-fold), transitional endoplasmic reticulum ATPase (VCP) (0.1-fold) and dihydropteridine reductase (QDPR) (0.55-fold). In Table 2 we show the number of known and predicted phosphoserine, phosphothreonine and phosphotyrosine residues confirming that all the proteins identified are well-known or putative phosphoproteins and that our methods are appropriate and reliable. Moreover, we have previously shown the phosphorylation levels of biliverdin reductase (BVR-A) in AD hippocampus by either Pro-Q diamond or Western blot with specific anti phospho- ser/thr/tyr antibodies. Both methods of analysis showed the same results, consistent with the robustness of our data [33].
Table 2.
Summary of the proteins identified by phosphoproteomics with aberrant phosphorylation in the AD versus control hippocampi. For each protein, the phosphorylation/protein expression values were averaged (n = 6) and expressed as fold phosphorylation compared to control. The p-value listed is the significance of altered phosphorylation levels relative to control samples with p< 0.05. Pb represents the probability of a false identity associated with each protein identification using the SEQUEST search algorithm. For the numbers of phosphorylated sites known we used Uniprot and Phosida databases, while for the number of phosphorylated sites predicted we used the NetphosK algorithm.
| PROTEIN | Swiss prot accession number |
Pb | # Of Peptides matched/s earched |
Fold AD/CTR |
P-Value | Number of phosphorylated sites: known/ (predicted) |
Biological process/molecular pathways |
|---|---|---|---|---|---|---|---|
| Alpha enolase | P06733 | 5e-010 | 4/4 | 2.50 | 0.033 | S 5/(8) T 2/(1) Y 3/(6) |
Glycolysis, energy metabolism |
| Gamma enolase | P09104 | 1e-014 | 6/6 | 29.6 | 0.038 | S 1/(6) T 0/(3) Y 2/(7) |
Glycolysis, Energy metabolism |
|
Glyceraldehyde-3- phosphate dehydrogenase |
P04406 | 3e-012 | 7/12 | 1.38 | 0.037 | S 6/(11) T 6/(3) Y 3/(4) |
Glycolysis, Energy metabolism, NO signals. |
| Creatine kinase B-type | P12277 | 4e-011 | 3/4 | 0.25 | 0.014 | S 3/(11) T 1/(6) Y 2/(4) |
Energy metabolism, creatine metabolic process |
|
NAD-dependent deacetylase sirtuin-2 |
Q8IXJ6 | 4e-012 | 6/9 | 0.63 | 0.049 | S 5/(14) T 0/(3) Y 0/(4) |
Cell cycle, Cell division, Mitosis |
|
Fructose-bisphosphate aldolase C |
P09972 | 1e-030 | 17/39 | 4.14 | 0.015 | S 1/(8) T 1/(4) Y 0/(2) |
Glycolysis, energy metabolism |
|
NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial |
O75489 | 2e-011 | 3/3 | 0.12 | 0.029 | S 0/(7) T 0/(5) Y 0/(2) |
Electron transport, Respiratory chain Transport |
|
6-phosphogluconate dehydrogenase, decarboxylating |
P52209 | 1e-030 | 9/11 | 10.1 | 0.001 | S 0/(13) T 0/(5) Y 0/(3) |
Cell anabolism. Pentose shunt, |
|
Glyoxylate reductase/hydroxypyruv ate reductase |
Q9UBQ7 | 1e-008 | 6/9 | 0.12 | 0.006 | S 0/(3) T 0/(6) Y 0/(1) |
Cell anabolism. glyoxylate metabolic process |
|
Dihydropteridine reductase |
P09417 | 4e-015 | 7/13 | 0.55 | 0.015 | S 0/(5) T 0/(4) Y 0/(0) |
Cell anabolism. Tetrahydrobiopterin biosynthesis, |
|
Glial fibrillary acidic protein |
P14136 | 1e-011 | 16/27 | 1.34 | 0.022 | S 0/(16) T 2/(5) Y 0/(3) |
Cell structure, astrocytic constituent of cytoskeleton |
|
Mitochondrial inner membrane protein |
Q16891 | 1e-011 | 10/14 | 0.20 | 0.035 | S 0/(32) T 0/(8) Y 0/(3) |
Cell structure, Mitochondrial structure |
|
Transitional endoplasmic reticulum ATPase |
P55072 | 2e-014 | 26/44 | 0.01 | 0.012 | S 14/(19) T 2/(9) Y 1/(3) |
Cellular trafficking, ER vesicles formation. |
|
Dihydropyrimidine related protein |
Q16555 | 8e-015 | 23/35 | 7.17 | 0.041 | S 5/(22) T 5/(7) Y 3/(7) |
Cell structure, Axonal growth, Differentiation, Neurogenesis |
| 1e-012 | 12/16 | 2.52 | 0.008 | ||||
| 1e-030 | 20/30 | 5.68 | 0.046 | ||||
|
Dual specificity mitogen activated protein kinase kinase 1 |
Q02750 | 5e-014 | 6/6 | 2.15 | 0.003 | S 4/(11) T 2/(4) Y 0/(1) |
Signaling. Activates ERK1 and ERK2 MAP kinases. |
|
Guanine nucleotide binding protein G(o) subunit alpha |
P09471 | 6e-015 | 11/22 | 16.4 | 0.050 | S 0/(10) T 0/(4) Y 0/(4) |
Signal Transducer |
|
Rab GDP dissociation inhibitor beta |
P50395 | 1e-030 | 22/34 | 0.61 | 0.024 | S 1/(9) T 0/(11) Y 1/(10) |
Signal transduction. Protein transport, regulation of GTPase activity, |
Figure 1.
Representative 2D phosphorylation maps of AD (top) and CTR (bottom) hippocampus. Gels were stained using Pro-Q Diamond fluorescent dye. The spots showing altered phosphorylation levels between AD and control are labeled. Relative change in phosphorylation, after normalization of the immunostaining intensities to the protein content, was significant for 17 spots.
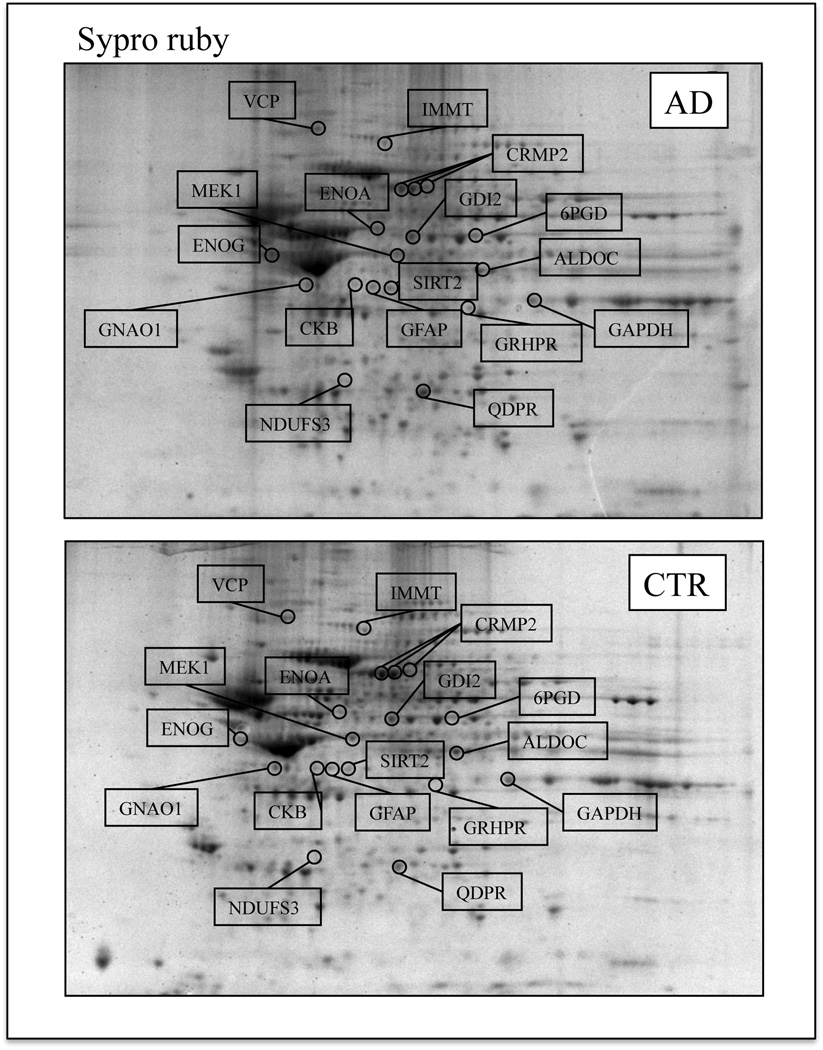
Figure 2.
