Abstract
The relative importance of the aerosol transmission route for influenza remains contentious. To determine the potential for influenza to spread via the aerosol route, we measured the size distribution of airborne influenza A viruses. We collected size-segregated aerosol samples during the 2009–2010 flu season in a health centre, a day-care facility and onboard aeroplanes. Filter extracts were analysed using quantitative reverse transcriptase polymerase chain reaction. Half of the 16 samples were positive, and their total virus concentrations ranged from 5800 to 37 000 genome copies m−3. On average, 64 per cent of the viral genome copies were associated with fine particles smaller than 2.5 µm, which can remain suspended for hours. Modelling of virus concentrations indoors suggested a source strength of 1.6 ± 1.2 × 105 genome copies m−3 air h−1 and a deposition flux onto surfaces of 13 ± 7 genome copies m−2 h−1 by Brownian motion. Over 1 hour, the inhalation dose was estimated to be 30 ± 18 median tissue culture infectious dose (TCID50), adequate to induce infection. These results provide quantitative support for the idea that the aerosol route could be an important mode of influenza transmission.
Keywords: influenza, bioaerosol, size distribution, aerosol transmission, emissions, deposition
1. Introduction
Influenza A viruses (IAVs) are transmitted through direct contact, indirect contact, large respiratory droplets and droplet nuclei (aerosols) that are left behind by the evaporation of larger droplets. The relative importance of each of these routes remains contentious. The aerosol transmission route has been particularly controversial since there is scant direct proof of infection mediated by virus-laden aerosols, partly owing to the difficulties in studies involving human subjects and partly owing to the challenges in detecting IAVs in ambient air [1–3].
Virus-laden aerosols may be released into air when infected people cough, sneeze, talk or breathe; however, the aerosols are quickly diluted by ambient air to extremely low concentrations [4]. In addition, the relatively insensitive culture methods to detect viruses, potential inactivation during aerosol sampling and inhibition of detection methods by airborne contaminants present challenges to the measurement of airborne IAVs [1,5]. Consequently, despite the rapid development of detection methods for IAVs in clinical and laboratory settings, there are still very few measurements of them in the airborne environment. Even fewer studies have determined the size of influenza virus-laden particles, which is important because it determines how long particles will remain suspended in air before being removed by gravitational settling or Brownian diffusion, and where they will deposit in the respiratory system.
Quantitative reverse transcriptase–polymerase chain reaction (qRT–PCR), based on the detection of viral RNA, affords a sensitive and rapid approach for quantifying low levels of viruses. Chen et al. [6] applied this method to detect IAVs in a live poultry market, but their sampling method did not discriminate by particle size. Using qRT–PCR, Blachere et al. [7] measured aerosolized influenza viruses in a hospital emergency department for six days. Eighty-one air samples were collected with a modified National Institute for Occupational Safety and Health two-stage cyclone sampler that separated the aerosols into greater than 4, 1–4 and less than 1 µm fractions, and IAV RNA was detected in 11 of the samples. They found that 46, 49 and 4 per cent of the IAVs were collected in each of the size ranges, respectively. A more extensive follow-up study by Lindsley et al. [8] reported that IAVs were detected on 10 out of 11 days, with 17 per cent out of 385 samples confirmed to contain IAV RNA. Of the detected IAV RNA, 42 per cent was associated with particles 4.1 µm or less.
Public places with a susceptible population and/or a high population density, such as hospitals, day-care centres and aeroplanes, may harbour high concentrations of pathogens. Of 218 surfaces (toys, nappy-changing areas, toilet seat tops, etc.) tested in 14 different day-care centres, Boone & Gerba [9] detected influenza viruses on 23 and 53 per cent of the samples during autumn and spring, respectively. Infected individuals on an aeroplane may spread the influenza virus to other passengers [10]. The Alaska Airlines outbreak [11] has been presented as proof of airborne influenza transmission: a jet with 54 persons aboard was delayed on the ground for 3 h (during which the aeroplane ventilation system was inoperative), and 72 per cent of the passengers who stayed on the aeroplane were infected by an influenza-contracted passenger within 72 h.
To evaluate the prevalence of airborne IAVs in high-risk, public spaces, we collected aerosol samples from a health centre, a day-care facility and onboard three commercial passenger aeroplane flights during the 2009–2010 flu season. Particles were divided into five size fractions, and IAVs in each were analysed using qRT–PCR. The indoor influenza virus emission strength, the deposition flux onto the wall surfaces and risk for airborne infection were then estimated using our experimental data.
2. Material and methods
2.1. Reference viruses
Reference strains of influenza A were from our collection at the Department of Biomedical Sciences and Pathology, Center for Molecular Medicine and Infectious Disease at Virginia Tech. Prototype strains used to develop the qPCR method were A/PR/8/34 (H1N1) and A/swine/Minnesota/1145/2007 (H3N2). These two strains were used to construct and test the qRT–PCR concentration standards.
