Abstract
The operating principles of complex regulatory networks are best understood with the help of mathematical modelling rather than by intuitive reasoning. Hereby, we study the dynamics of the mitotic exit (ME) control system in budding yeast by further developing the Queralt's model. A comprehensive systems view of the network regulating ME is provided based on classical experiments in the literature. In this picture, Cdc20–APC is a critical node controlling both cyclin (Clb2 and Clb5) and phosphatase (Cdc14) branches of the regulatory network. On the basis of experimental situations ranging from single to quintuple mutants, the kinetic parameters of the network are estimated. Numerical analysis of the model quantifies the dependence of ME control on the proteolytic and non-proteolytic functions of separase. We show that the requirement of the non-proteolytic function of separase for ME depends on cyclin-dependent kinase activity. The model is also used for the systematic analysis of the recently discovered Cdc14 endocycles. The significance of Cdc14 endocycles in eukaryotic cell cycle control is discussed as well.
Keywords: mathematical model, mitotic exit, budding yeast, oscillation, endocycles
1. Introduction
Cell cycle progression is largely regulated by protein phosphorylation and dephosphorylation catalysed by protein-kinases and protein-phosphatases [1]. Budding yeast has a single essential cyclin-dependent protein-kinase (called Cdc28 or Cdk1), which is activated by Cln- and Clb-type cyclins during the cycle. Cdk1–Cln complexes regulate the budding process while Cdk1–Clbs control DNA replication and mitosis in the chromosome cycle. Clb5–6 are the partners of Cdk1 during S phase, while Clb1–4 are the mitotic cyclins with Clb2 being the most important one [2].
Cdk1–Clb2 activity reaches its maximum during metaphase, when it contributes to the activation of the Polo-kinase (Cdc5) [3]. Subsequent mitotic progression (anaphase and mitotic exit (ME)) requires continuous dephosphorylation of mitotic phospho-proteins, which is achieved by the downregulation of Cdk1 activity and upregulation of Cdc14, a Cdk-counteracting phosphatase [4]. Both of these processes are dependent on the activation of the anaphase-promoting complex (or cyclosome, APC/C), an ubiquitin-ligase that promotes the destruction of the B-type cyclins (Clbs) and securin. While Clb destruction reduces Cdk1 activity, securin degradation is required for sister chromatid separation. Securin (Pds1 in budding yeast) is an inhibitor of separase (Esp1 in budding yeast), a protease specialized in the cleavage of sister chromatid cohesion. As Pds1 is degraded, Esp1 is activated, and cells transit into anaphase. Besides triggering sister chromatid separation, separase also leads to the activation of Cdc14 phosphatase. The phosphatase is kept inactive in the nucleolus by Net1 throughout the cell cycle until anaphase. After the onset of anaphase, Net1 is phosphorylated by two different signalling cascades, both of them activated by separase. The FEAR (Cdc Fourteen Early Anaphase Release) network acts first, whereas MEN (Mitotic Exit Network) is responsible for later release of Cdc14 [5]. FEAR includes the kinases Cdk1 and Cdc5 and the phosphatase PP2ACdc55. MEN centres around a GTPase (Tem1) regulated by its GAP (Bub2-Bfa1) and GEF (Lte1) and a kinase-cascade (Cdc15, Dbf2-Mob1).
Separase activates FEAR and MEN through its non-proteolytic and proteolytic functions, respectively [4,6–8]. The proteolytic function of separase causes spindle elongation by cohesin cleavage, which activates MEN by bringing Tem1 together with its activator Lte1 [6,9]. The proteolytic function of separase can be mimicked by inducing TEV protease in the presence of TEV-cleavable cohesins [10]. In contrast, the non-proteolytic function of separase is not reproduced by the ectopic induction of TEV and it is required for the downregulation of PP2ACdc55 phosphatase—an inhibitor of both FEAR and MEN [11].
A mathematical model with reduced number of components was developed previously for the budding yeast ME transition from metaphase (i.e. high Cdk1 activity) to G1 (i.e. low Cdk1 activity state) [11,12]. Queralt's model (QM) centres around the non-proteolytic function of separase (Esp1), which triggers a positive feedback in the activation of MEN by FEAR-induced release of Cdc14. Because of a reduced number of components in QM, only a limited number of experiments could be studied. In order to overcome this limitation, we have supplemented QM with other cell cycle regulators involved in the ME process. A disadvantage of the extended model is that it becomes very complex and loses the elegancy of the simple model. Furthermore, this complexity comes with large number of kinetic parameters whose values are unknown. On the other hand, the complex behaviour of real biological systems cannot be captured to any great extent by simple models, and the extended model does a better job in this respect.
We have expanded QM in a stepwise manner by adding more and more components and tested each extension with data from several relevant experimental situations. The kinetic parameters were determined by matching the temporal patterns of simulations with the experimental data. Finally, the parametrized model is used to analyse the molecular mechanisms of the recently discovered Cdc14 endocycles [13,14].
2. Model description
The model proposed here incorporates all the essential features of QM along with other key proteins and interactions controlling ME. At the heart of QM is the antagonism between Cdh1–APC and Cdk1–Clb2, with Cdh1 being the only G1 regulator in the model [11,12]. The budding yeast stoichiometric Cdk inhibitor (Sic1) is another important G1 regulator that acts in parallel with Cdh1 and is included in the present model. Sic1 synthesis is driven by the transcription factor, Swi5 [15,16]. Nuclear localization of Swi5 is inhibited by Cdk1 phosphorylation and promoted by the Cdc14 phosphatase [17]. Ubiquitin-dependent degradation of Sic1 requires prior phosphorylation by Cdk1–Clb or Cdk1–Cln complexes [18]. As Sic1 binds to the Cdk1–Clb complexes and inhibits their activities [19,20], Sic1 and Cdk1–Clbs have an antagonistic relationship. In contrast, Cln kinases are resistant to Sic1 inhibition [21].
Only the mitotic cyclin, Clb2, was included in the QM. Here, we have included Clb5 as well, which regulates many proteins in the exit process and which is degraded during ME [22]. Moreover, we have also distinguished in the model the Cln kinases, which are resistant to Sic1 inhibition and thus very different from Clb2 and Clb5 kinases. The transcription factors (TFs) for Clb5 (MBF) and Clns (SBF) are not separated in the present model (called SBF/MBF), because they are regulated very similarly [23]. Both TFs are inhibited after the ‘Start’ transition1 [23] and in the model SBF/MBF is inhibited by Cdk1–Clb complexes.
In contrast to the QM that assumed constitutive Clb2 synthesis, Clb2 synthesis is governed by an auto-activation loop [24,25] in which Clb2 activates its own TF, the Fkh2–Mcm1 Ndd1 complex [23], called Mcm1 in the model.
The QM describes Cdc14 release as a consequence of phosphorylation-dependent inactivation of Net1 by Cdk1–Clb2 and MEN kinases [26,27]. Net1 phosphorylation by Polo (Cdc5) has been incorporated into the present model, as being dependent on priming phosphorylation by Cdk1–Clb2 [28] or MEN kinases [14]. We assume that only the double-phosphorylated form of Net1 (phosphorylated by Cdk1–Clb2 or MEN and then by Polo) readily dissociates from Cdc14. Net1 phosphorylations are counteracted by PP2ACdc55 and Cdc14, which promote Cdc14 recapture by dephosphorylation of Net1. A detailed scheme of Net1 phosphorylation is illustrated in the electronic supplementary material.
The non-proteolytic function of separase (PP2ACdc55 downregulation) in QM [11] was supplemented with the separase proteolytic function, whereby it directly activates Tem1. By its proteolytic function (cohesin cleavage), separase induces spindle elongation, which brings MEN GTPase (Tem1) in the vicinity of its activating GEF (Lte1) [6].
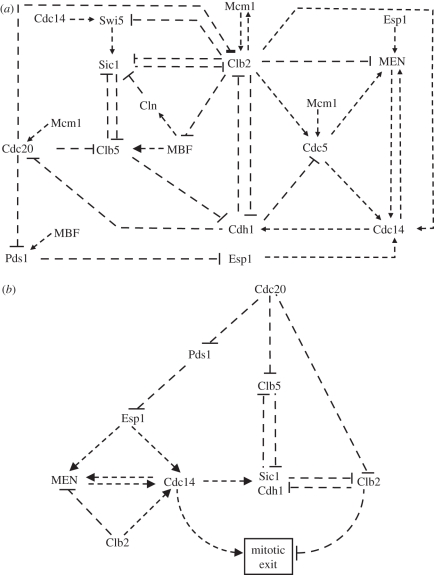
In addition to the nucleolar and nuclear forms of Cdc14 already present in the QM, we have now introduced a cytoplasmic form of Cdc14. The Net1 : Cdc14 complex is restricted to the nucleolus and Net1 double-phosphorylation releases Cdc14 into the nucleus. The shuttle of Cdc14 between nucleus and cytoplasm is regulated by MEN in the model as suggested in the literature [29,30]. All the reactions described so far constitute the wiring diagram of the ME model and are shown in figure 1a.