Representative 2D protein expression maps of AD (top) and CTR (bottom) hippocampus. Gels were stained using Sypro Ruby fluorescent dye. The spots showing altered phosphorylation levels between AD and control are labeled.
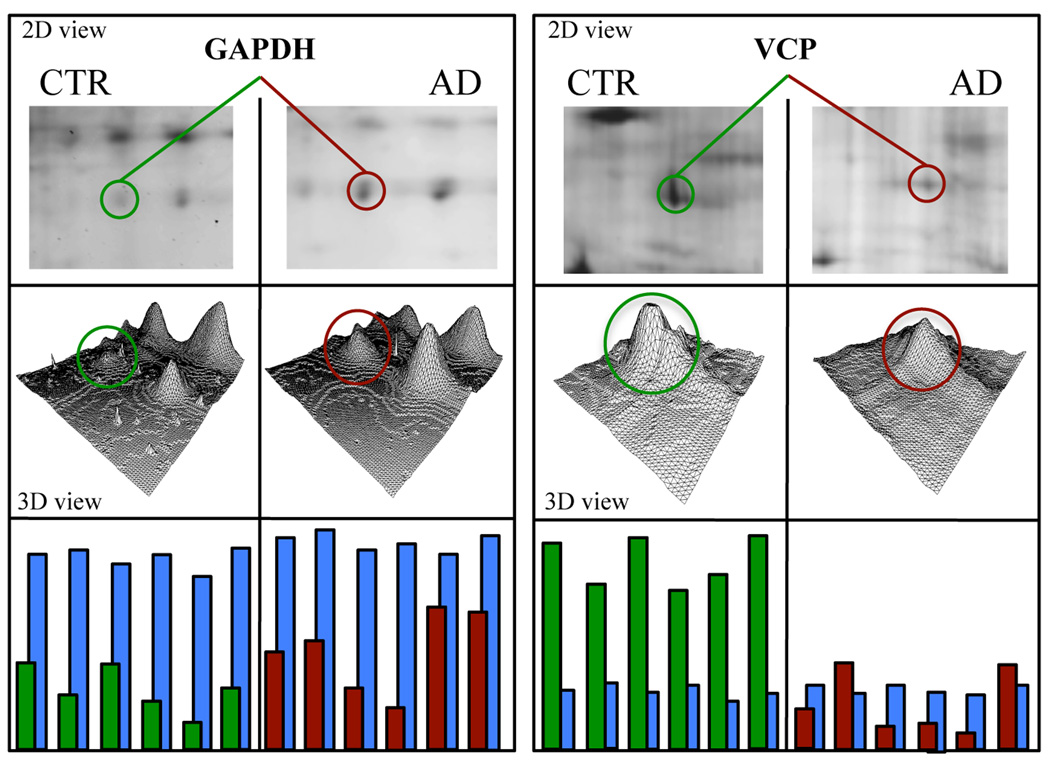
Figure 3.
Representative enlarged images of selected spots from CTR and AD hippocampus showing the altered phosphorylation of two selected proteins: GAPDH and tER ATPase. In the figure are shown the position of the respective protein spot in the gel in the 2D image, 3D density graphs elaborated by PD-Quest software, and the histograms with phosphorylation (green and red) and expression (blue) intensity bars of each of the 6 samples per group.
DISCUSSION
Increasing evidence indicates that aberrant protein phosphorylation occurs in AD, resulting from the concomitant alteration of protein kinase and phosphatase machinery [34, 35]. In AD brain, protein kinases such as glycogen synthase kinase cyclin-dependent kinase 5, dual-specific tyrosine regulated kinase and mitogen-activated protein kinases (MAPKs) are increased in expression and/or activity, whereas decreased activity was observed for protein phosphatases such as PP1, PP2A and PP5 [36, 37]. The hyperphosphorylation of Tau is one of the most extensively studied abnormal phosphorylation in neurodegenerative disease and has been a major focus of research and drug development [38]. The level of Tau phosphorylation is regulated dynamically by several kinases and phosphatases and any imbalance between these enzymes cause Tau hyperphosphorylation affecting its biological function [39–42]. Recent studies suggest that alterations of phosphorylation occurs on several brain proteins other that tau affecting their functionality and contributing to AD progression [34]. Altered phosphorylation of neurofilaments, microtubule-associated proteins 1B, APP, PS1, syntaxin 1 and amphiphysin II, have been associated with AD pathogenesis [10, 43, 44].
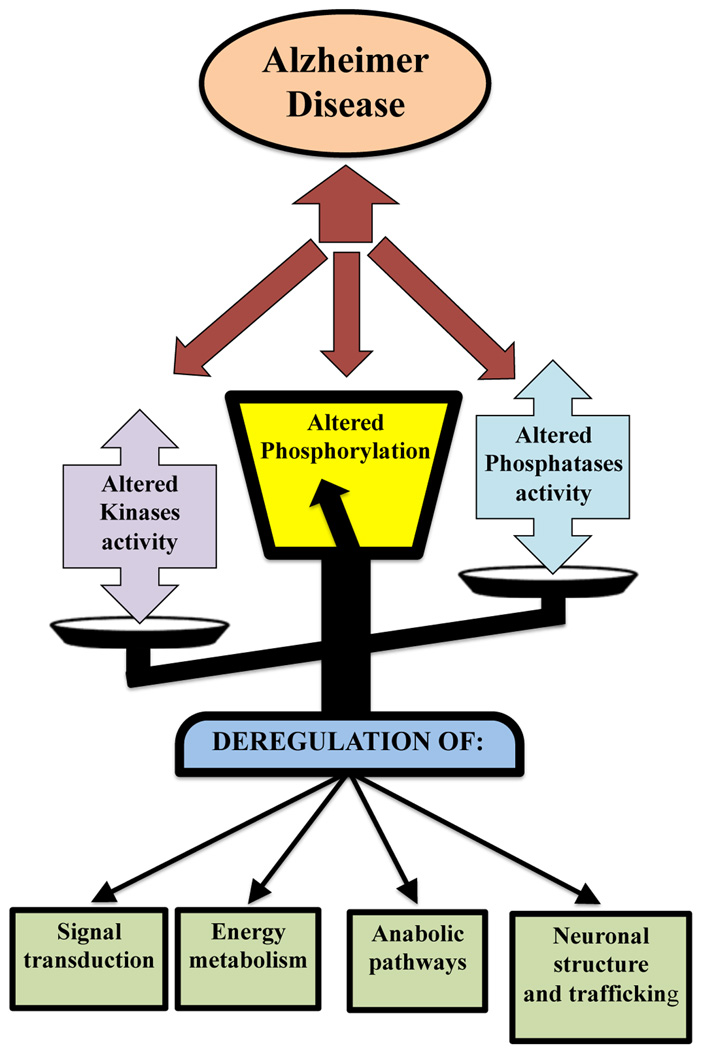
The use of proteomics coupled with Pro-Q Diamond staining allow the identification of phosphorylated proteins and the corresponding changes in their phosphorylation levels (normalized to expression levels). This is the first study on the identification of brain proteins with quantitative modification of ser/thr/tyr phosphorylation in AD hippocampus compared to normal brain. We recently reported the phosphorylation levels of BVR-A in AD hippocampus confirming the specificity of the of the Pro-Q diamond staining by immunochemical methods [33]. We referred to Uniprot and Phosida internet database (www.uniprot.org; 141.61.102.18/phosida/index.aspx) [45, 46] to obtain detailed information on type and numbers of phosphorylated residues of the protein identified in this work. Some of the proteins identified are not documented as phosphoproteins. For this reason, NetPhosK (www.cbs.dtu.dk/services/NetPhosK), a software tool for potential phosphorylation sites recognition, was also consulted [47]. We grouped the identified proteins based on their biological and functional activity to link phosphoprotein alteration with the possible failure of molecular pathways in AD (Figure 4).