2.2. Detection of viral genome
2.2.1. Viral genomic RNA extraction
Influenza virus RNA collected on the filters was extracted using a Trizol–chloroform-based method modified from a protocol reported elsewhere [12,13]. Briefly, the filter was rolled and put into a 2 ml microcentrifuge tube containing 250 µl of phosphate-buffered saline (PBS) supplemented with 20 µg of glycogen (Ambion, TX, USA), 15 µg of glycoblue (Ambion) and 50 ng of human genomic DNA (Cat. no. 636401, Clontech Laboratories, Inc., CA, USA). A volume of 750 µl of Trizol LS (Invitrogen, CA, USA) was added, and the sample was vortexed thoroughly and incubated at room temperature for 10 min. The sample was then briefly centrifuged, and the supernatant was transferred to a 1.5 ml microcentrifuge tube, to which 230 µl of chloroform was added (Sigma-Aldrich, MO, USA). The sample was briefly vortexed, incubated at room temperature for 5 min and then centrifuged at 2100g for 5 min. The colourless upper aqueous phase was carefully transferred to a new 1.5 ml tube containing 600 µl of isopropanol (Sigma-Aldrich) for RNA precipitation for 1 h. Then, the RNA was pelleted by centrifuging for 12 min at 20 000g and was washed with 600 µl of 75 per cent ethanol. The RNA was finally dissolved in 20 µl of diethylpyrocarbonate-treated water (Sigma-Aldrich) and immediately converted to complementary DNA (cDNA) or stored at −80°C until use.
2.2.2. Reverse transcription
cDNA was generated with a TaqMan Reverse Transcription Reagents Kit (N8080234, Applied Biosystems, CA, USA) according to the manufacturer's instructions. A 20 µl reaction mixture was made with a final concentration of 1× TaqMan RT buffer, 5.5 mM of Mg2+, 500 µM of each dNTP, 2.5 µM of RT random hexamer primers, 0.4 U µl−1 of RNase inhibitor and 1.25 U µl−1 of MultiScribe Reverse Transcriptase, plus 7.7 µl of RNA. cDNA synthesis was carried out on a thermal cycler (1000-Series Thermal Cycling Platform, Bio-Rad, USA) at 25°C for 10 min, 48°C for 30 min and 95°C for 5 min.
2.2.3. Quantifying standard and standard curve preparation
The cDNA standard solution was constructed by ligation of the targeted gene fragment in a pCR2.1-TOPO vector according to the instructions of the TOPO TA Cloning Kit (Invitrogen). Two sets of IAV primers, one reported by Ward et al. [14] and the other by van Elden et al. [15], are widely used to detect the M1 protein gene of IAVs [6,14,16]. The primers by Ward et al. [14] have proved to be applicable for the currently circulating A (H3N2), seasonal A (H1N1) and pandemic A (H1N1) strains [17,18]. The genomic regions amplified by these two sets of primers are partially overlapping (table 1). We used the forward primer reported by Ward et al. [14] and the reverse primer reported by van Elden et al. [15] to amplify a segment that spans both genomic regions. The amplicon obtained for cloning was a 262 bp segment by RT–PCR from stocks of A/swine/Minnesota/1145/2007 (H3N2). The ligation plasmids were transformed into competent E. coli cells, and recombinant bacteria were selected on kanamycin-containing LB agar. Positive inserts were amplified by M13 primers embedded within the pCR2.1-TOPO vector according to the manufacturer's protocol. The resulting PCR products were sequenced and confirmed to be the target IAV gene fragment. The PCR products were quantified by a Molecular Imager Gel Doc XR system (Bio-Rad) and were used as the cDNA standard for qPCR. A standard stock solution was prepared at a concentration of 1010 genome copies µl−1. It was tested and confirmed to quantify successfully both the H3N2 and the H1N1 influenza virus strains.
Table 1.
Primers and probes of influenza A virus.
| primer or probe | sequence | positiona | reference |
|---|---|---|---|
| INFA-1 | 5′GGA CTG CAG CGT AGA CGC TT3′ | 241–260 | [15] |
| INFA-2 | 5′CAT CCT GTT GTA TAT GAG GCC CAT3′ | 406–429 | |
| INFA-3 | 5′CAT TCT GTT GTA TAT GAG GCC CAT3′ | 406–429 | |
| INFA probe | 5′CTC AGT TAT TCT GCT GGT GCA CTT GCC A3′ | 373–400 | |
| senseA | 5′AAG ACC AAT CCT GTC ACC TCT GA3′ | 168–190 | [14] |
| antisenseA | 5′CAA AGC GTC TAC GCT GCA GTC C3′ | 241–262 | |
| influenza A probe | 5′TTT GTG TTC ACG CTC ACC GT3′ | 208–227 |
aAligned with A/PR/8/34 (H1N1) segment 7 (M gene).
The calibration curve was generated using serial 10-fold dilutions of the standard solution from 107 to 10 genome copies µl−1 in triplicate. A standard curve was generated each time that field samples were quantified, and the amount of IAV genome in field samples was determined according to the linear regression of cycle threshold (Ct) values against the known log concentrations (C0). Autoclaved ultrapure water (NANOpure Ultrapure Water System, Barnstead/Thermolyne, IA, USA) was used as a qPCR negative control during each run.