Figure 1.
The network controlling mitotic exit (ME) in budding yeast. (a) Biochemical and genetic interactions among cell cycle regulators controlling ME. Arrows and blunt-ended lines represent activation and inhibition, respectively. See text for more details. (b) The hierarchy of the ME controlling network. Cdc20–APC, at the top of the hierarchy, promotes ME via three different pathways defined by its substrates Pds1 (securin), Clb5 and Clb2. The ME process is amplified by Cdc14–MEN-positive feedback and the double-negative feedbacks between Cdk1–Clb kinases and their negative regulators (Sic1 and Cdh1).
We are using nonlinear, ordinary differential equations (ODEs) to describe the time rate of change of components in the regulatory network following our previously applied principles [11,24,25], which are explained in the electronic supplementary material. Nonlinearities, which are important for the dynamic behaviour, are introduced by Michaelis–Menten kinetics describing protein phosphorylation/dephosphorylation [31] and by stoichiometric inhibitors of Cdk1–Clb (Sic1) and Cdc14 (Net1). Of course, the validity of Michaelis–Menten kinetics could be questioned in many cases, but most of them are essential for the expected dynamic behaviour.
3. Results
3.1. The logic of the mitotic exit process in the budding yeast
Based on the data in the literature, we provide the following systems view of the ME process in the budding yeast (figure 1b). We place Cdc20–APC at the top of hierarchy in the network controlling the ME process (figure 1b). In the absence of Cdc20, cells are blocked in the metaphase of mitosis with short metaphase spindles. Further mitotic progression and ME requires Cdc20–APC-dependent poly-ubiquitylation of three proteins which are degraded by the proteasome. The three crucial substrates of Cdc20–APC are securin (Pds1 in budding yeast), the S phase (Clb5) and the M phase (Clb2) cyclins [1], which we use to define three converging pathways important for mitotic progression and exit. Along the first pathway, Pds1 degradation activates separase (Esp1 in budding yeast) that brings about anaphase [32]. Separase is also required for the activation of MEN [6] and Cdc14 phosphatase [8], both of them being essential for timely ME in the budding yeast. This part of the network is called the Cdc14 pathway. The two other pathways are defined by the degradation of mitotic (Clb2) and S phase (Clb5) cyclins and are named accordingly. Both of these cyclins in complex with Cdk1 are phosphorylating their target proteins, which need to be dephosphorylated for ME (figure 1b). The three pathways (Cdc14, Clb5 and Clb2) converge on substrates (like the TFs Swi5 and MBF) that are phosphorylated by Cdk1–cyclin complexes and dephosphorylated by Cdc14 [33]. As the extent of dephosphorylation of these TFs is a measure of the ratio of Cdc14 activity to Cdk1 activity, we use dephosphorylation of TFs as indicators of the ME process. Cdh1 and Sic1 proteins, which are also among substrates of the Cdk1–cyclin and Cdc14 [34], work as amplifiers of the exit process. These two proteins feed back negatively on the Clb5 and Clb2 pathways by downregulating the corresponding kinase activities (Sic1 inhibits both Cdk1–Clb2 and Cdk1–Clb5, while Cdh1–APC promotes Clb2 degradation), thereby creating double-negative feedback loops [24]. As a consequence of these antagonistic relationships, both Sic1 and Cdh1 behave like switches being high and active in G1 and low and inactive during S/G2/M phases of the cycle [24,25].
In the following, we analyse the process of ME in budding yeast by numerical simulation of different experimental situations reported in the literature. These simulations serve as a test and validation for the ME model to yield experimentally observed phenotypes. These simulations help us to quantify the role and contribution of individual proteins and their interaction in controlling ME (i.e. estimating kinetic parameters). We classify the ME experiments in the literature into two categories. In the first set of experiments, Cdc20 is deprived together with one or more of its substrates. In the second set of experiments, Cdc20 is re-induced in the presence of its non-degradable substrate. Both sets of experiments are compared with the simulation of Cdc20 block and release described in the next session.
3.2. Cdc20 block and release experiment
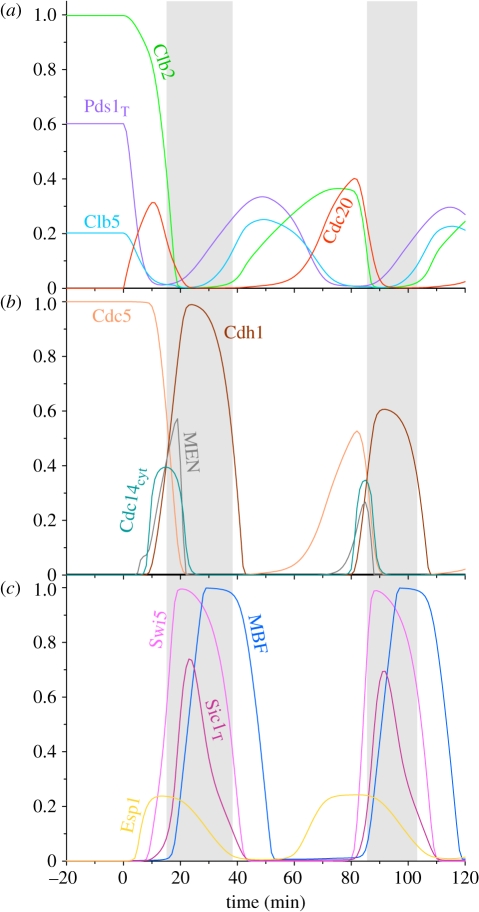
In the model, cells reach a steady state during the Cdc20 block, where the levels of all the APC/C substrates (Pds1, Clb5, Clb2 and Cdc5) are high (figure 2). In contrast, Cdc14, Cdh1, Esp1, MBF, Swi5 and MEN are all inactive, and Sic1 level is low. The stable steady state in the absence of Cdc20 is shown in figure 2 between −20 and 0 min, after which Cdc20 has been re-induced (at time zero).2 After Cdc20 induction, the rising Cdc20–APC activity causes an abrupt drop in the level of all Cdc20–APC substrates in the following order: Pds1, Clb5 and Clb2 (figure 2a). Pds1 degradation activates Esp1, which together with Clb2- and Polo-kinases promotes Cdc14 activation through the FEAR network. The rising Cdc14 to Clb2 activity ratio activates MEN, which induces further Cdc14 release (positive feedback) and its spread into the cytoplasm. Cytoplasmic Cdc14 activates Swi5 and Cdh1 by dephosphorylation, and the Swi5 TF promotes Sic1 synthesis. Cdh1–APC and Sic1 downregulate Clb2 activity by degradation and stoichiometric inhibition, and the cell exits from mitosis.
Figure 2.
Simulation of Cdc20 block and release experiments. The model was simulated without Cdc20 synthesis (ks20′ = ks20 = 0) which defines a stable steady state corresponding to metaphase block. The metaphase state is shown for 20 min, after which Cdc20 synthesis is induced and the simulation of mitotic cycles are continued for 120 min. The three panels show the fluctuation of different cell cycle regulators during Cdc20 block and release. The grey, shaded area corresponds to the G1 phase of the cell cycle on this and following figures as well.
After exiting from mitosis, cells enter into the next cycle, indicated in the model by the activation of MBF/SBF TF and disappearance of Sic1. In the new cycle, both S and M phase cyclins re-accumulate, and Clb2 activates its own TF, Mcm1, which induces Cdc5 and Cdc20 synthesis as well. Observe that in cycling cells (when Cdc20 is present) the Clb2 peak is only about half of the level (approx. 0.4 arb. units) that is reached during the Cdc20 block (1 arb. units), a feature of the model that is consistent with the experimental data [35,36].
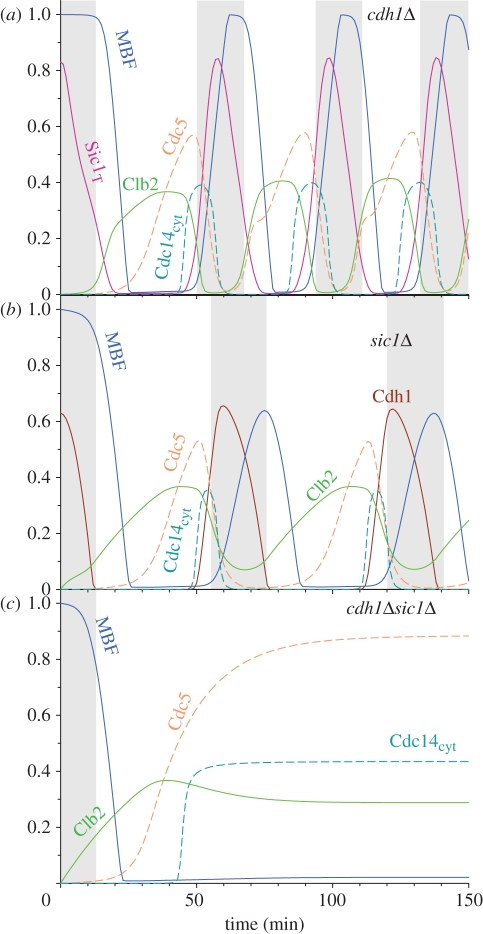
Cdc20 activation promotes ME through the three pathways shown in figure 1b. The Clb5 and Clb2 pathways are further inhibited by the activation of Cdh1–APC and Sic1. However, neither Cdh1 [37] nor Sic1 [21] are essential for ME, as their single deletions produce viable cells. The model captures these features of the budding yeast cell cycle, as both cdh1Δ and sic1Δ cells show cell cycle oscillations with short G1 (figure 3a,b). However, cdh1Δ sic1Δ double-mutants cannot cycle, and the model predicts that they are blocked with Cdc14 released into the cytoplasm and partial Cdk1 activity maintained (figure 3c). This cell cycle state could correspond to a late mitotic block [37] or an early G1 block [38]. This conclusion is sensitive to the Cdc20-dependent Clb2 degradation rate constant (kdclb2′). If the rate of degradation is increased, the model shows continuous cycling, which might correspond to disordered cycles observed by Lu & Cross [13].