Figure 4.
Representative scheme of pathologic implications of aberrant phosphorylation in AD.
Signal transduction
The MAP kinase cascade plays a central role in integrating the signals from a diverse group of extracellular stimuli and proto-oncogenes to the nucleus where activation of specific transcription factors elicits cellular responses. Map kinase kinase 1 (MEK1) is a dual-specificity protein kinase that functions in a mitogen activated protein kinase cascade. Factors that activate MEK1 include growth factors like NGF, cytokines, chemokines, and phorbol ester, resulting in cellular proliferation and survival. Phosphorylation and autophosphorylation have been shown to play a central role in the activity of protein kinases such as MEK 1/2. Active forms of MEK1/2 and ERK1/2 were found to be co-distributed with the progressive accumulation of neurofibrillary changes in several AD brain regions comprise hippocampus [48–50]. The neurofibrillary pathology positive for active forms of MEK1/2 and ERK1/2 was also found to occur prior to the deposition of A β [48, 49]. Among the pharmacological agents used to inhibit several kinase cascades, only MEK-ERK1/2 inhibition significantly attenuated A β 42 oligomer-induced toxicity [51].
G proteins and their receptors (GPCRs) form one of the most prevalent signaling systems in mammalian cells, regulating systems as diverse as sensory perception, cell growth and hormonal regulation. The alpha subunit and the beta/gamma dimer go on to activate distinct downstream effectors, such as adenylyl cyclase, phosphodiesterases, phospholipase C, and ion channels [52].
GDI2 constitute a small family of highly conserved proteins. Rab-GDP dissociation inhibitor is a member of the GDP dissociation inhibitor (GDI) family that controls the recycling of the Rab GTPases involved in membrane trafficking of molecules between cellular organelles. GDIs slow the rate of dissociation of GDP from rab proteins and release GDP from membrane-bound rabs [53, 54]. We previously found a decrease of Con-A-associated GDI levels in AD hippocampus [55].
Altered phosphorylation levels of proteins involved in signal transduction pathways could impair their functionality leading to profound consequences on neuronal survival.
Energy metabolism
A significant fraction of the proteins identified in this study with altered phosphorylation are involved in energy metabolism such as glycolysis or the pentose shunt. Previous proteomics studies from our lab reported significant increase in the oxidation of proteins involved in glycolysis and the TCA cycle, and a significant decrease in TCA enzyme levels in AD brain [56, 57]. In the current study we show changes in phosphorylation of four glycolytic proteins in AD hippocampus: alpha- and gamma-enolase, fructose bisphosphate aldolase and glyceraldehyde 3-phoshate dehydrogenase. All of these enzymes are well known phosphoproteins.
GAPDH, as a key enzyme in glycolysis, catalyzes the NAD-mediated oxidative phosphorylation of glyceraldehyde 3-phosphate to 1,3- diphosphoglycerate. However, it is also involved in several biological functions, including the organization of the cytoskeleton and regulation of endocytosis, binding and transport of tRNA, and regulation of translation, transcription, replication, DNA repair, cell proliferation, and apoptosis [58, 59]. GAPDH could be phosphorylated by several kinases such as PKC or AMPK and it can undergo autophosphorylation [60, 61]. Previously it has been reported that GAPDH is the GABA receptor endogenous kinase, providing a direct functional link between glycolysis and neurotransmission [62].
Enolase is a key glycolytic enzyme that catalyzes the dehydration of 2-phosphoglycerate to phosphoenolpyruvate, which in turn is converted by pyruvate kinase to pyruvate for entry the TCA cycle. Alpha-enolase is found in almost all human tissues, whereas gamma-enolase is only found in neurons and neuroendocrine tissues. Recently, accumulating evidence revealed that, in addition to its innate glycolytic function, enolase plays an important role in several biological and physiological processes [63], including activation of the ERK 1/2-associated pro-survival pathway. Consistent with our present results, alpha and gamma enolase were found with altered phosphorylation in the hippocampus of an AD mouse model [64]. Previous studies demonstrated that both GAPDH and enolase are oxidatively modified in AD brain samples compared with CTR [65].
ALDOC catalyzes the reversible aldol cleavage of fructose-1,6-biphosphate and fructose 1-phosphate to dihydroxyacetone phosphate and either glyceraldehyde-3-phosphate or glyceraldehyde, respectively. ALDOC is expressed specifically in the hippocampus and Purkinje cells of the brain and its hyperphosporylation related to AD progression has not been observed yet.
We found the altered phosphorylation of the iron-sulfur protein (IP) components 3 of mitochondrial NADH:ubiquinone oxidoreductase (complex I). Complex I is the "entry enzyme complex" of oxidative phosphorylation in the mitochondria, located in the inner mitochondrial membrane where it catalyzes the transfer of electrons from NADH to coenzyme Q (CoQ). A generalized depression of all electron transport chain complexes occurs in AD brain mitochondria [66][67]. Defects in the expression of mitochondrial subunits of NADH dehydrogenase enzyme complex in the AD brain were reported [68], and there are evidences that Complex I defects may play a role in the etiology of neurodegenerative disease possibly because of the formation of reactive oxygen species. Of course in AD brain extensive oxidative stress is found [12, 56].
The creatine kinase BB system is the most important immediate energy buffering and transport system in neuronal tissue with kinase activity toward creatine and phosphocreatine. CKs are primary targets of oxidative damage, and is also well established that oxidative modification of CK-BB decreases its activity in AD and other neurodegenerative diseases [69–71]. Hemmer et al. proposed CK autophosphorylation [72] and in vitro phosphorylation of CK BB by protein kinase C has been shown. The observations of altered CK activity in the AD brain are supported by a number of reports in the literature [73, 74] contributing to decreased neurotransmission and neuronal loss observed in the AD brain [73, 74].
Taken together, these results suggest that the altered phosphorylation of proteins involved in energy metabolism might be one of the main events associated with AD leading to reduced activity of metabolic pathways and therefore decreased ATP production, as is well known in AD.
Anabolic pathways
Dihydropteridine reductase (QDPR) participates in the pathway that recycles tetrahydrobiopterin (BH4) an essential cofactor for the synthesis of dopamine, noradrenaline, adrenaline and 5-hydroxytryptophan. Barford et al. and Jeeps et al. have described disturbed BH4 metabolism in several neurological diseases, including AD, [75, 76]. Katoh et al. showed that QDPR undergo phosphorylation by Ca2+-activated kinases consistent with our data [77].