2.2.4. Quantitative polymerase chain reaction
The qPCR assay was performed in 96-well reaction plates (MicroAmp Optical, Applied Biosystems) on a 7300 Real Time PCR System (Applied Biosystems). Two sets of primers [14,15] were tested using an SYBR Green PCR Master Mix Kit (Applied Biosystems). The qPCR mixture consisted of a final concentration of 1× SYBR Green Master Mix, 200 nM of each primer, 5 µl of cDNA and autoclaved ultrapure water to bring the qPCR reaction volume to 25 µl. Cycling conditions were one cycle of AmpliTaq Gold enzyme activation at 95°C for 10 min, 40 cycles of denaturation of DNA at 95°C for 15 s and annealing and extension at 60°C for 1 min. The amplification was followed by a melting curve analysis with a dissociation stage from 60°C to 95°C.
The standard and Ward's primers were further tested with a TaqMan One-Step RT–PCR Master Mix Reagents Kit (Applied Biosystems), and the influenza A probe (6-FAM-5′ TTT GTG TTC ACG CTC ACC GT 3′- Black Hole Quencher 1) [14] was used. One-step RT–PCR was performed in 25 µl consisting of a final concentration of 1× Master Mix without UNG, 1× MultiScribe and RNase Inhibitor Mix (0.25 and 0.4 U µl−1, respectively), 900 nM of each primer and 225 nM of the influenza A probe, plus 3 µl of viral RNA. The reaction mixture was held at 48°C for 30 min for cDNA synthesis, 95°C for 10 min for AmpliTaq Gold enzyme activation and 40 two-step cycles followed (95°C for 15 s for denaturation and 60°C for 1 min for primer annealing and extension). All qPCR assays were run in triplicate.
2.3. Virus spike recovery experiments
Virus spike recovery experiments were conducted to test the recovery efficiencies of the viral genome from the filters used to collect ambient particle samples and the PBS buffer used for RNA extraction. Two polytetrafluoroethylene (PTFE) filters, 25 and 37 mm in diameter (Cat. no. 225-1708 and 225-1709, SKC Inc., PA, USA), were used for sample collection. The H1N1 virus stock was diluted 2 × 10−2 with autoclaved ultrapure water and used as a spiking solution. Filters were spiked with 50 µl of virus solution (2.5 µl per droplet, 20 droplets total for the 25 mm filter, and 5 µl per droplet, 10 droplets total for the 37 mm filter). Because the 37 mm filter was especially hydrophobic, the droplet volume had to be increased to 5 µl for it to be taken up from a pipette. For the PBS buffer, 50 µl of the virus solution was spiked into a 2 ml microcentrifuge tube containing 200 µl of PBS buffer (referred to as ‘PBS control’ hereafter). To test for possible decay of the virus with time, 50 µl of the virus solution was added into a 2 ml microcentrifuge tube without PBS (referred to as ‘decay control’ hereafter). All samples were placed in a biological safety cabinet for 2 h, allowing the virus solution droplets on the filters to dry out. The temperature in the cabinet was maintained at approximately 20°C. Aliquots of 50 µl of the same virus solution used for spiking were stored at 4°C for quantification of the spiked amount. All samples were supplemented with PBS to a final volume of 250 µl and subjected to viral RNA extraction as described above. All tests were conducted in duplicate.
2.4. Field sample collection
2.4.1. Sampling locations
Samples were collected from a health centre at Virginia Tech, a day-care centre in Blacksburg, Virginia, and aeroplanes corresponding to three cross-country flights between Roanoke and San Francisco. The health centre samples were collected from a waiting room, which is a semi-open space about 8.5 × 5 m. The mean indoor temperature (±s.d.) was 22.0 ± 1.0°C, with a mean relative humidity of 34.5 ± 11.4%. Design room air exchange rates (AERs) were 8–12 air changes h−1 (ACH). The day-care centre samples were collected in two toddlers' rooms and a babies' room. Each of the toddlers' rooms is about 8 × 4 m and holds 16 children plus four adults, and the babies' room is about 8 × 3.5 m and holds 12 children and four adults. The mean indoor temperature in the toddlers' rooms was 22.8 ± 1.7°C, with a mean relative humidity of 40.6 ± 5.1%. The mean indoor temperature in the babies' room was 25.1 ± 1.1°C, with a mean relative humidity of 32.9 ± 2.0%. The mean temperature in the aeroplanes (between Roanoke and San Francisco with a stopover) was 23.6 ± 3.1°C, with a mean relative humidity of 27.1 ± 11.9%. The ventilation systems were operating properly during all sampling periods.
2.4.2. Sample collection
A total of 16 samples were collected between 10 December 2009 and 22 April 2010, of which nine were collected from the health centre, four from the day-care centre and three from aeroplanes. A cascade impactor (Sioutas Cascade Impactor, SKC Inc.) and a pump running at 9 l min−1 (Leland Legacy, SKC Inc.) were used to collect the samples over 6–8 h. The impactor consists of four stages that allow the separation and collection of airborne particles in five size ranges: greater than 2.5, 1.0–2.5, 0.5–1.0, 0.25–0.5 and less than 0.25 µm. Particles larger than each cut-point were collected on 25 mm PTFE filters (Cat. no. 225-1708, SKC Inc.); those smaller than the 0.25 µm cut-point of the last stage were collected on a 37 mm PTFE after-filter (Cat. no. 225-1709, SKC Inc.).