Figure 3.
The effect of Cdh1 and Sic1 on ME. Simulations were started from a low Cdk1 activity G1-like state (grey, shaded area) and Cdh1 activation (kdcdh′ and kdcdh) or the rate of Sic1 synthesis (kssic′, kssic) were set to zero, which correspond to cdh1Δ (a) and sic1Δ (b). The combined effect of Cdh1 and Sic1 deletions (cdh1Δ sic1Δ) which blocks ME is shown in (c).
3.3. Suppression of Cdc20 requirement
Although Cdc20 is essential for mitotic progression and ME during a normal cycle, its requirement can be suppressed by the simultaneous deletion of the substrates whose degradations are the initial events along the three pathways triggered by Cdc20–APC activation. In the following, the stability of Cdc20 block is analysed when different Cdc20–APC substrates are deleted.
Shirayama et al. [22] showed that if Pds1 is deleted besides Cdc20 (cdc20Δ pds1Δ double mutant), the Cdc14 pathway is activated but cells are still blocked in mitosis. cdc20Δ pds1Δ double-mutants are only one step further in mitotic progression than the Cdc20 block [39] because they are blocked after anaphase in another stable steady state. The transition from cdc20Δ to cdc20Δ pds1Δ steady state is shown in electronic supplementary material, figure S1. According to the model, Esp1 activates Cdc14 which is released into the cytoplasm. Cytoplasmic Cdc14 dephosphorylates Cdh1–APC partially, causing Clb2 degradation to about 60 per cent of the value characteristic of the Cdc20 block. But this Clb2 level is still higher than the peak value during normal mitosis. Moreover, Clb5 is not degraded at all in the absence of Cdc20. The cumulative Cdk1–Clb kinase activities block the full activation of Cdh1–APC and the accumulation of Sic1; therefore cells cannot exit mitosis.
Overexpression of separase has similar effects to the deletion of Pds1. Esp1 overexpression [8] titrates out securin and causes the release of Cdc14. This leads to a late mitotic block in the model very similar to the cdc20Δ pds1Δ double-mutants (see electronic supplementary material, figure S2). Actually, after many hours of Esp1 overexpression some cells divide after exiting mitosis [8,29], but for simplicity we consider cdc20Δ ESP1op as a stable, late mitotic block. Bifurcation analysis of the QM showed that the stable steady state occupied by this double mutant is very close to the saddle-node bifurcation point for ME [12]. Therefore, a small decrease in one of the cyclins (Clb2 or Clb5) under separase overexpression can bring about ME in the model, but we decide to ignore these details.
Based on the Pds1 deletion (pds1Δ) and separase overexpression (ESP1op) experiments, we confirm that hyper-activation of the Cdc14 pathway is not sufficient to trigger ME in cells deprived of Cdc20.
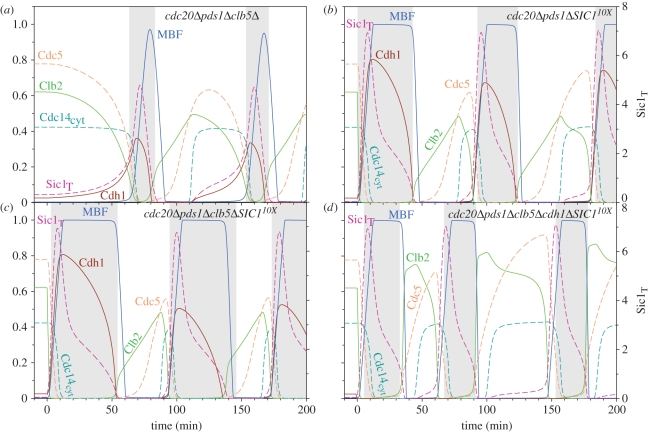
However, the stable, late mitotic block of cdc20Δ pds1Δ cells can be challenged further by removing another Cdc20 substrate from the network. By deletion of Clb5 (cdc20Δ pds1Δ clb5Δ triple mutant), the synergetic effect between the Clb5 and Cdc14 pathways releases the mitotic block [22]. In the simulation of the cdc20Δ pds1Δ clb5Δ triple mutant, Cdh1–APC gets activated and promotes the degradation of Clb2 and Polo (figure 4a), because we attribute a strong inhibition of Cdh1–APC by Cdk1–Clb5. As the lack of Cdc20 in the cdc20Δ pds1Δ clb5Δ triple mutant is compensated by Cdh1-dependent Clb2 destruction, Cdh1 becomes essential for cell cycling [22,40], and the model is consistent with this observation (see electronic supplementary material, figure S3a).
Figure 4.
Suppression of lack of Cdc20 activity by different genetic backgrounds: (a) cdc20Δ pds1Δ clb5Δ, (b) cdc20Δ pds1Δ SIC110x, (c)cdc20Δ pds1Δ clb5Δ SIC110X, (d)cdc20Δ pds1Δ clb5Δ cdh1Δ SIC110X. All simulations were started from cdc20Δ pds1Δ late mitotic steady state as initial conditions. In clb5Δ, the rate of unregulated and regulated Clb5 synthesis were halved, while in cdh1Δ, the activation of Cdh1 (kdcdh′ and kdcdh) was set to zero. In SIC110X, the rate of Swi5-dependent Sic1 synthesis was increased 10-fold (kssic′ = 2).
Similarly, separase overexpression in the absence of Clb5 (cdc20Δ clb5Δ ESP1op) triggers ME [8], and the model postdicts that this triple mutant can cycle in the absence of Cdc20–APC (see electronic supplementary material, figure S2b).
The function of the Clb5 pathway does not necessarily require Clb5 degradation. It can also be stimulated by overexpression of Sic1, the stoichiometric inhibitor of Cdk1–Clb5, which helps cdc20Δ pds1Δ to exit mitosis and cycle normally [40]. Indeed, the model simulates such an experimental situation where endogenous Sic1 is overexpressed by 10-fold in cdc20Δ pds1Δ double-mutants (figure 4b). When expressed at physiological level (as in cdc20Δ pds1Δ double-mutant), Sic1 cannot accumulate in the presence of Clb5 as Cdk1–Clb5 readily phosphorylates Sic1 and causes its rapid degradation. However, strong Sic1 overexpression overcomes the block on ME caused by Cdk1–Clb5 and allows Sic1 accumulation [40]. In the model, cdc20Δ pds1Δ cdh1Δ SIC110X cells cannot exit mitosis (electronic supplementary material, figure S3b) suggesting that cdc20Δ pds1Δ SIC110X cells are dependent on Cdh1–APC activity to reduce Clb2 level as observed in experiments [40].
The model is also capable of simulating the combined effect of Clb5 deletion and Sic1 overexpression in a cdc20Δ pds1Δ background (cdc20Δ pds1Δ clb5Δ SIC110X; figure 4c), which is known to produce viable cells [40]. The model captures the emergent property of the quadruple mutant, which is not characteristic for any triple mutants simulated before. In the quadruple mutant, Cdh1 becomes dispensable for ME and viability since Clb2 kinase activity can be downregulated by Sic1 only (see cdc20Δ pds1Δ clb5Δ cdh1Δ SIC110X simulation on figure 4d). These observations are consistent with the experimental findings of Thornton & Toczynski [40], who showed that cdc20Δpds1Δclb5Δ cdh1Δ SIC110X cells cycle in the absence of any APC activity.
3.4. The role of separase in Cdc14 activation
As explained above, Cdc14 activation is induced by two networks, FEAR and MEN, and separase controls both of them [4,34]. The FEAR network is considered non-essential for Cdc14 activation, as FEAR mutants can exit mitosis but only after a slight delay [7]. By contrast, MEN is an essential pathway as MEN mutants, like cdc15Δ, block late in mitosis [41].
In the absence of MEN (cdc15Δ), the model predicts that separase-induced Cdc14 release (cdc20Δ ESP1op cdc15Δ or cdc20Δ pds1Δ cdc15Δ; see electronic supplementary material, figure S4) is nuclear only and solely driven by the non-proteolytic function of separase. As recent data have suggested that the non-proteolytic function of separase is not essential [29], we have decided to explore the role of the two functions further.