6-Phosphogluconate dehydrogenase (6PGD) is an oxidative carboxylase that catalyzes the decarboxylating reduction of 6-phosphogluconate into ribulose 5-phosphate in the presence of NADP. This reaction is a component of the hexose mono-phosphate shunt and pentose phosphate pathways (PPP) essential for formation of the cellular reducing agent, NADPH [78]. G6PD is considered an early marker of oxidative stress because it is capable of responding rapidly to the increased demand of NADPH required for the maintenance of the cellular redox state. Phosphogluconate dehydrogenase activity is up-regulated in pertinent cortical regions of AD brains as a compensatory reaction to highly oxidative environments [79]. Phosphorylation of 6PGD has been previously demonstrated [80].
In addition to 6PGD, glyoxylate reductase also is involved in oxidoreductase reactions specifically those acting on the CH-OH group of donors with NAD+ or NADP+ as acceptors. The glyoxylate cycle describes an important subset of reactions involved in biosynthesis of carbohydrates from fatty acids or two-carbon precursors which enter the system as acetyl-coenzyme A [81].
We speculate that alteration in these systems due to changes in phosphorylation observed in the current study in AD could affect important intermediary metabolic pathways and be detrimental to neurons.
Neuronal structure and trafficking
CRMP2 is involved in the formation, outgrowth and guidance of neurites. CRMP2 has been shown to be phosphorylated by cyclin-dependent kinase 5, glycogen synthase kinase 3β and by Rho kinase II, thereby mediating neurite retraction [82–84]. Phosphorylation of CRMP2 disturbs its association with tubulin heterodimers that cannot be transported to the plus ends of microtubules for assembly. CRMP2 localizes with neurofibrillary tangles in human AD cortex and is considered an early event in the pathogenesis of the disease [85, 86]. Interestingly, CRMP2 is hyperphosphorylated at the Cdk5 and GSK3 sites like tau protein, in brain tissue from human AD subjects and in some mouse models of AD [85]. CRMP2 is also an oxidatively modified protein in AD brain [87].
The mitochondrial inner membrane protein (IMMT) is an integral component of the inner mitochondrial membrane with role in the regulation of the architecture of mitochondrial cristae. Down-regulation of IMMT resulted in a drastic change in the organization of the inner membrane. A study in cortical brain samples of fetal Down syndrome showed a double-fold reduction of IMMT, highlighting its importance for normal mitochondrial function [88]. IMMT is phosphorylated at several serine residues [89].
GFAP is the major constituent of glial intermediate filaments that form the cytoskeleton of mature astrocytes. Its expression is greatly increased during chronic inflammation in the brain. The self-assembly and degradation of GFAP is strictly regulated by phosphorylation and dephosphorylation, and soluble phosphorylated GFAP is thought to increase maintenance of the equilibrium with polymerized nonphosphorylated GFAP [90, 91]. Hyperphosphorylation of GFAP has been detected in rat hippocampus after ischemic insult [92]. Phosphorylation has been suggested to be protective against the degradation of GFAP [90] and during neurodegeneration may reflect increased neuroinflammation.
The transitional endoplasmic reticulum ATPase (VCP) is an essential component of a complex of proteins involved in the ATP-dependent formation of transition vesicles from the transitional endoplasmic reticulum [93]. VCP is also involved in a wide variety of other cellular activities, including Golgi reassembly after mitotic division, endoplasmic reticulum- (ER-) associated degradation (ERAD), ubiquitin-proteasome-mediated proteolysis. VCP is a mediator of ER stress-induced cell death [94]. It is a phosphoprotein and several ser/thr/tyr residues are known to be involved in its activity [95]. Several studies showed its involvement in the neurodegeneration process characterized by protein misfolding and aggregation, in particular linked to frontotemporal dementia [96].
Sirtuins possess either histone deacetylase or mono-ribosyltransferase activity and are a conserved class found in organisms ranging from bacteria to humans. Phosphorylation, on several residues, plays a key role in regulating the biological function of a human sirtuin 2 [97], controlling its expression levels and activity [98]. Sirtuin 1, the most closely related homologue of Sirtuin 2, is phosphorylated on several sites by cyclinB/Cdk1, and this phosphorylation regulates its deacetylase activity and affects cell proliferation [99]. Furthermore, SIRT1 also induces the Notch signaling pathway, involved in repairing neuronal damage in the brain [100]. SIRT2 knockdown mice showed impaired cell motility by deacetylating α -tubulin, influencing microtubule dynamics and inhibiting neurite outgrowth [97, 101, 102]. SIRT2 may interact with and modulate other cytoskeletal proteins that participate in axonal transport deficits characteristic in neurons exhibiting AD symptoms [103].
Altered phosphorylation of these structural proteins, as observed in the present study, could be related to the known altered neuronal architecture and the altered neurotransmission in AD.
Several hypotheses have been proposed to clarify the altered phosphorylation events in AD. A role has been suggested for insulin signaling that appears impaired in AD brain. The binding of insulin to insulin receptor leads to its autophosphorylation and activation of its kinase activity, and this in turn phosphorylates different substrates activating various downstream signaling partners and signaling cascade like MAPK [104]. In other recent studies has been suggested that the impairment of brain glucose metabolism contributes to the pathogenesis of Alzheimer’s disease also through altered phosphorylation. Impaired brain glucose metabolism decreases HBP flux and, consequently, decreases O-GlcNAcylation, leading to abnormal phosphorylation [105]. Interestingly, O-linked glycosylation occurs on cytoplasmic and nucleoplasmic proteins through the addition of a single N-acetylglucosamine (O-GlcNAc) to Ser/Thr residues. Several studies suggest that nucleoplasmic and cytoplasmic O-linked glycosylation work in concert with phosphorylation to regulate proteins [106, 107]. A reciprocal relationship between O-GlcNAcylation and phosphorylation has been observed in many proteins and the eminent example of this fact is the alteration of Tau o-glycosylation/phosphorylation status during AD with the decrease of the former and the increase of the latter [108]. Liu et al showed that total levels of glycosylation decrease in cerebrum but not in the cerebellum in AD, linking this status with the pathologic condition [108]. We previously analyzed the glycosylation status of brain proteins in AD through fractionation of the proteome with ConA and WGA lectins that allow the identification of proteins with N- and O- linked glycosylation [55, 109]. Comparing this current study with our previous investigation, we observe only for gamma enolase the interchange between O-glycosylation and phosphorylation in AD hippocampus [109]. Given the pleiotropic nature of gamma enolase [63], this interplay may be profound for AD.
CONCLUSIONS
Our proteomics study shows that altered phosphorylation in AD samples compared to CTR occurs on proteins that are involved in energy metabolism and ATP production, signal transduction and neuronal structure. Alterations in functions of such proteins with altered phosphorylation status conceivably could compromise various neuronal functions, though additional studies would be necessary to prove this notion. Indeed, a more specific and detailed picture of AD altered phosphorylation events would need additional studies focused on the effects on the molecular pathways involved and the primary mechanisms controlling the activities of both protein kinases and phosphatases. Within this frame it is expected that the list of altered phosphorylated proteins, protein kinases and protein phosphatases in AD would continue to rise, increasing the importance of altered phosphorylation in the pathogenesis of AD. A potential approach for the treatment of AD over the past decade has been focused on abnormal Tau hyperphosphorylation by inhibiting the activity of Tau kinases or by increasing phosphatase activity in the brain [34]. Hence, targeting protein kinase/phosphatase might represent a promising treatment strategy for AD as has already been suggested in cancer treatments. In this perspective, a better understanding of phosphorylation events is hoped to eventually lead to the development of novel therapeutic strategies for AD.