In the health centre, the sampler was placed on a desk (approx. 0.5 m high) around which patients sit while waiting; in the day-care centre, the sampler was placed on a shelf (approx. 1.0 m high); and on aeroplanes, it was placed near the seat pocket (less than 0.5 m high). Temperature and relative humidity were recorded every 2 min during sampling (OM-73, Omega Engineering, Inc., USA). After each sampling period, the impactor was washed with 10 per cent bleach, cleaned with ultrapure water and autoclaved (121°C, 30 min). New filters were loaded into the impactor, left overnight before sampling and used as device blank controls. Only those with results confirmed to have no detectable influenza virus RNA in the device blank controls were adopted.
3. Results
3.1. Quantitative reverse transcriptase–polymerase chain reaction
We tested two sets of influenza virus primers that have been widely cited in the literature [4,6,14–16]. Both sets of primers proved to be adequate; they were able to specifically amplify the target gene segments from the H1N1 and H3N2 strains, as indicated by the dissociation curve (data not shown). We tested the efficiency of primers to detect a field isolate of the H3N2 and pandemic H1N1 strains. Ward's primers achieved better qPCR efficiency (91 versus 61%) and a lower detection limit (100 genome copies per reaction versus 1000 genome copies per reaction) than did van Elden's. Hence, we used Ward's primers in subsequent experiments. TaqMan qRT–PCR showed that with Ward's primers, the detection limit was 10 genome copies per reaction, with an efficiency around 100 per cent and R2 > 0.99 for our samples.
3.2. Virus spike recovery experiments
According to qRT–PCR results, each spiked sample contained approximately 2.4 × 107 genome copies of the H1N1 virus. The recovery efficiencies were calculated by dividing the amount of virus detected by the number spiked into each sample (filter, PBS control or decay control). Results are reported in table 2. The viral genome recovery efficiencies were 40.5 per cent from the 25 mm filter (p = 0.00011), 62.0 per cent from the 37 mm filter (p = 0.0058), 86.6 per cent from the PBS-control samples (p = 0.077) and 91.2 per cent from the decay-control samples (p = 0.23). The control experiments showed that PBS had no significant adverse effect on the viral genome, and the natural decay of virus genome was insignificant within a 2 h period. By contrast, the recovery efficiencies from the two filters were significantly less than 100 per cent. During the RNA extraction step, only 800–900 µl of Trizol lysate was retrieved for phase separation by chloroform, with 100–200 µl of lysate retained by the filter. This loss accounts for a portion of the incomplete recovery from the filter.
Table 2.
Virus recovery efficiency from PTFE filters and control solutions (virus solution in PBS and virus solution only) spiked with 2.4 ± 0.1 × 107 genome copies. Samples were incubated for 2 h and then analysed by qRT–PCR. Recovery efficiencies were significantly less than 100% with both filters.
| sample | amount of virus recovered (genome copies) |
recovery efficiency (%) | |
|---|---|---|---|
| mean | s.d. | ||
| 25 mm filter | 9.6 × 106 | 2.2 × 106 | 41a |
| 37 mm filter | 1.5 × 107 | 3.1 × 106 | 62a |
| PBS | 2.1 × 107 | 2.4 × 106 | 87 |
| virus solution only | 2.2 × 107 | 2.6 × 106 | 91 |
aRecovery efficiency significantly less than 100%.
3.3. Concentrations of airborne influenza A viruses
Between 10 December 2009 and 22 April 2010, we collected 16 samples, listed in table 3. Half of the samples were confirmed to contain aerosolized IAVs: 33 per cent of the health centre samples (three of nine), 75 per cent of the day-care centre samples (three of four) and 67 per cent of the aeroplane samples (two of three). Concentrations in all of the field and laboratory blanks were below the detection limit. In the samples containing detectable amounts of IAVs, the average concentration was 1.6 ± 0.9 × 104 genome copies m−3.
Table 3.
Average ambient relative humidity and temperature and total airborne influenza A virus concentration in each of 16 samples. Humidity and temperature were recorded every 2 min, and each sample was collected using a cascade impactor for 6–8 h. IAV RNA was extracted from filters and quantified by qRT–PCR. It was not detected in half of the samples. In the samples containing detectable amounts of IAV, the average concentration was 1.6 ± 0.9 × 104 genome copies m−3.