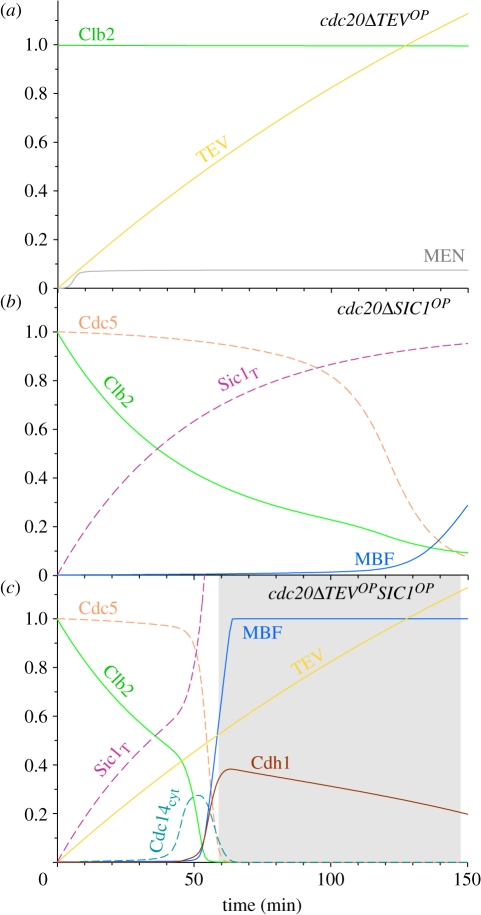
Experimentally, the proteolytic (MEN activating) and non-proteolytic (FEAR activating) roles of separase could be distinguished by ectopic TEV protease expression. TEV can activate Tem1 via the degradation of cohesins engineered with TEV-cleavage sites, but it lacks the non-proteolytic functions of separase [8]. TEV overexpression in Cdc20 block fails to activate Cdc14 and initiate ME [8,29], and the model reproduces this fundamental feature of ME in budding yeast (figure 5a). Why does an operational Tem1 not suffice to fully activate MEN and drive Cdc14 release? Although Tem1 is activated by TEV-induced spindle elongation, MEN activation is still blocked in the model by Clb2-dependent inhibition of Cdc15 [42]. In other words, the comparison of simulations between separase and TEV overexpressions in Cdc20 block situations, supports the notion that high Clb2 levels inhibit MEN and this inhibition could be manifested through a Cdk1 target protein in MEN, which in our case is Cdc15 [7]. Furthermore, it also highlights the significance of the positive feedback loop between MEN and Cdc14, which gets activated by the initial Cdc14 release induced by the non-proteolytic function of separase to overcome the Clb2 inhibition.
Figure 5.
Efficient exit from the cdc20Δ stable mitotic state requires both inhibition of Cdk1–Clbs and activation of Cdc14. These three simulations provide descriptions of the biochemical changes happening in Lu & Cross [29] experiments (their fig. 1). (a) A 10-fold TEV expression over separase level can only induce partial MEN activation but no Cdc14 release and ME in Cdc20 block. (b) Non-degradable Sic1 overexpressed (kssic = 0.02, kssic′ = 0.2, kdsic = 0.02, kdsic′ = kdsic′′ = kdsic′′′ = 0) in a Cdc20 block brings about ME after a long time because Cdc14 is not activated. (c) If the TEV protease is overexpressed together with non-degradable Sic1, Cdc14 gets activated as well and induces ME much earlier.
As the non-proteolytic function of separase is required to oppose the inhibitory effect that Clb2 exerts on MEN, it is not surprising that the non-proteolytic role of Esp1 can be bypassed by reducing Cdk1 activity, for example by the overexpression of Sic1, in Cdc20-arrested cells. However, this can be achieved only by the overexpression of the non-degradable (non-phosphorylable) form of Sic1 [29], because the phosphorylable form is rapidly destroyed by the high Cdk1 activities present in Cdc20-arrested cells according to our model prediction. The expression of non-degradable Sic1 (SIC1nd) brings about ME, but only after a long time (figure 5b and [29]). According to the model, ME induced by non-degradable Sic1 is not accompanied by Cdc14 activation (figure 5b). If non-degradable Sic1 overexpression is combined with TEV induction in the model, ME becomes as efficient (figure 5c) as in wild type (WT), in agreement with Lu & Cross [29]. In this setting, the role of Clb5 is less important, as shown by the fact that cells arrested in metaphase by Cdc20 depletion and lacking Clb5 but overexpressing TEV (cdc20Δ clb5Δ TEVop) cannot exit mitosis ([8] and electronic supplementary material, figure S5). We conclude that non-degradable Sic1 releases the ME block through inhibition of Cdk1–Clb2 rather than through Cdk1–Clb5. We predict with the model that MEN gets activated and Cdc14 is released in cdc20ΔTEVop only if Clb2 activity is reduced as in cdc20ΔTEVopSIC1ndop. According to model simulations, MEN-induced Cdc14 release is responsible for the acceleration of ME in cdc20ΔTEVopSIC1ndop (figure 5c) when compared with cdc20ΔSIC1ndop (figure 5b).
In summary, model simulations suggest that the essential nature of non-proteolytic separase function to activate MEN at high Clb2 levels could be compensated by the drop of Clb2-kinase activity either through Clb2 degradation or Sic1-dependent inhibition.
3.5. Cdc20 block and release in the presence of non-degradable substrates
Above, we have investigated which substrate(s) of Cdc20–APC need to be removed in the absence of Cdc20 for cells to exit mitosis. Here, we address the same problem using an opposite approach, namely which substrate(s) are sufficient to prevent ME if they are rendered resistant to degradation by Cdc20–APC.
Cdc20 deprivation and re-induction synchronize the ME process by activating the three exit pathways through substrate degradation (figure 1b). Each pathway can be turned off during this process by making Cdc20 substrates non-degradable, which allows us to analyse the necessary role of individual pathways in the exit process. Notice here that Cdc20–APC is operational and active, as opposed to the mutants analysed above.
3.6. Non-degradable securin
The Cdc14 pathway can be turned off by expressing a form of securin (Pds1) that carries a mutated destruction box [11,43]. Non-degradable Pds1 blocks both anaphase and ME (see electronic supplementary material, figure S6). Cdc14 is not activated and Clb2 is only partially degraded because of the lack of full Cdh1 activation. If the proteolytic function of separase is provided to these cells by TEV protease expression, cells exit mitosis [29] and the model is consistent with this experiment (figure 6a). Notice that the model does not keep track of the premature cohesin cleavage caused by constitutive TEV expression, and thus suggests that these cells can cycle normally, which is possibly not the case. Similar results are obtained with the deletion of Bub2 (see electronic supplementary material, figure S6) as both TEV expression and Bub2 deletion ectopically activate the Tem1 GTPase.
Figure 6.
The effect of non-degradable Cdc20 substrates on Cdc20-induced ME. Expression of securin (Pds1) with mutated destruction box (PDS1-mdb) blocks Cdc20-induced ME (see electronic supplementary material, figure S6a). However, simultaneous expression of TEV protease restores ME in the presence of PDS1-mdb (a). Blocking Cdc20–APC-dependent Clb5 degradation in the model simulates Clb5 without its destruction box, which does not interfere with Cdc20-induced ME (b). By setting the model parameter Clb2-kd to 0.5, we simulate the effect of KEN- and destruction box-deficient (dbΔ) Clb2 on mitotic progression (c).
These simulations (PDS1-mdb and PDS1-mdb TEVop) are consistent with the view that the proteolytic function of separase is both necessary and sufficient for ME while the non-proteolytic (FEAR) function is dispensable. However, notice that this conclusion is drawn from Cdc20 block and release experiments, where Clb2 is partially degraded by Cdc20–APC and thus Cdk inhibition on MEN is weakened. As a consequence, the non-proteolytic function of Esp1 is not needed to trigger the positive feedback loop in MEN activation.
In contrast to FEAR, MEN is required for Cdc20-induced ME, and in the absence of MEN Cdc14 release is only transient [7]. Simulation of the MEN (cdc15Δ) mutant with the model yields a transient nuclear release of Cdc14 (electronic supplementary material, figure S7). This transient Cdc14 release is caused by an incoherent feed-forward loop [11] by which Cdc20–APC both triggers and inhibits Cdc14 release. On the one hand, Cdc20–APC activates separase through securin degradation which, via the inhibition of PP2ACdc55, leads to Net1 phosphorylation. This is even further increased as both Cdk1–Clb2 and Cdc5 are high. As a consequence, Cdc14 is rapidly released to the nucleus. On the other hand, Cdc20–APC also promotes Clb2 degradation, and in the absence of MEN, the reduced level of Clb2 is insufficient to keep Net1 phosphorylated. Hence, Cdc14 gets re-sequestered to the nucleolus. Because Cdc14 does not reach cytoplasm, the activation (dephosphorylation) of the cytoplasmic forms of Swi5 and Cdh1 is compromised.