Supplementary Material
TABLE 3.
In the first four columns expression and phosphorylation intensity raw data in AD and CTR samples are reported. In the fifth and sixth columns phosphorylation data are normalized to expression. In the last column phosphorylation difference between CTR and AD is reported.
| PROTEIN | Expression Density (arbitrary unit) |
Phosphorylation density (arbitrary unit) |
Phospho/exp | Fold Ph/exp | ||||
|---|---|---|---|---|---|---|---|---|
| CTR | AD | CTR | AD | CTR | AD | AD/CTR | ||
| ENOA | 766±55 | 853±62 | 206±108 | 571±44 | 0.27 | 0.67 | 2.50 | |
| ENOG | 4851±119 | 4454±225 | 97±72 | 3652±332 | 0.02 | 0.82 | 29.6 | |
| GAPDH | 5883±221 | 6102±339 | 2294±194 | 3295±344 | 0.39 | 0.54 | 1.38 | |
| CKB | 699±202 | 548±165 | 1076±95 | 208±197 | 1.54 | 0.38 | 0.25 | |
| SIRT2 | 854±87 | 903±129 | 196±98 | 126±34 | 0.23 | 0.14 | 0.63 | |
| ALDOC | 2791±46 | 2882±166 | 223±87 | 922±84 | 0.08 | 0.32 | 4.14 | |
| NDUFS3 | 719±112 | 884±52 | 187±101 | 27±56 | 0.26 | 0.03 | 0.12 | |
| 6PGD | 2093±173 | 1865±119 | 63±24 | 578±44 | 0.03 | 0.31 | 10.1 | |
| GRHPR | 1572±158 | 1345±175 | 597±93 | 67±86 | 0.38 | 0.05 | 0.12 | |
| QDPR | 3626±223 | 3412±246 | 2429±211 | 1262±329 | 0.67 | 0.37 | 0.55 | |
| GFAP | 1447±66 | 1564±137 | 274±112 | 391±76 | 0.19 | 0.25 | 1.34 | |
| IMMT | 1296±107 | 1348±124 | 285±49 | 54±25 | 0.22 | 0.04 | 0.20 | |
| VCP | 1256±178 | 1489±303 | 9551±1052 | 1130±229 | 7.16 | 0.78 | 0.11 | |
| CRMP2 | 2987±258 | 3011±221 | 358±75 | 2679±195 | 0.12 | 0.89 | 7.17 | |
| 2889±231 | 2832±229 | 433±104 | 2407±133 | 0.15 | 0.85 | 5.68 | ||
| 1108±78 | 1254±187 | 288±126 | 840±117 | 0.26 | 0.67 | 2.52 | ||
| MEK1 | 1683±276 | 1788±231 | 336±64 | 769±139 | 0.20 | 0.43 | 2.15 | |
| GNAO1 | 1053±108 | 986±155 | 53±39 | 897±118 | 0.05 | 0.91 | 16.4 | |
| GDI2 | 1218±162 | 1164±133 | 1242±190 | 722±89 | 1.02 | 0.62 | 0.61 | |
AKNOWLEDGEMENTS
This work was supported in part by a NIH grant to D.A.B. [AG-05119]. F.D.D. was supported by a Fellowship from Istituto Pasteur – Fondazione Cenci Bolognetti. We are grateful to the Neuropathology Core of the University of Kentucky Alzheimer's Disease Clinical Center for providing well-characterized specimens for this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no financial/commercial conflicts of interests with the results of this study.
REFERENCES
- 1.Walsh CT. Posttranslational modification of proteins: expanding nature's inventory. Greenwood Village, CO: Roberts and Company Publishers; 2006. [Google Scholar]
- 2.Zhou H, Elisma F, Denis NJ, Wright TG, Tian R, Hou W, et al. Analysis of the subcellular phosphoproteome using a novel phosphoproteomic reactor. J Proteome Res. 2010;9:1279–1288. doi: 10.1021/pr900767j. [DOI] [PubMed] [Google Scholar]
- 3.Cohen P. The origins of protein phosphorylation. Nat Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 4.Ruan L, Wang GL, Chen Y, Yi H, Tang CE, Zhang PF, et al. Identification of tyrosine phosphoproteins in signaling pathway triggered TGF-a by using functional proteomics technology. Med Oncol. 2010 doi: 10.1007/s12032-009-9394-6. [DOI] [PubMed] [Google Scholar]
- 5.Grimsrud PA, Swaney DL, Wenger CD, Beauchene NA, Coon JJ. Phosphoproteomics for the masses. ACS Chem Biol. 2010;5:105–119. doi: 10.1021/cb900277e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers LD, Foster LJ. Phosphoproteomics--finally fulfilling the promise? Mol Biosyst. 2009;5:1122–1129. doi: 10.1039/b905580k. [DOI] [PubMed] [Google Scholar]
- 7.Lemeer S, Heck AJ. The phosphoproteomics data explosion. Curr Opin Chem Biol. 2009;13:414–420. doi: 10.1016/j.cbpa.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 8.Morales MA, Watanabe R, Laurent C, Lenormand P, Rousselle JC, Namane A, et al. Phosphoproteomic analysis of Leishmania donovani pro- and amastigote stages. Proteomics. 2008;8:350–363. doi: 10.1002/pmic.200700697. [DOI] [PubMed] [Google Scholar]
- 9.Gannon J, Staunton L, O'Connell K, Doran P, Ohlendieck K. Phosphoproteomic analysis of aged skeletal muscle. Int J Mol Med. 2008;22:33–42. [PubMed] [Google Scholar]
- 10.Xia Q, Cheng D, Duong DM, Gearing M, Lah JJ, Levey AI, et al. Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J Proteome Res. 2008;7:2845–2851. doi: 10.1021/pr8000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovacech B, Zilka N, Novak M. New age of neuroproteomics in Alzheimer's disease research. Cell Mol Neurobiol. 2009;29:799–805. doi: 10.1007/s10571-009-9358-6. [DOI] [PubMed] [Google Scholar]
- 12.Markesbery WR. Oxidative stress hypothesis in Alzheimer's disease. Free Radic Biol Med. 1997;23:134–147. doi: 10.1016/s0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- 13.Selkoe DJ. Amyloid beta-protein and the genetics of Alzheimer's disease. J Biol Chem. 1996;271:18295–18298. doi: 10.1074/jbc.271.31.18295. [DOI] [PubMed] [Google Scholar]
- 14.Kowall NW, McKee AC, Yankner BA, Beal MF. In vivo neurotoxicity of beta-amyloid [beta(1–40)] and the beta(25–35) fragment. Neurobiol Aging. 1992;13:537–542. doi: 10.1016/0197-4580(92)90053-z. [DOI] [PubMed] [Google Scholar]
- 15.Pike CJ, Cummings BJ, Cotman CW. beta-Amyloid induces neuritic dystrophy in vitro: similarities with Alzheimer pathology. Neuroreport. 1992;3:769–772. doi: 10.1097/00001756-199209000-00012. [DOI] [PubMed] [Google Scholar]
- 16.Butterfield DA. beta-Amyloid-associated free radical oxidative stress and neurotoxicity: implications for Alzheimer's disease. Chem Res Toxicol. 1997;10:495–506. doi: 10.1021/tx960130e. [DOI] [PubMed] [Google Scholar]
- 17.Lauderback CM, Hackett JM, Huang FF, Keller JN, Szweda LI, Markesbery WR, et al. The glial glutamate transporter, GLT-1, is oxidatively modified by 4-hydroxy-2-nonenal in the Alzheimer's disease brain: the role of Abeta1–42. J Neurochem. 2001;78:413–416. doi: 10.1046/j.1471-4159.2001.00451.x. [DOI] [PubMed] [Google Scholar]
- 18.Markesbery WR. Neuropathological criteria for the diagnosis of Alzheimer's disease. Neurobiol Aging. 1997;18:S13–S19. doi: 10.1016/s0197-4580(97)00064-x. [DOI] [PubMed] [Google Scholar]
- 19.Glenner GG, Wong CW. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 20.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selkoe DJ. Cell biology of protein misfolding: the examples of Alzheimer's and Parkinson's diseases. Nat Cell Biol. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 22.Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci U S A. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee VM, Balin BJ, Otvos L, Jr, Trojanowski JQ. A68: a major subunit of paired helical filaments and derivatized forms of normal Tau. Science. 1985;251:675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- 24.Nukina N, Ihara Y. Proteolytic fragments of Alzheimer's paired helical filaments. J Biochem. 1985;98:1715–1718. doi: 10.1093/oxfordjournals.jbchem.a135443. [DOI] [PubMed] [Google Scholar]