| date | location | RH (%) |
temperature (°C) |
total IAV concentration (genome copies m−3) |
|||
|---|---|---|---|---|---|---|---|
| mean | s.d. | mean | s.d. | mean | s.d. | ||
| 10 Dec 2009 | health centre | 16.98 | 1.71 | 21.17 | 0.25 | 1.6 × 104 | 1.2 × 104 |
| 17 Dec 2009 | day-care | 42.93 | 2.85 | 20.71 | 0.36 | 1.6 × 104 | 1.0 × 104 |
| 30 Dec 2009 | aeroplane | n.a.a | n.a. | n.a. | n.a. | n.d.b | n.d. |
| 26 Jan 2010 | health centre | 21.72 | 0.89 | 20.75 | 0.54 | n.d. | n.d. |
| 27 Jan 2010 | day-care | 32.93 | 2.03 | 25.12 | 1.06 | 3.7 × 104 | 2.0 × 103 |
| 9 Feb 2010 | health centre | 25.15 | 0.80 | 20.86 | 0.56 | 5.8 × 103 | 6.2 × 102 |
| 11 Feb 2010 | health centre | 18.77 | 1.13 | 20.51 | 0.68 | n.d. | n.d. |
| 22 Mar 2010 | aeroplane | 29.25 | 13.86 | 25.11 | 3.47 | 1.4 × 104 | 1.0 × 103 |
| 24 Mar 2010 | aeroplane | 25.06 | 8.69 | 21.85 | 1.37 | 1.1 × 104 | 1.1 × 103 |
| 30 Mar 2010 | health centre | 29.81 | 4.83 | 22.16 | 0.85 | n.d. | n.d. |
| 31 Mar 2010 | day-care | 43.61 | 1.58 | 21.82 | 0.85 | 1.6 × 104 | 1.1 × 102 |
| 6 Apr 2010 | health centre | 54.66 | 5.00 | 23.15 | 0.75 | 1.5 × 104 | 1.7 × 103 |
| 8 Apr 2010 | health centre | 49.33 | 1.15 | 22.71 | 0.37 | n.d. | n.d. |
| 9 Apr 2010 | day-care | 52.12 | 0.82 | 22.02 | 1.01 | n.d. | n.d. |
| 20 Apr 2010 | health centre | 31.70 | 1.07 | 21.84 | 0.86 | n.d. | n.d. |
| 22 Apr 2010 | health centre | 32.53 | 2.29 | 21.91 | 0.75 | n.d. | n.d. |
aNot available owing to a logging error.
bNo detectable influenza A virus genome.
3.4. Virus-laden particle size distribution
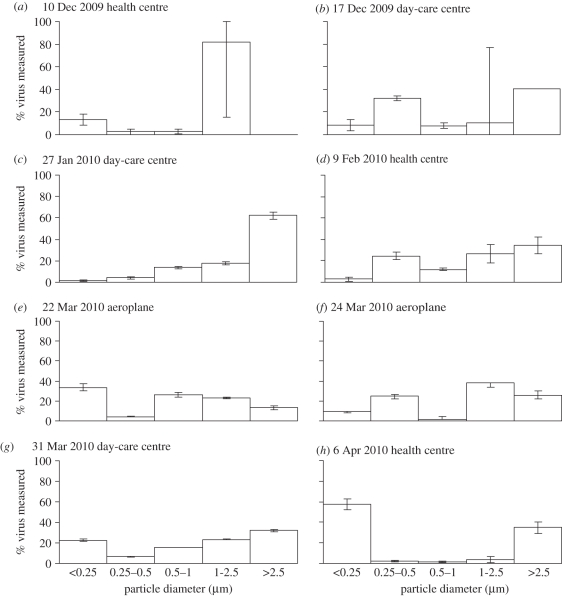
The cascade impactor separated particles into five size fractions: greater than 2.5, 1.0–2.5, 0.5–1.0, 0.25–0.5 and less than 0.25 µm. The amounts of virus found in each fraction, summed over all samples, were 36, 28, 11, 10 and 15 per cent, respectively. As shown in figure 1, the virus-laden particle size distributions of the eight positive samples were diverse, and no obvious trend was observed. In some cases, the virus was relatively evenly distributed across the different particle sizes, while in others, it was found predominantly in the smallest and largest, or just the largest, size fractions.
Figure 1.
Airborne IAV particle size distributions in each positive sample (date and location shown at top). Aerosol samples were collected over 6–8 h in each location using a cascade impactor with cut-point diameters of 0.25, 0.5, 1.0 and 2.5 µm. The y-axis indicates the percentage of total virus genome copies found in each size range. In seven of the eight cases, the majority of viruses were associated with fine particles smaller than 2.5 µm, which can remain suspended for hours, but there were no obvious trends in size distributions across different samples.
3.5. Indoor influenza virus emission and deposition flux by modelling
3.5.1. Indoor influenza A virus emission rate
To estimate the emission source strength of IAVs in airborne particles, we developed a mass-balance model. The model assumes that well-mixed, steady-state conditions apply at each sampling site: in a room of volume V (m3), air flows in and out through the heating, ventilating and air-conditioning system with a flow rate of Q (m3 h−1). Aerosolized viruses are generated by the occupants with an emission rate of E (genome copies h−1), disperse into ambient air and become well mixed immediately upon release. Assuming that the virus concentration in the air entering the room is zero, the indoor virus concentration is maintained at C (genome copies m−3) and the outlet virus concentration is also C, we establish the mass balance for the modelled room:
| 3.1 |
where C, Cin and Cout are, respectively, the virus concentrations in the room, in the inflow and in the outflow (genome copies m−3), and t is time (h).
At steady state,
| 3.2 |
where EV is the emission rate in genome copies m−3 h−1 and λ is the AER.