The lack of MEN activity (e.g. cdc15Δ) can be compensated by the absence of Net1 (net1Δ), i.e. double-mutant cells (cdc15Δ net1Δ) are alive [26]. The model is consistent with this observation (electronic supplementary material, figure S7b). In the absence of Net1, all the Cdc14 is released into the nucleus, where it can dephosphorylate and activate Swi5 and Cdh1 which are shuttling between the cytoplasm and the nucleus.
3.7. Non-degradable Clb5
The Clb5 destruction pathway can be compromised by non-degradable Clb5. As Clb5 promotes Sic1 degradation and keeps Cdh1 inactive by phosphorylation, Clb5 stabilization delays ME in the model (figure 6b) consistent with the experiments [44]. In the simulations, Sic1 appears only after the first exit from mitosis, but not during subsequent G1 phases (not shown). The persistent Clb5–Cdk1 activity and the absence of its stoichiometric inhibitor (Sic1) might be the reason of experimentally observed replication-licensing problems and loss of viability [38,44].
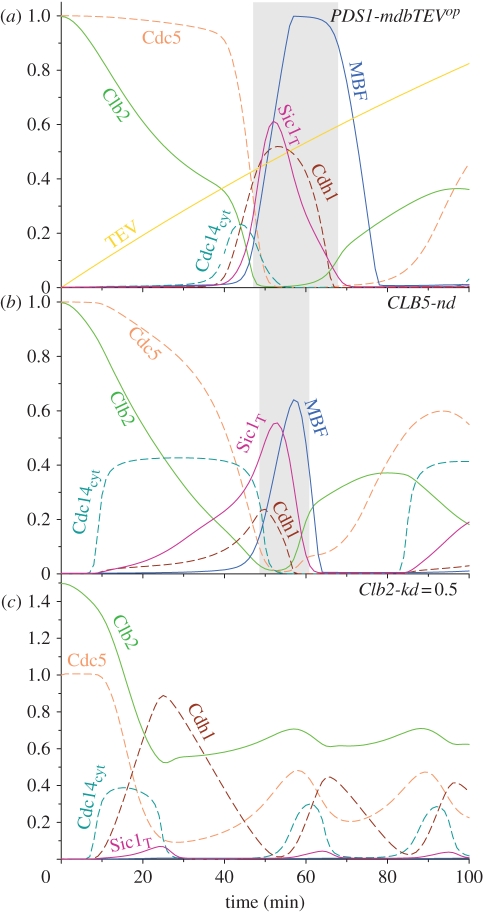
3.8. Non-degradable Clb2
Recently, two laboratories have analysed the effect of non-degradable Clb2 on ME and Cdc14 release [13,14]. With the model, we address the experiments performed by Lu & Cross [13]. Cells were blocked in metaphase by methionine-repressible Cdc20 (MET3-Cdc20), and a non-degradable Clb2 (Clb2-kd, lacking both destruction and KEN boxes) was induced transiently to different levels. The expression level of non-degradable Clb2 was estimated by the GFP signal attached to Clb2 and its value was related to the Clb2 peak value during the WT cycle.3 The expression of non-degradable Clb2 was terminated at the time of Cdc20 re-induction and the effect of different levels of Clb2-kd on mitotic progression was analysed in single cells. Figure 6c shows the simulation of these experiments with Clb2-kd expression (Clb-kd = 0.5) roughly equal to the peak value during the normal mitotic cycle in the model. After Cdc20 induction, Cdc14 is activated, while Cdc5 and endogenous Clb2 are degraded by activated Cdh1–APC. Observe that the total (the sum of non-degradable and endogenous) Clb2 level drops to the value of Clb2-kd (0.5) and it shows small amplitude oscillations after that. In the model, although the oscillation of cell cycle components is similar to normal Cdc20 block and release experiments, the amplitude of some network components (e.g. the Swi5 and MBF TFs) is much smaller than during normal ME. The small peak of the dephosphorylated form of these TFs indicates a block of ME. Despite blocking ME, Cdc14 continues to oscillate, a phenomenon called Cdc14 endocycles by Lu & Cross [13]. In the following section, we will analyse in detail the Cdc14 endocycles.
3.9. Molecular details of the Cdc14 oscillations with non-degradable Clb2
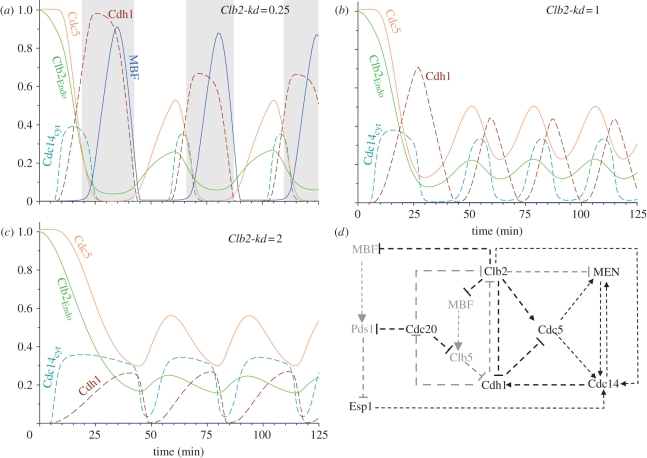
The experiments of Lu & Cross [13] for different levels of non-degradable Clb2 (Clb2-kd) (0.25, 1 and 2) were reproduced by numerical simulations (figure 7). After Cdc20 re-induction, the model shows oscillations at every level of non-degradable Clb2 analysed,4 and the Clb2-kd values influence only the period of the oscillations without any significant effect on the amplitude (see electronic supplementary material, figure S8). Increasing Clb2-kd values shorten the period of the oscillation until it becomes more or less constant above a threshold value of Clb2-kd as observed by Lu & Cross [13]. For Clb2-kd expression levels below the Clb2 peak in the mitotic cycle (Clb2-kd = 0.25), all the cell cycle regulators oscillate with significant amplitudes (figure 7a). Therefore proteins are effectively dephosphorylated at the end of mitosis, cells exit from M phase, divide and can start a new cycle [35]. At Clb2-kd levels higher than mitotic Clb2 peak values, the amplitude of oscillation of some cell cycle regulators is reduced to low levels by high Clb2 kinase activities and the period of oscillation shortens. The components with reduced amplitude of oscillations include the TFs, Swi5 and MBF, all repressed by high Clb2 kinase activity. As Swi5 is inhibited, the stoichiometric inhibitor (Sic1) cannot accumulate during the oscillations at high Clb2-kd. SBF/MBF inhibition explains the lack of Cln, Pds1 and Clb5 peaks for Clb2-kd levels higher than 0.4. However, other cell cycle regulators, like Cdc20, Cdc14, Cdh1, Cdc5 and MEN, keep on oscillating with large amplitudes at every level of Clb2-kd thus driving the Cdc14 endocycles. In order to find out the essential requirements for Cdc14 endocycles, we have systematically analysed the role of individual components (nodes) and interactions (edges) in the control of Cdc14 oscillations. These data are summarized in figure 7d, where the non-essential nodes and edges are indicated by grey colour.
Figure 7.
Cdc20 block and release at different levels of non-degradable Clb2 (Clb2-kd). Clb2-kd levels, smaller than peak value during normal mitosis (e.g. 0.25) permit mitotic cycles (a). At higher Clb2-kd values (1 and 2), Cdc14 endocycles are observable (b,c). The positive and negative interaction among cell cycle regulators create positive and negative feedback loops that drive cell cycle oscillations. The cell cycle regulators and interactions labelled using black colours are required for Cdc14 endocycles at high Clb2-kd levels. In contrast, the components and interactions labelled using grey colours are dispensable (d).
We show that the cell cycle regulators required for the Cdc14 endocycles fall into categories: some of the essential components are parts of the oscillatory mechanism while others are responsible for the right conditions for the ‘clock’ to oscillate. Cdc20–APC and separase are the only two components belonging to this later category. The essential role of Cdc20–APC is explained by its function in the degradation of both securin and Clb5 whose activity inhibits the activation of Cdh1, an essential clock component (see later). Once these Cdc20 substrates are degraded, Cdc20 is not required anymore for the oscillations [14] and the model is consistent with these observations (simulations not shown). A non-degradable Clb5 blocks the Cdc14 endocycles with Cdc14 released and Cdh1 inactivated (electronic supplementary material, figure S9a). Securin also needs to be degraded because it is an inhibitor of separase which is required for both FEAR and MEN activation. The degradation of the third Cdc20 substrate (Clb2) is not required for Cdc14 endocycles and the model oscillates with and without endogenous, degradable Clb2. In fact, it is rather the opposite, because non-degradable Clb2 is a necessary requirement for Cdc14 endocycles.