- 25.Selkoe DJ, Ihara Y, Salazar FJ. Alzheimer's disease: insolubility of partially purified paired helical filaments in sodium dodecyl sulfate and urea. Science. 1982;215:1243–1245. doi: 10.1126/science.6120571. [DOI] [PubMed] [Google Scholar]
- 26.Wischik CM, Novak M, Thogersen HC, Edwards PC, Runswick MJ, Jakes R, et al. Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc Natl Acad Sci U S A. 1988;85:4506–4510. doi: 10.1073/pnas.85.12.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryu SI, Kim WK, Cho HJ, Lee PY, Jung H, Yoon TS, et al. Phosphoproteomic analysis of AML14.3D10 cell line as a model system of eosinophilia. J Biochem Mol Biol. 2007;40:765–772. doi: 10.5483/bmbrep.2007.40.5.765. [DOI] [PubMed] [Google Scholar]
- 28.Kang TH, Bae KH, Yu MJ, Kim WK, Hwang HR, Jung H, et al. Phosphoproteomic analysis of neuronal cell death by glutamate-induced oxidative stress. Proteomics. 2007;7:2624–2635. doi: 10.1002/pmic.200601028. [DOI] [PubMed] [Google Scholar]
- 29.Steinberg TH, Agnew BJ, Gee KR, Leung WY, Goodman T, Schulenberg B, et al. Global quantitative phosphoprotein analysis using Multiplexed Proteomics technology. Proteomics. 2003;3:1128–1144. doi: 10.1002/pmic.200300434. [DOI] [PubMed] [Google Scholar]
- 30.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 31.Thongboonkerd V, McLeish KR, Arthur JM, Klein JB. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 2002;62:1461–1469. doi: 10.1111/j.1523-1755.2002.kid565.x. [DOI] [PubMed] [Google Scholar]
- 32.Stasyk T, Morandell S, Bakry R, Feuerstein I, Huck CW, Stecher G, et al. Quantitative detection of phosphoproteins by combination of two-dimensional difference gel electrophoresis and phosphospecific fluorescent staining. Electrophoresis. 2005;26:2850–2854. doi: 10.1002/elps.200500026. [DOI] [PubMed] [Google Scholar]
- 33.Barone E, Di Domenico F, Cenini G, Sultana R, Cini C, Preziosi P, et al. Biliverdin reductase-A protein levels and activity in the brains of subjects with Alzheimer disease and mild cognitive impairment. Biochim Biophys Acta. 2011;1812:480–487. doi: 10.1016/j.bbadis.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung SH. Aberrant phosphorylation in the pathogenesis of Alzheimer's disease. BMB Rep. 2009;42:467–474. doi: 10.5483/bmbrep.2009.42.8.467. [DOI] [PubMed] [Google Scholar]
- 35.Stoothoff WH, Johnson GV. Tau phosphorylation: physiological and pathological consequences. Biochim Biophys Acta. 2005;1739:280–297. doi: 10.1016/j.bbadis.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 36.Tian Q, Wang J. Role of serine/threonine protein phosphatase in Alzheimer's disease. Neurosignals. 2002;11:262–269. doi: 10.1159/000067425. [DOI] [PubMed] [Google Scholar]
- 37.Tanimukai H, Grundke-Iqbal I, Iqbal K. Up-regulation of inhibitors of protein phosphatase-2A in Alzheimer's disease. Am J Pathol. 2005;166:1761–1771. doi: 10.1016/S0002-9440(10)62486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong CX, Liu F, Grundke-Iqbal I, Iqbal K. Dysregulation of protein phosphorylation/dephosphorylation in Alzheimer's disease: a therapeutic target. J Biomed Biotechnol. 2006;2006:31825. doi: 10.1155/JBB/2006/31825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kopke E, Tung YC, Shaikh S, Alonso AC, Iqbal K, Grundke-Iqbal I. Microtubule-associated protein tau. Abnormal phosphorylation of a non-paired helical filament pool in Alzheimer disease. J Biol Chem. 1993;268:24374–24384. [PubMed] [Google Scholar]
- 40.Bretteville A, Ando K, Ghestem A, Loyens A, Begard S, Beauvillain JC, et al. Two-dimensional electrophoresis of tau mutants reveals specific phosphorylation pattern likely linked to early tau conformational changes. PLoS One. 2009;4:e4843. doi: 10.1371/journal.pone.0004843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong CX, Grundke-Iqbal I, Iqbal K. Dephosphorylation of Alzheimer's disease abnormally phosphorylated tau by protein phosphatase-2A. Neuroscience. 1994;61:765–772. doi: 10.1016/0306-4522(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 42.Liu F, Grundke-Iqbal I, Iqbal K, Gong CX. Contributions of protein phosphatases PP1, PP2A, PP2B and PP5 to the regulation of tau phosphorylation. Eur J Neurosci. 2005;22:1942–1950. doi: 10.1111/j.1460-9568.2005.04391.x. [DOI] [PubMed] [Google Scholar]
- 43.Standen CL, Brownlees J, Grierson AJ, Kesavapany S, Lau KF, McLoughlin DM, et al. Phosphorylation of thr(668) in the cytoplasmic domain of the Alzheimer's disease amyloid precursor protein by stress-activated protein kinase 1b (Jun N-terminal kinase-3) J Neurochem. 2001;76:316–320. doi: 10.1046/j.1471-4159.2001.00102.x. [DOI] [PubMed] [Google Scholar]
- 44.Verdile G, Gandy SE, Martins RN. The role of presenilin and its interacting proteins in the biogenesis of Alzheimer's beta amyloid. Neurochem Res. 2007;32:609–623. doi: 10.1007/s11064-006-9131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steen H, Jebanathirajah JA, Rush J, Morrice N, Kirschner MW. Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol Cell Proteomics. 2006;5:172–181. doi: 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 48.Veeranna, Kaji T, Boland B, Odrljin T, Mohan P, Basavarajappa BS, et al. Calpain mediates calcium-induced activation of the erk1,2 MAPK pathway and cytoskeletal phosphorylation in neurons: relevance to Alzheimer's disease. Am J Pathol. 2004;165:795–805. doi: 10.1016/S0002-9440(10)63342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pei JJ, Braak H, An WL, Winblad B, Cowburn RF, Iqbal K, et al. Up-regulation of mitogen-activated protein kinases ERK1/2 and MEK1/2 is associated with the progression of neurofibrillary degeneration in Alzheimer's disease. Brain Res Mol Brain Res. 2002;109:45–55. doi: 10.1016/s0169-328x(02)00488-6. [DOI] [PubMed] [Google Scholar]
- 50.Swatton JE, Sellers LA, Faull RL, Holland A, Iritani S, Bahn S. Increased MAP kinase activity in Alzheimer's and Down syndrome but not in schizophrenia human brain. Eur J Neurosci. 2004;19:2711–2719. doi: 10.1111/j.0953-816X.2004.03365.x. [DOI] [PubMed] [Google Scholar]
- 51.Chong YH, Shin YJ, Lee EO, Kayed R, Glabe CG, Tenner AJ. ERK1/2 activation mediates Abeta oligomer-induced neurotoxicity via caspase-3 activation and tau cleavage in rat organotypic hippocampal slice cultures. J Biol Chem. 2006;281:20315–20325. doi: 10.1074/jbc.M601016200. [DOI] [PubMed] [Google Scholar]
- 52.Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 2002;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 53.Shisheva A, Chinni SR, DeMarco C. General role of GDP dissociation inhibitor 2 in membrane release of Rab proteins: modulations of its functional interactions by in vitro and in vivo structural modifications. Biochemistry. 1999;38:11711–11721. doi: 10.1021/bi990200r. [DOI] [PubMed] [Google Scholar]