Typical AERs are 13 ACH in hospitals, 9 ACH in schools and 4 ACH in commercial offices [19]. The AER in commercial aircraft is usually higher with a typical value of 15 ACH [20]. Therefore, we adopt an AER of 10 ± 5 ACH to estimate the IAV emission rate in these public places, and for a measured indoor virus concentration of 1.6 ± 0.9×104 genome copies m−3, the emission rate EV is 1.6 ± 1.2 × 105 genome copies m−3 h−1.
3.5.2. Influenza A virus deposition on surfaces by Brownian motion
Applying the well-mixed model, we assume the virus-laden particles are evenly distributed throughout the room, except in a thin boundary layer alongside each wall surface, across which the virus-laden particles diffuse by Brownian motion and finally deposit onto the surface. The deposition flux can be calculated according to Fick's law:
| 3.3 |
where Jd is the virus flux to the surface (genome copies m−2 s−1), D is the diffusion coefficient (m2 s−1) and x is the distance to the wall (m). The diffusion coefficient can be calculated by the Einstein–Stokes equation:
| 3.4 |
where k is the Boltzmann constant (1.38 × 10−23 m2 kg s−2 K−1), T is temperature (K), μ is the viscosity of air at standard conditions (1.81 × 10−5 kg m−1 s−1), dp is particle diameter (m) and Cc is the Cunningham slip correction factor [21]. The concentration gradient can be estimated as the ratio of the concentration in the core of the room to the thickness of the diffusion boundary layer, given by the modified Einstein equation:
| 3.5 |
where t is the residence time of particles in the room, or 360 s in this case.
We sum over all particle sizes and use the midpoint diameter of each range, assuming a minimum of 0.1 µm for the smallest one and a maximum of 10 µm for the largest one, to calculate the diffusion coefficients. Based on our measurements, the total diffusive flux of viruses to indoor surfaces is 13 ± 7 genome copies m−2 h−1. This flux is sufficiently small that it can be neglected in the previous mass–balance model used to estimate the virus emission rate.
4. Discussion
4.1. Influenza A virus concentrations and size distributions in indoor facilities
To our knowledge, there have been only a few studies on the presence of airborne IAVs in a healthcare environment, and no airborne IAV detection has been reported in day-care centres or onboard passenger aeroplanes. Blachere et al. [7] reported airborne IAV concentrations in a health centre ranging from 460 to 16 278 median tissue culture infectious dose (TCID50)-equivalent RNA particles for an entire sample. The sampling time and flow rate were 3–5 h and 3.5 l min−1, respectively, and if we assume a total sample volume of 840 l of air (4 h at 3.5 l min−1), then the corresponding concentrations were 5.5 × 102 to 1.9 × 104 TCID50-equivalent RNA particles m−3. Their PCR was calibrated in TCID50 by using serial dilutions of a live-attenuated influenza virus quantified in TCID50 ml−1. Lindsley et al. [8], in a more detailed study in the same clinic, detected 1.2 ± 4.4 pg RNA m−3 in examination rooms, 1.1 ± 3.0 pg RNA m−3 in procedure rooms and 0.3 ± 4.3 pg RNA m−3 in a waiting room. These values can be converted to 5.0 ± 18.5 TCID50 m−3, 4.6 ± 12.6 TCID50 m−3 and 1.3 ± 18.1 TCID50 m−3, respectively, using the ratio of approximately 4.2 TCID50 FluMist vaccine pg−1 RNA reported in the study.
The ratio of viral particles to TCID50 can vary greatly depending on types of viruses (even for strains of the same type), culture methods and conditions (e.g. culturing cells, media and harvest time). For influenza viruses, this ratio has been reported to be in the range of hundreds to thousands: Fabian et al. [12] established a ratio of 300 copies  ; Ward et al. [14] determined that 1000 genome copies ml−1 corresponded to 1 TCID50 ml−1; and Poon et al. [22] estimated that 1 TCID50 of A/California/04/2009 (H1N1) contained approximately 5000 copies of the M gene. In our experiment, 1 PFU of A/PR/8/34 (H1N1) stock was equivalent to 3 × 103 genome copies, or approximately 2.1 × 103 genome copies per TCID50 according to the relationship between TCID50 and PFU [23], and the ratio for the pandemic A/California/04/2009 (H1N1) strain was determined to be 452 ± 84 copies/TCID−150. Based on this ratio (i.e. 452), our results from the health centre correspond to airborne IAV concentrations of 12.8–81.9 TCID50 m−3, one to two orders of magnitude lower than those observed by Blachere et al. [7] but slightly higher than those reported by Lindsley et al. [8]. With respect to size distributions, we found a larger fraction of total genome copies to be associated with fine particles: 80 per cent with particles smaller than 2.5 µm versus 53 per cent [7] and 42 per cent [8] with particles smaller than 4.1 µm.