Cdc14 phosphatase is an essential component of the oscillator, because in the absence of Cdc14 activity, Cdh1 cannot be activated and the oscillations fail (simulations not shown). As a consequence, Cdh1–APC is also required for the oscillations and without Cdh1 cells are blocked in a stable steady state with Cdc14 released in the cytoplasm, similar to the CLB5-nd block (electronic supplementary material, figure S9b). The crucial Cdh1 substrate for the oscillation to work is Cdc5. Model simulation with a non-degradable Cdc5 (destruction box delete, kdpolo′ = 0) blocks the oscillation similar to cdh1Δ and CLB5-nd (electronic supplementary material, figure S9c). These three components define a negative feedback loop (Cdc14 → Cdh1 —| Cdc5 → Cdc14), which is shown to be required for Cdc14 endocycles [13,14]. Interestingly, another negative feedback loop through Cdc20 (Cdc20 —| Clb5 —| Cdh1 —| Cdc20) is not required for Cdc14 endocycles. If Cdc20 is not degraded by Cdh1, Cdc20 stays high and constant, but Cdc14 still oscillates periodically (not shown).
The cell cycle control network has a double-negative feedback loop, a mutual antagonism between Cdh1–APC and Cdk1–Clb2, which can create a switch-like behaviour similar to a positive feedback loop. This double-negative feedback loop is not essential for the Cdc14 endocycles which persists in the absence of endogenous (degradable) Clb2 when all the Cdh1-mediated Clb2 degradation is blocked by Clb2-kd. In contrast, we predict with the model that the other leg of this feedback loop (Cdh1 inhibition by Clb2-kinase) plays an essential role during Cdc14 endocycles, because Cdk1–Clb2 is the only kinase that can inactivate Cdh1 after Cdc14 inactivation. In the absence of Clb2-dependent inactivation of Cdh1 (kpcdh′ = 0), the oscillation is blocked with partially inactivated Cdh1 after the first activation of Cdc14. We also find that Cdh1 inhibition by Clb2 is a major determinant of the period of Cdc14 oscillations. The stronger the inhibition of Cdh1 by Clb2, the shorter is the period of the endocycles. Eventually, if Cdh1 inhibition by Clb2 is too strong, the oscillations are blocked.
Furthermore, MEN inactivation (e.g. cdc15Δ) also blocks Cdc14 endocycles in the model (electronic supplementary material, figure S10a). MEN activates Cdc14 and regulates its cellular localization as well. In return, Cdc14 activates MEN by dephosphorylating Cdc15, which is inhibited by Cdk1–Clb2 phosphorylation. This positive feedback is essential for the Cdc14 oscillations to overcome the inhibition of Cdc15 by non-degradable Clb2. We observe that if Clb2 inhibition on Cdc15 is weaker, then the Cdc14–Cdc15 positive feedback becomes less important. However, the function of MEN in controlling cytoplasmic Cdc14 localization is essential for endocycles.
Net1 phosphorylation increases owing to the activation of Esp1, which downregulates the Net1 dephosphorylating phosphatase, PP2ACdc55. This non-proteolytic function of Esp1 causes FEAR-dependent Cdc14 release, which is required for the endocycles to activate MEN under high Cdk1–Clb2 condition (electronic supplementary material, figure S10b). In contrast, we find that Net1 phosphorylation by either MEN or Cdk1–Clb2 kinases is dispensable for the endocycles, assuming that the other one of them is phosphorylating Net1.
4. Discussion
Cell cycle regulation in budding yeast has been a subject of intensive experimental and theoretical studies. However, the continuous appearance of new experimental data requires a steady refinement of mathematical models proposed to explain the regulatory network. In this work, we have revisited the control of ME in budding yeast to keep track of new experimental observations ranging from single to quintuple mutations.
We demonstrate by modelling how ME is orchestrated by Cdc20–APC, which functions as a critical node controlling both the kinase and phosphatase branches required for ME. The role and contribution of individual network branches (figure 1b) were analysed based on their ability
— to suppress the Cdc20 requirement for ME when they are hyperactivated or
— to block Cdc20-induced ME when they are inactivated.
We show that Cdc20–APC-mediated degradation of Clb2 is insufficient to drive ME, but the ability of Cdc20–APC to degrade its other substrates, Pds1 and Clb5, ensures ME by reducing the kinase-to-phosphatase ratio in favour of Cdh1 and Sic1 activation. Although Cdc20–APC is able to reduce Cdk1–Clb2 kinase activity directly by degrading Clb2 and Clb5, its ability to upregulate Cdc14 phosphatase is indirect through FEAR and MEN. Cdc20–APC by degrading Pds1 relieves the inhibition on Esp1, which activates both FEAR and MEN through its non-proteolytic and proteolytic functions. Using the model, we propose that Esp1 activates the positive feedback on Cdc15 through its non-proteolytic function and activates Tem1 through its proteolytic function, thereby activating the two important MEN components. In the absence of the non-proteolytic function, cells exit mitosis after a delay if Clb2 is degraded, indicating that non-proteolytic function of separase is dispensable. On the other hand, we demonstrate that the non-proteolytic separase function becomes essential for ME in the absence of Cdc20-mediated Clb2 degradation. Under this circumstance, we show that Clb2 inhibition on Cdc15 is strong, therefore the positive feedback on Cdc15 must be turned on by non-proteolytic separase-dependent Cdc14 release. These mechanisms underline the differences in phenotypes of cdc20Δ TEVop (figure 5a), cdc20Δ TEVop SIC1op (figure 5c) and PDS1-mdb TEVop (figure 6a), where TEV mimics only the proteolytic function of the Esp1. Furthermore, the loss of both separase functions, e.g. in PDS1-mdb cells, blocks ME completely. Thus, Esp1 through its dual functions couples anaphase to ME through Cdc14 activation.
In the model, expression of non-degradable Clb2 above a certain threshold blocks ME and induces Cdc14 endocycles, as observed by recent experiments [13,14]. These Cdc14 endocycles are interesting because certain regulators of ME oscillate independent of Clb2 oscillations and cell cycle progression. The regulatory network responsible for Cdc14 endocycles in our model is essentially the same as proposed by Lu & Cross [13] and Manzoni et al. [14]. However, we emphasize that our model explains the Cdc14 endocycles together with other classical experiments on ME in budding yeast, which is not a trivial accomplishment. Among other things, we had to assume a weak inhibition by Clb2–Cdk1 on Cdh1–APC in order to get periodic activation of Cdh1 at high Clb2 levels.
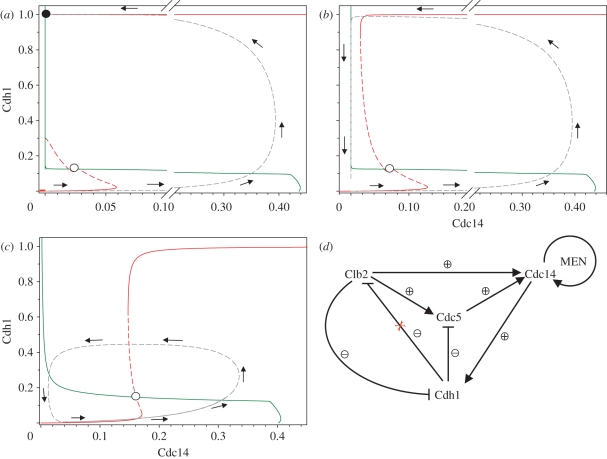
Despite these similarities, our interpretation of the significance of Cdc14 endocycles is different from the proposal of Lu & Cross [13]. In our view, the budding yeast cell cycle is characterized by two one-way switches: one controlling Start, and the other controlling ME transition [45]. These one-way switches are the consequence of antagonistic interactions between Cdk1–Clbs and their negative regulators (Cdh1–APC and Sic1). Cells turn on these irreversible switches with ‘helper’ molecules, which are regulated by negative feedback loops [45]. The helper molecules for ME in budding yeast are Cdc20 and the Cdc14 phosphatase. In figure 8a, we illustrate the irreversible ME switch on a phaseplane calculated with the present model. The phaseplane shows two characteristic curves, called nullclines (or balance curves), along which one or other dynamical variables is not changing its value. The Cdh1 nullcline (red curve) is S-shaped because of the antagonistic interactions mentioned above. The Cdc14 nullcline (green curve) has a sigmoid shape because Cdc14 is released (active) only when Cdh1–APC is inactive and therefore its substrates (Clb2 and Cdc5) are high. At the intersection of nullclines, all the dynamic variables of the cell cycle control system are in steady state. In figure 8a (daughter cell), there is an unstable (open circle) and a stable (filled circle) steady state on the phaseplane. Mitotic progression starts near the origin (both Cdh1 and Cdc14 are close to zero) and it follows the trajectory (dashed grey line) calculated by model simulation. The final state is a stable steady state corresponding to G1 phase of the cell cycle, where Cdh1 is kept active without any apparent Cdc14 activity. The phaseplane and the trajectory were calculated by setting all starter kinase (Cln) activities to zero; therefore this picture corresponds to a small budding yeast daughter cell, which has to grow before it activates MBF and begins to synthesize Clns. This phaseplane portrait supports our theory [45] about an underlying irreversible switch in ME of the budding yeast.
Figure 8.