- 54.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Owen JB, Di Domenico F, Sultana R, Perluigi M, Cini C, Pierce WM, et al. Proteomics-determined differences in the concanavalin-A-fractionated proteome of hippocampus and inferior parietal lobule in subjects with Alzheimer's disease and mild cognitive impairment: implications for progression of AD. J Proteome Res. 2009;8:471–482. doi: 10.1021/pr800667a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Butterfield DA, Reed T, Newman SF, Sultana R. Roles of amyloid beta-peptide-associated oxidative stress and brain protein modifications in the pathogenesis of Alzheimer's disease and mild cognitive impairment. Free Radic Biol Med. 2007;43:658–677. doi: 10.1016/j.freeradbiomed.2007.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butterfield DA, Sultana R. Redox proteomics identification of oxidatively modified brain proteins in Alzheimer's disease and mild cognitive impairment: insights into the progression of this dementing disorder. J Alzheimers Dis. 2007;12:61–72. doi: 10.3233/jad-2007-12107. [DOI] [PubMed] [Google Scholar]
- 58.Butterfield DA, Hardas SS, Lange ML. Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer's disease: many pathways to neurodegeneration. J Alzheimers Dis. 2010;20:369–393. doi: 10.3233/JAD-2010-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- 60.Tisdale EJ. Glyceraldehyde-3-phosphate dehydrogenase is phosphorylated by protein kinase Ciota /lambda and plays a role in microtubule dynamics in the early secretory pathway. J Biol Chem. 2002;277:3334–3341. doi: 10.1074/jbc.M109744200. [DOI] [PubMed] [Google Scholar]
- 61.Nagradova NK. Study of the properties of phosphorylating D-glyceraldehyde-3-phosphate dehydrogenase. Biochemistry (Mosc) 2001;66:1067–1076. doi: 10.1023/a:1012472627801. [DOI] [PubMed] [Google Scholar]
- 62.Laschet JJ, Minier F, Kurcewicz I, Bureau MH, Trottier S, Jeanneteau F, et al. Glyceraldehyde-3-phosphate dehydrogenase is a GABAA receptor kinase linking glycolysis to neuronal inhibition. J Neurosci. 2004;24:7614–7622. doi: 10.1523/JNEUROSCI.0868-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Butterfield DA, Lange ML. Multifunctional roles of enolase in Alzheimer's disease brain: beyond altered glucose metabolism. J Neurochem. 2009;111:915–933. doi: 10.1111/j.1471-4159.2009.06397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takano M, Otani M, Sakai A, Kadoyama K, Matsuyama S, Matsumoto A, et al. Use of a phosphosensor dye in proteomic analysis of human mutant tau transgenic mice. Neuroreport. 2009;20:1648–1653. doi: 10.1097/WNR.0b013e328333b0e0. [DOI] [PubMed] [Google Scholar]
- 65.Sultana R, Poon HF, Cai J, Pierce WM, Merchant M, Klein JB, et al. Identification of nitrated proteins in Alzheimer's disease brain using a redox proteomics approach. Neurobiol Dis. 2006;22:76–87. doi: 10.1016/j.nbd.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 66.Parker WD, Jr, Parks J, Filley CM, Kleinschmidt-DeMasters BK. Electron transport chain defects in Alzheimer's disease brain. Neurology. 1994;44:1090–1096. doi: 10.1212/wnl.44.6.1090. [DOI] [PubMed] [Google Scholar]
- 67.Aksenov MY, Tucker HM, Nair P, Aksenova MV, Butterfield DA, Estus S, et al. The expression of several mitochondrial and nuclear genes encoding the subunits of electron transport chain enzyme complexes, cytochrome c oxidase, and NADH dehydrogenase, in different brain regions in Alzheimer's disease. Neurochem Res. 1999;24:767–774. doi: 10.1023/a:1020783614031. [DOI] [PubMed] [Google Scholar]
- 68.Chandrasekaran K, Hatanpaa K, Rapoport SI, Brady DR. Decreased expression of nuclear and mitochondrial DNA-encoded genes of oxidative phosphorylation in association neocortex in Alzheimer disease. Brain Res Mol Brain Res. 1997;44:99–104. doi: 10.1016/s0169-328x(96)00191-x. [DOI] [PubMed] [Google Scholar]
- 69.Sultana R, Perluigi M, Butterfield DA. Oxidatively modified proteins in Alzheimer's disease (AD), mild cognitive impairment and animal models of AD: role of Abeta in pathogenesis. Acta Neuropathol. 2009;118:131–150. doi: 10.1007/s00401-009-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perluigi M, Poon HF, Maragos W, Pierce WM, Klein JB, Calabrese V, et al. Proteomic analysis of protein expression and oxidative modification in r6/2 transgenic mice: a model of Huntington disease. Mol Cell Proteomics. 2005;4:1849–1861. doi: 10.1074/mcp.M500090-MCP200. [DOI] [PubMed] [Google Scholar]
- 71.Castegna A, Aksenov M, Aksenova M, Thongboonkerd V, Klein JB, Pierce WM, et al. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic Biol Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 72.Hemmer W, Furter-Graves EM, Frank G, Wallimann T, Furter R. Autophosphorylation of creatine kinase: characterization and identification of a specifically phosphorylated peptide. Biochim Biophys Acta. 1995;1251:81–90. doi: 10.1016/0167-4838(95)00083-7. [DOI] [PubMed] [Google Scholar]
- 73.David S, Shoemaker M, Haley BE. Abnormal properties of creatine kinase in Alzheimer's disease brain: correlation of reduced enzyme activity and active site photolabeling with aberrant cytosol-membrane partitioning. Brain Res Mol Brain Res. 1998;54:276–287. doi: 10.1016/s0169-328x(97)00343-4. [DOI] [PubMed] [Google Scholar]
- 74.Aksenov M, Aksenova M, Butterfield DA, Markesbery WR. Oxidative modification of creatine kinase BB in Alzheimer's disease brain. J Neurochem. 2000;74:2520–2527. doi: 10.1046/j.1471-4159.2000.0742520.x. [DOI] [PubMed] [Google Scholar]
- 75.Barford PA, Blair JA, Eggar C, Hamon C, Morar C, Whitburn SB. Tetrahydrobiopterin metabolism in the temporal lobe of patients dying with senile dementia of Alzheimer type. J Neurol Neurosurg Psychiatry. 1984;47:736–738. doi: 10.1136/jnnp.47.7.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeeps CM, Silcox A, Lloyd B, Clayton BE. Dihydropteridine reductase activity in dried blood spots: effects of aging and senile dementia of the Alzheimer type. J Clin Pathol. 1986;39:199–203. doi: 10.1136/jcp.39.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katoh S, Sueoka T, Yamamoto Y, Takahashi SY. Phosphorylation by Ca2+/calmodulin-dependent protein kinase II and protein kinase C of sepiapterin reductase, the terminal enzyme in the biosynthetic pathway of tetrahydrobiopterin. FEBS Lett. 1994;341:227–232. doi: 10.1016/0014-5793(94)80462-1. [DOI] [PubMed] [Google Scholar]
- 78.Toews ML, Kanji MI, Carper WR. 6-Phosphogluconate dehydrogenase. Purification and kinetics. J Biol Chem. 1976;251:7127–7131. [PubMed] [Google Scholar]