; Ward et al. [14] determined that 1000 genome copies ml−1 corresponded to 1 TCID50 ml−1; and Poon et al. [22] estimated that 1 TCID50 of A/California/04/2009 (H1N1) contained approximately 5000 copies of the M gene. In our experiment, 1 PFU of A/PR/8/34 (H1N1) stock was equivalent to 3 × 103 genome copies, or approximately 2.1 × 103 genome copies per TCID50 according to the relationship between TCID50 and PFU [23], and the ratio for the pandemic A/California/04/2009 (H1N1) strain was determined to be 452 ± 84 copies/TCID−150. Based on this ratio (i.e. 452), our results from the health centre correspond to airborne IAV concentrations of 12.8–81.9 TCID50 m−3, one to two orders of magnitude lower than those observed by Blachere et al. [7] but slightly higher than those reported by Lindsley et al. [8]. With respect to size distributions, we found a larger fraction of total genome copies to be associated with fine particles: 80 per cent with particles smaller than 2.5 µm versus 53 per cent [7] and 42 per cent [8] with particles smaller than 4.1 µm.
For the three positive samples from the day-care centre, the total concentrations ranged from 1.6 × 104 to 3.7 × 104 genome copies m−3, half of which were associated with particles greater than 2.5 µm and the other half with smaller particles. The average concentration in the day-care centre was nearly two times higher than that in the health centre. Considering that children are the primary susceptible population of influenza, the difference is not surprising. In addition, the IAV size distributions in the day-care centre differed from those in the health centre: a larger portion of genome copies was found in particles greater than 2.5 µm (50 versus 20%). This discrepancy could originate from the ways that viruses were released (from coughing, sneezing, talking or breathing) and/or differences in the droplet size distribution of different age groups [24]. Viruses were probably released from latent subjects in the day-care centre (children are sent home as soon as symptoms are apparent), whereas those released in the health centre are assumed to come from symptomatic patients. Whether there are any differences between virus-laden particles released at different stages of infection and between hosts of different age warrants further investigation.
Virus concentrations of the two positive aeroplane samples were very similar (1.4 × 104 and 1.1 × 104 genome copies m−3), and virus-laden particles were relatively evenly distributed across each size fraction (figure 1e,f). It is possible that the diverse ages of aeroplane passengers evened out the difference observed in particular groups such as college students in the university health centre and children in the day-care centre.
Although this discussion has focused on the positive results, half of the samples were negative for IAVs. These negative results could be attributable to inhibitors to qRT–PCR, as observed by Chen et al. [6]. However, since the RNA extraction method adopted in this study has the potential to eliminate such inhibitions [4], it is more likely that in these instances, there were no infected individuals in the sampling locations or that concentrations were below the detection limit. The fact that the total virus concentration in the measurable samples ranged over a factor of only six, rather than orders of magnitude, seems surprising but could possibly be explained by the presence of only one or two infected individuals and similar AERs in each setting.
While the qPCR method is a powerful tool for determining the presence of viral genomic material, it does not indicate whether the virus is viable or not. Therefore, the results presented here are an upper limit on the concentration of viable viruses. On the other hand, the recovery efficiency of viruses spiked onto filters was roughly 50 per cent across the two types of filters used in this study, so the reported concentration of genome copies may be underrepresented by a factor of approximately 2. However, the true recovery efficiency is unknown because sample collection by impacting particles onto filters is not equivalent to spiking them onto filters from solution. Additionally, viral RNA may be subject to decay during extended sampling times. One important question to address in the future that could not be answered by qPCR is, ‘Are the viruses found across different sizes of particles equally viable, or are those in one size fraction more so?’.
4.2. Risk of airborne influenza A virus infection
Assuming a uniform airborne IAV concentration of 1.6 ± 0.9 × 104 copies m−3 air (corresponding to 35.4 ± 21.0 TCID50 m−3 air) and a adult breathing rate of 20 m3 d−1 [25], we estimate the inhalation doses during exposures of 1 h (for example, the duration of a clinical visit), 8 h (a workday) and 24 h to be 30 ± 18, 236 ± 140 and 708 ± 419 TCID50, respectively. Compared with the human infectious dose 50 per cent (ID50) by aerosols of 0.6–3 TCID50 [26], these doses are adequate to induce infection. In most instances, the measured concentration of airborne IAVs could be either over- or underestimated based on the sensitivity of the qRT–PCR assay. However, it is not our intent to imply that all the estimated amounts of airborne viral particles are infectious. Our results allow an accurate estimate of exposure to viral particles in air.
While illustrative, this calculation is subject to several limitations. First, the conversion from genome copies to TCID50 is based on the ratio determined with the laboratory strain A/California/04/2009 (H1N1) rather than with samples from the field, where infectivity may decay owing to environmental factors such as temperature, humidity and UV radiation. Therefore, a more sensitive method that can determine the infectivity of IAVs is needed to assess the exposure risk more accurately.
Second, the exposure doses calculated above are cumulative over 1–24 h, while the ID50 measured by Alford et al. [26] is based on inoculations completed within 1 min. In reality, viruses depositing within the respiratory tract will be cleared by mucociliary action instead of simply accumulating at the deposition sites [27]. Therefore, the exposure doses calculated in this study provide only the simplest estimation of the amount inhaled. Models taking into account particle deposition efficiency within the respiratory tract as well as host defence mechanisms are needed to estimate the infection risk more accurately. The deposition efficiency of particles can range from less than 1 per cent to nearly 100 per cent, depending on particle size, density, airway geometry and the individual's breathing pattern [21].