Phaseplane analysis of the budding yeast cell cycle and Cdc14 endocycles. The Cdc14 (green) and Cdh1 (red) nullclines were calculated as one-parameter bifurcation diagrams with the model using XPP/AUT and combined to create a phaseplane portraits. All phaseplanes were calculated in anaphase when Cdc20–APC has completely degraded Pds1 and Clb5. (a) Small daughter cell (kscln = kscln′ = ksclb5 = ksclb5′ = kiswi = kpcdh = 0). (b) Mother cell. (c) Cdc14 endocycles (Clb2kd = 1). (d) Cdc14 endocycles are the consequence of the ‘two-hit’ mechanism [14] controlling Cdc14 release. As both Clb2 and Cdc5 are required for Cdc14 release, there are two negative feedback loops. If Clb2 is made non-degradable (Clb2-kd), the outside negative feedback loop (Clb2 → Cdc14 → Cdh1 —| Clb2) is broken but the inside one with Polo-kinase (Cdc5 → Cdc14 → Cdh1 —| Cdc5) is still functional. The inside negative feedback together with the MEN–Cdc14 positive feedback drives the Cdc14 endocycles.
The situation for mother cells (figure 8b) is different, because they are larger than daughter cells. Therefore, they bypass the growth requirement for activation of Cln synthesis and do not stop at a stable G1 steady state. The TF (called MBF in the model) responsible for Cln synthesis gets activated as soon as Cdh1-dependent Clb2 degradation relieves MBF from Clb2 inhibition. Because of the spontaneous, growth-independent activation of Cln synthesis, Cdh1 is activated only transiently in mother cells. The one way-switch characteristic of small daughter cells becomes a relaxation oscillator in large mother cells.
Cdc14 endocycles are observed in the presence of non-degradable Clb2 (Clb2-kd), in which case the Cdh1 nullclines is shifted to the right in both mother and daughter cells (figure 8c). Only an unstable steady state is left; therefore, Clb2-kd-expressing cells follow a limit cycle oscillation. However, the oscillations that underlie Cdc14 endocycles are much different from the limit cycles characteristic of mother cells because high activity of Cdk1–Clb2 represses many cell cycle regulators (Sic1, Swi5, Cln, MBF; electronic supplementary material, figure S11). For these reasons, Cdc14 endocycles are uncoupled from normal progression through DNA replication–division cycle.
Based on our analysis, we consider Cdc14 endocycles as a consequence of redundant negative feedback loops in Cdc14 release (figure 8d): Cdc14 release requires both Clb2 and Cdc5 kinase activities, and both kinases are targeted for degradation by Cdc14-activated Cdh1–APC. Clb2-kd-expressing cells are stuck in mitosis, but after Cdc20 induction, Cdc14 and later Cdh1 get activated. The cell undergoes anaphase but it cannot exit mitosis because of the high activity of Clb2. Cdc5 gets degraded by Cdh1, which causes re-sequestration of Cdc14 and Cdh1 inactivation. But the other negative feedback loop involving Clb2 degradation cannot be engaged. Clb2-kd-blocked cells find themselves in an unusual situation not characteristic of the normal cell cycle: most of the events of mitosis are completed but they cannot exit mitosis. As synthesis of Cdc5 is kept high during mitosis by high Cdk1–Clb2 activity, Polo is re-synthesized when Cdh1 is inactive thereby causing another round of Cdc14 activation. Repetition of these steps creates the Cdc14 endocyles.
Negative feedback controls are common in both biologically evolved and human-engineered systems. Negative feedback is used to prevent large sustained activation of components, but it is susceptible to oscillations if the feedback loop is sufficiently time-delayed. Lu & Cross [13] proposed that the Cdc5-dependent negative feedback responsible for Cdc14 endocycles evolved as an independent oscillator. By contrast, we view Cdc14 endocycles as a consequence of an unusual cell cycle block caused by stabilization of Clb2. We emphasize that this provides an ample setting for the redundant delayed negative feedback loops in the system to oscillate and these oscillations might not have a physiological role.
To further illustrate this point, we observe that the model contains another negative feedback loop that can potentially drive Cdc14 oscillations: Cdc14 → Cdh1 —| Cdc20 —| Pds1 —| Esp1 → Cdc14. This negative feedback loop is dependent on re-accumulation of Pds1 in the presence of Clb2-kd. With our present parameter values, this alternative negative feedback loop is suppressed because Pds1 synthesis (SBF/MBF-dependent) is inhibited by high Clb2-kinase activity. If Pds1 synthesis were less dependent on MBF or if Pds1 turnover were 10 times faster, the alternative negative feedback loop could oscillate independent of Cdc5 degradation (electronic supplementary material, figure S12).
Our model describes Cdc14 endocycles in the presence of non-degradable Clb2, but it fails to explain periodic Cdc14 release in G1 phase. The interpretation of these features of Cdc14 endocycles requires further experimentation and modelling efforts. To this end, we propose a potential mechanism for the Cdc14 endocycles in G1 phase, if Cdc5 is present (electronic supplementary material, figure S13). This fluctuation of Cdc14 in G1 is dependent on the periodic activation of MEN, and it requires another negative feedback loop, by which Cdc14 inactivates MEN. This new negative feedback loop is in addition to the one already in our model: Cdc14 inactivates MEN by Cdh1-dependent degradation of Cdc5 [11,46]. If Cdc14 also inactivates Tem1 in a Cdc5-independent manner, Cdc14 is periodically released in G1 phase lacking Clbs activity (electronic supplementary material, figure S13). This potential mechanism provides another example for redundant feedback loops employed by the ME control system.
In summary, the model developed in this work complements several important experimental findings and offers quantitative insights into the ME regulation. We have also observed few experimental situations where the model fails (electronic supplementary material, table S3). Despite these limitations, the model still serves as a platform for further analysis of structural stability (the form of governing equations) and kinetic parameter sensitivity. However, in future work, these limitations of the model should be investigated in more detail.
5. Methods
The regulatory network in figure 1 was translated into a set of ordinary differential and algebraic equations (electronic supplementary material), which were solved numerically using XPPAUT, available from http://www.math.pitt.edu/∼bard/xpp/xpp.html. For more technical details see electronic supplementary material.
Acknowledgements
We thank Kathy Chen, Kim Nasmyth, John J. Tyson and all our group members for discussions. This work has been supported from grants of BBSRC and the European Community's Seventh Framework Programme (UniCellSys and MitoSys). P.F. and A.R. are supported by the PhD programmes of the Instituto Gulbenkian de Ciência (Portugal) and by Boehringer Ingelheim Fonds, respectively.
Footnotes
‘Start’ is the point in the cell cycle where cells become committed to bud formation, DNA synthesis and mitosis. Start is marked by activation of Cln kinases.
In practice, Cdc20 is induced with a constant rate of transcription (i.e. MET3–Cdc20 after methionine deprivation). In contrast, in the model, the rate of Cdc20 synthesis is regulated by Cdk1–Clb2 similar to the wild-type protein.
Clb2 mitotic peak value is assigned to 1 by Lu & Cross [13], and it corresponds to 0.4 in our case.
Cdc14 endocycles disappear at Clb2 values higher than 2.25 in the model, which corresponds to more than 5.5 in Lu & Cross's [13] terminology.