- 79.Martins RN, Harper CG, Stokes GB, Masters CL. Increased cerebral glucose-6-phosphate dehydrogenase activity in Alzheimer's disease may reflect oxidative stress. J Neurochem. 1986;46:1042–1045. doi: 10.1111/j.1471-4159.1986.tb00615.x. [DOI] [PubMed] [Google Scholar]
- 80.Dallocchio F, Matteuzzi M, Bellini T. Non-enzymic protein phosphorylation. Phosphorylation of 6-phosphogluconate dehydrogenase by acyl phosphates. Biochem J. 1982;203:401–404. doi: 10.1042/bj2030401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yokota A, Haga S, Kitaoka S. Purification and some properties of glyoxylate reductase (NADP+) and its functional location in mitochondria in Euglena gracilis z. Biochem J. 1985;227:211–216. doi: 10.1042/bj2270211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fujisawa H, Ohtani-Kaneko R, Naiki M, Okada T, Masuko K, Yudoh K, et al. Involvement of post-translational modification of neuronal plasticity-related proteins in hyperalgesia revealed by a proteomic analysis. Proteomics. 2008;8:1706–1719. doi: 10.1002/pmic.200700928. [DOI] [PubMed] [Google Scholar]
- 83.Cole AR, Soutar MP, Rembutsu M, van Aalten L, Hastie CJ, McLauchlan H, et al. Relative resistance of Cdk5-phosphorylated CRMP2 to dephosphorylation. J Biol Chem. 2008;283:18227–18237. doi: 10.1074/jbc.M801645200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 85.Petratos S, Li QX, George AJ, Hou X, Kerr ML, Unabia SE, et al. The beta-amyloid protein of Alzheimer's disease increases neuronal CRMP-2 phosphorylation by a Rho-GTP mechanism. Brain. 2008;131:90–108. doi: 10.1093/brain/awm260. [DOI] [PubMed] [Google Scholar]
- 86.Kanamori T, Matsukawa N, Kobayashi H, Uematsu N, Sagisaka T, Toyoda T, et al. Suppressed phosphorylation of collapsin response mediator protein-2 in the hippocampus of HCNP precursor transgenic mice. Brain Res. 2010;1355:180–188. doi: 10.1016/j.brainres.2010.07.081. [DOI] [PubMed] [Google Scholar]
- 87.Castegna A, Aksenov M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, et al. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part II: dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 88.Myung JK, Gulesserian T, Fountoulakis M, Lubec G. Deranged hypothetical proteins Rik protein, Nit protein 2 and mitochondrial inner membrane protein, Mitofilin, in fetal Down syndrome brain. Cell Mol Biol (Noisy-le-grand) 2003;49:739–746. [PubMed] [Google Scholar]
- 89.Zhao X, Leon IR, Bak S, Mogensen M, Wrzesinski K, Hojlund K, et al. Phosphoproteome analysis of functional mitochondria isolated from resting human muscle reveals extensive phosphorylation of inner membrane protein complexes and enzymes. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M110.000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takemura M, Gomi H, Colucci-Guyon E, Itohara S. Protective role of phosphorylation in turnover of glial fibrillary acidic protein in mice. J Neurosci. 2002;22:6972–6979. doi: 10.1523/JNEUROSCI.22-16-06972.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Inagaki M, Nakamura Y, Takeda M, Nishimura T, Inagaki N. Glial fibrillary acidic protein: dynamic property and regulation by phosphorylation. Brain Pathol. 1994;4:239–243. doi: 10.1111/j.1750-3639.1994.tb00839.x. [DOI] [PubMed] [Google Scholar]
- 92.Valentim LM, Michalowski CB, Gottardo SP, Pedroso L, Gestrich LG, Netto CA, et al. Effects of transient cerebral ischemia on glial fibrillary acidic protein phosphorylation and immunocontent in rat hippocampus. Neuroscience. 1999;91:1291–1297. doi: 10.1016/s0306-4522(98)00707-6. [DOI] [PubMed] [Google Scholar]
- 93.Wu XQ, Lefrancois S, Morales CR, Hecht NB. Protein-protein interactions between the testis brain RNA-binding protein and the transitional endoplasmic reticulum ATPase, a cytoskeletal gamma actin and Trax in male germ cells and the brain. Biochemistry. 1999;38:11261–11270. doi: 10.1021/bi990573s. [DOI] [PubMed] [Google Scholar]
- 94.Song C, Wang Q, Li CC. Characterization of the aggregation-prevention activity of p97/valosin-containing protein. Biochemistry. 2007;46:14889–14898. doi: 10.1021/bi700499j. [DOI] [PubMed] [Google Scholar]
- 95.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, et al. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weihl CC. Valosin Containing Protein Associated Fronto-Temporal Lobar Degeneration: Clinical Presentation, Pathologic Features and Pathogenesis. Curr Alzheimer Res. 2011 doi: 10.2174/156720511795563773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- 98.Pandithage R, Lilischkis R, Harting K, Wolf A, Jedamzik B, Luscher-Firzlaff J, et al. The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. J Cell Biol. 2008;180:915–929. doi: 10.1083/jcb.200707126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, et al. Phosphorylation regulates SIRT1 function. PLoS One. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 102.Dent EW, Gertler FB. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 2003;40:209–227. doi: 10.1016/s0896-6273(03)00633-0. [DOI] [PubMed] [Google Scholar]
- 103.Anekonda TS, Reddy PH. Neuronal protection by sirtuins in Alzheimer's disease. J Neurochem. 2006;96:305–313. doi: 10.1111/j.1471-4159.2005.03492.x. [DOI] [PubMed] [Google Scholar]
- 104.Deng Y, Li B, Liu Y, Iqbal K, Grundke-Iqbal I, Gong CX. Dysregulation of insulin signaling, glucose transporters, O-GlcNAcylation, and phosphorylation of tau and neurofilaments in the brain: Implication for Alzheimer's disease. Am J Pathol. 2009;175:2089–2098. doi: 10.2353/ajpath.2009.090157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu F, Shi J, Tanimukai H, Gu J, Grundke-Iqbal I, Iqbal K, et al. Reduced O-GlcNAcylation links lower brain glucose metabolism and tau pathology in Alzheimer's disease. Brain. 2009;132:1820–1832. doi: 10.1093/brain/awp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hart GW. Dynamic O-linked glycosylation of nuclear and cytoskeletal proteins. Annu Rev Biochem. 1997;66:315–335. doi: 10.1146/annurev.biochem.66.1.315. [DOI] [PubMed] [Google Scholar]
- 107.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 108.Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong CX. O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer's disease. Proc Natl Acad Sci U S A. 2004;101:10804–10809. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Di Domenico F, Owen JB, Sultana R, Sowell RA, Perluigi M, Cini C, et al. The wheat germ agglutinin-fractionated proteome of subjects with Alzheimer's disease and mild cognitive impairment hippocampus and inferior parietal lobule: Implications for disease pathogenesis and progression. J Neurosci Res. 2010;88:3566–3577. doi: 10.1002/jnr.22528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.