References
- 1.Morgan D. O. 2007. The cell cycle: principles of control. London, UK: New Science Press [Google Scholar]
- 2.Bloom J., Cross F. R. 2007. Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell. Biol. 8, 149–160 10.1038/nrm2105 (doi:10.1038/nrm2105) [DOI] [PubMed] [Google Scholar]
- 3.Mortensen E. M., Haas W., Gygi M., Gygi S. P., Kellogg D. R. 2005. Cdc28-dependent regulation of the Cdc5/Polo kinase. Curr. Biol. 15, 2033–2037 10.1016/j.cub.2005.10.046 (doi:10.1016/j.cub.2005.10.046) [DOI] [PubMed] [Google Scholar]
- 4.Queralt E., Uhlmann F. 2008. Cdk-counteracting phosphatases unlock mitotic exit. Curr. Opin. Cell. Biol. 20, 661–668 10.1016/j.ceb.2008.09.003 (doi:10.1016/j.ceb.2008.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Amours D., Amon A. 2004. At the interface between signaling and executing anaphase–Cdc14 and the FEAR network. Genes Dev. 18, 2581–2595 10.1101/gad.1247304 (doi:10.1101/gad.1247304) [DOI] [PubMed] [Google Scholar]
- 6.Bardin A. J., Visintin R., Amon A. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102, 21–31 10.1016/S0092-8674(00)00007-6 (doi:10.1016/S0092-8674(00)00007-6) [DOI] [PubMed] [Google Scholar]
- 7.Stegmeier F., Visintin R., Amon A. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108, 207–220 10.1016/S0092-8674(02)00618-9 (doi:10.1016/S0092-8674(02)00618-9) [DOI] [PubMed] [Google Scholar]
- 8.Sullivan M., Uhlmann F. 2003. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat. Cell. Biol. 5, 249–254 10.1038/ncb940 (doi:10.1038/ncb940) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereira G., Hofken T., Grindlay J., Manson C., Schiebel E. 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell. 6, 1–10 10.1016/S1097-2765(00)00002-2 (doi:10.1016/S1097-2765(00)00002-2) [DOI] [PubMed] [Google Scholar]
- 10.Uhlmann F., Wernic D., Poupart M. A., Koonin E. V., Nasmyth K. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103, 375–386 10.1016/S0092-8674(00)00130-6 (doi:10.1016/S0092-8674(00)00130-6) [DOI] [PubMed] [Google Scholar]
- 11.Queralt E., Lehane C., Novak B., Uhlmann F. 2006. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125, 719–732 10.1016/j.cell.2006.03.038 (doi:10.1016/j.cell.2006.03.038) [DOI] [PubMed] [Google Scholar]
- 12.Toth A., Queralt E., Uhlmann F., Novak B. 2007. Mitotic exit in two dimensions. J. Theor. Biol. 248, 560–573 10.1016/j.jtbi.2007.06.014 (doi:10.1016/j.jtbi.2007.06.014) [DOI] [PubMed] [Google Scholar]
- 13.Lu Y., Cross F. R. 2010. Periodic cyclin-Cdk activity entrains an autonomous Cdc14 release oscillator. Cell 141, 268–279 10.1016/j.cell.2010.03.021 (doi:10.1016/j.cell.2010.03.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzoni R., Montani F., Visintin C., Ciliberto A., Visintin R. 2010. Oscillations in Cdc14 release and sequestration reveal the circuit underlying mitotic exit. J. Cell. Biol. 190, 209–222 10.1083/jcb.201002026 (doi:10.1083/jcb.201002026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knapp D., Bhoite L., Stillman D. J., Nasmyth K. 1996. The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol. Cell. Biol. 16, 5701–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toyn J. H., Johnson A. L., Donovan J. D., Toone W. M., Johnston L. H. 1997. The Swi5 transcription factor of Saccharomyces cerevisiae has a role in exit from mitosis through induction of the cdk-inhibitor Sic1 in telophase. Genetics 145, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moll T., Tebb G., Surana U., Robitsch H., Nasmyth K. 1991. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66, 743–758 10.1016/0092-8674(91)90118-I (doi:10.1016/0092-8674(91)90118-I) [DOI] [PubMed] [Google Scholar]
- 18.Verma R., Annan R. S., Huddleston M. J., Carr S. A., Reynard G., Deshaies R. J. 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278, 455–460 10.1126/science.278.5337.455 (doi:10.1126/science.278.5337.455) [DOI] [PubMed] [Google Scholar]
- 19.Donovan J. D., Toyn J. H., Johnson A. L., Johnston L. H. 1994. P40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 8, 1640–1653 10.1101/gad.8.14.1640 (doi:10.1101/gad.8.14.1640) [DOI] [PubMed] [Google Scholar]
- 20.Schwob E., Bohm T., Mendenhall M. D., Nasmyth K. 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79, 233–244 10.1016/0092-8674(94)90193-7 (doi:10.1016/0092-8674(94)90193-7) [DOI] [PubMed] [Google Scholar]
- 21.Mendenhall M. D., al-Jumaily W., Nugroho T. T. 1995. The Cdc28 inhibitor p40SIC1. Prog. Cell Cycle Res. 1, 173–185 [DOI] [PubMed] [Google Scholar]
- 22.Shirayama M., Toth A., Galova M., Nasmyth K. 1999. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 402, 203–207 10.1038/46080 (doi:10.1038/46080) [DOI] [PubMed] [Google Scholar]
- 23.Amon A., Tyers M., Futcher B., Nasmyth K. 1993. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74, 993–1007 10.1016/0092-8674(93)90722-3 (doi:10.1016/0092-8674(93)90722-3) [DOI] [PubMed] [Google Scholar]
- 24.Chen K. C., Csikasz-Nagy A., Gyorffy B., Val J., Novak B., Tyson J. J. 2000. Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol. Biol. Cell 11, 369–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K. C., Calzone L., Csikasz-Nagy A., Cross F. R., Novak B., Tyson J. J. 2004. Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell 15, 3841–3862 10.1091/mbc.E03-11-0794 (doi:10.1091/mbc.E03-11-0794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shou W., et al. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97, 233–244 10.1016/S0092-8674(00)80733-3 (doi:10.1016/S0092-8674(00)80733-3) [DOI] [PubMed] [Google Scholar]
- 27.Visintin R., Hwang E. S., Amon A. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398, 818–823 10.1038/19775 (doi:10.1038/19775) [DOI] [PubMed] [Google Scholar]
- 28.Azzam R., Chen S. L., Shou W., Mah A. S., Alexandru G., Nasmyth K., Annan R. S., Carr S. A., Deshaies R. J. 2004. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science 305, 516–519 10.1126/science.1099402 (doi:10.1126/science.1099402) [DOI] [PubMed] [Google Scholar]
- 29.Lu Y., Cross F. 2009. Mitotic exit in the absence of separase activity. Mol. Biol. Cell. 20, 1576–1591 10.1091/mbc.E08-10-1042 (doi:10.1091/mbc.E08-10-1042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohl D. A., Huddleston M. J., Collingwood T. S., Annan R. S., Deshaies R. J. 2009. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J. Cell. Biol. 184, 527–539 10.1083/jcb.200812022 (doi:10.1083/jcb.200812022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldbeter A., Koshland D. E., Jr 1981. An amplified sensitivity arising from covalent modification in biological systems. Proc. Natl Acad. Sci. USA 78, 6840–6844 10.1073/pnas.78.11.6840 (doi:10.1073/pnas.78.11.6840) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasmyth K., Peters J. M., Uhlmann F. 2000. Splitting the chromosome: cutting the ties that bind sister chromatids. Science 288, 1379–1385 10.1126/science.288.5470.1379 (doi:10.1126/science.288.5470.1379) [DOI] [PubMed] [Google Scholar]
- 33.Visintin R., Craig K., Hwang E. S., Prinz S., Tyers M., Amon A. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 2, 709–718 10.1016/S1097-2765(00)80286-5 (doi:10.1016/S1097-2765(00)80286-5) [DOI] [PubMed] [Google Scholar]
- 34.Stegmeier F., Amon A. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38, 203–232 10.1146/annurev.genet.38.072902.093051 (doi:10.1146/annurev.genet.38.072902.093051) [DOI] [PubMed] [Google Scholar]
- 35.Drapkin B. J., Lu Y., Procko A. L., Timney B. L., Cross F. R. 2009. Analysis of the mitotic exit control system using locked levels of stable mitotic cyclin. Mol. Syst. Biol. 5, 328. 10.1038/msb.2009.78 (doi:10.1038/msb.2009.78) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeong F. M., Lim H. H., Padmashree C. G., Surana U. 2000. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol. Cell. 5, 501–511 10.1016/S1097-2765(00)80444-X (doi:10.1016/S1097-2765(00)80444-X) [DOI] [PubMed] [Google Scholar]
- 37.Schwab M., Lutum A. S., Seufert W. 1997. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90, 683–693 10.1016/S0092-8674(00)80529-2 (doi:10.1016/S0092-8674(00)80529-2) [DOI] [PubMed] [Google Scholar]
- 38.Wasch R., Cross F. R. 2002. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418, 556–562 10.1038/nature00856 (doi:10.1038/nature00856) [DOI] [PubMed] [Google Scholar]
- 39.Lim H. H., Goh P. Y., Surana U. 1998. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr. Biol. 8, 231–234 10.1016/S0960-9822(98)70088-0 (doi:10.1016/S0960-9822(98)70088-0) [DOI] [PubMed] [Google Scholar]
- 40.Thornton B. R., Toczyski D. P. 2003. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell. Biol. 5, 1090–1094 10.1038/ncb1066 (doi:10.1038/ncb1066) [DOI] [PubMed] [Google Scholar]
- 41.Jaspersen S. L., Charles J. F., Tinker-Kulberg R. L., Morgan D. O. 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 9, 2803–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaspersen S. L., Morgan D. O. 2000. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr. Biol. 10, 615–618 10.1016/S0960-9822(00)00491-7 (doi:10.1016/S0960-9822(00)00491-7) [DOI] [PubMed] [Google Scholar]
- 43.Cohen-Fix O., Koshland D. 1999. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev. 13, 1950–1959 10.1101/gad.13.15.1950 (doi:10.1101/gad.13.15.1950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan M., Holt L., Morgan D. O. 2008. Cyclin-specific control of ribosomal DNA segregation. Mol. Cell. Biol. 28, 5328–5336 10.1128/MCB.00235-08 (doi:10.1128/MCB.00235-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyson J. J., Novak B. 2008. Temporal organization of the cell cycle. Curr. Biol. 18, R759–R768 10.1016/j.cub.2008.07.001 (doi:10.1016/j.cub.2008.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visintin C., Tomson B. N., Rahal R., Paulson J., Cohen M., Taunton J., Amon A., Visintin R. 2008. APC/C-Cdh1-mediated degradation of the Polo kinase Cdc5 promotes the return of Cdc14 into the nucleolus. Genes Dev. 22, 79–90 10.1101/gad.1601308 (doi:10.1101/gad.1601308) [DOI] [PMC free article] [PubMed] [Google Scholar]