Third, the assumption of uniform concentrations throughout a room is complicated when accounting for true dispersion patterns from a point source and the existence of non-homogeneous ventilation. Finally, only IAVs were measured in this study, so it did not account for infection risk owing to the influenza B virus (IBV). However, according to the nationwide Weekly Influenza Surveillance by the US World Health Organization and National Respiratory and Enteric Virus Surveillance System [28], the IBV accounted for only 0–3.3% of samples that tested positive for influenza viruses during the period 4 October 2009 to 10 April 2010. Therefore, underestimation owing to exclusion of IBVs may be negligible.
On the other hand, the virus deposition flux to surfaces was estimated to be only 13 ± 7 genome copies m−2 h−1. Over an 8 h workday, 106 ± 60 genome copies m−2 could accumulate on surfaces. This analysis suggests that all surfaces, not just those that come into direct contact with an infected host, can harbour influenza viruses, although they are not expected to survive beyond 2–3 days [29]. However, the amount deposited on surfaces via Brownian motion seems unlikely to produce infectious doses, as the surface–hand–nasal mucosa route requires transferring at least 104 TCID50 from the surface [29]. This estimation does not account for deposition owing to gravitational settling, which is important for larger particles and can result in higher deposition fluxes in the vicinity of emission.
5. Conclusions
The concentrations and size distributions of airborne influenza viruses were measured in a health centre, a day-care facility and aeroplanes by qRT–PCR. During the 2009–2010 flu season, 50 per cent of the samples collected (8/16) contained IAVs with concentrations ranging from 5800 to 37 000 genome copies m−3. On average, 64 per cent of virus-laden particles were found to be associated with particles smaller than 2.5 µm, which can remain airborne for hours. Modelling of virus concentrations indoors suggests a source strength of 1.6 ± 1.2 × 105 genome copies m3 h−1 and a deposition flux onto surfaces of 13 ± 7 genome copies m−2 h−1. Doses of 30 ± 18, 236 ± 140 and 708 ± 419 TCID50 were estimated for 1, 8 and 24 h exposures, respectively. As a whole, these results provide quantitative support for the possibility of airborne transmission of influenza.
Acknowledgements
This material is based in part upon work supported by the National Science Foundation under Grant Number CBET-0547107. We thank A. Pruden for sharing laboratory space and equipment, J. Petruska for much assistance, K. Charoensiri of the university health centre and a day-care centre in Blacksburg for allowing sampling in their facilities.
References
- 1.Tellier R. 2009. Aerosol transmission of influenza A virus: a review of new studies. J. R. Soc. Interface 6, S783–S790 10.1098/rsif.2009.0302.focus (doi:10.1098/rsif.2009.0302.focus) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brankston G., Gitterman L., Hirji Z., Lemieux C., Gardam M. 2007. Transmission of influenza A in human beings. Lancet Infect. Dis. 7, 257–265 10.1016/S1473-3099(07)70029-4 (doi:10.1016/S1473-3099(07)70029-4) [DOI] [PubMed] [Google Scholar]
- 3.Tellier R. 2006. Review of aerosol transmission of influenza A virus. Emerg. Infect. Dis. 12, 1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabian P., Mcdevitt J. J., Lee W.-M., Houseman E. A., Milton D. K. 2009. An optimized method to detect influenza virus and human rhinovirus from exhaled breath and the airborne environment. J. Environ. Monit. 11, 314–317 10.1039/b813520g (doi:10.1039/b813520g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fabian P., Mcdevitt J. J., Houseman E. A., Milton D. K. 2009. Airborne influenza virus detection with four aerosol samplers using molecular and infectivity assays: considerations for a new infectious virus aerosol sampler. Indoor Air 19, 433–441 10.1111/j.1600-0668.2009.00609.x (doi:10.1111/j.1600-0668.2009.00609.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen P.-S., et al. 2009. Quantification of airborne influenza and avian influenza virus in a wet poultry market using a filter/real-time qPCR method. Aerosol Sci. Technol. 43, 290–297 10.1080/02786820802621232 (doi:10.1080/02786820802621232) [DOI] [Google Scholar]
- 7.Blachere F. M., et al. 2009. Measurement of airborne influenza virus in a hospital emergency department. Clin. Infect. Dis. 48, 438–440 10.1086/596478 (doi:10.1086/596478) [DOI] [PubMed] [Google Scholar]
- 8.Lindsley W. G., et al. 2010. Distribution of airborne influenza virus and respiratory syncytial virus in an urgent care medical clinic. Clin. Infect. Dis. 50, 693–698 10.1086/650457 (doi:10.1086/650457) [DOI] [PubMed] [Google Scholar]
- 9.Boone S. A., Gerba C. P. 2005. The occurrence of influenza A virus on household and day care center fomites. J. Infect. 51, 103–109 10.1016/j.jinf.2004.09.011 (doi:10.1016/j.jinf.2004.09.011) [DOI] [PubMed] [Google Scholar]
- 10.Wagner B., Coburn B., Blower S. 2009. Calculating the potential for within-flight transmission of influenza A (H1N1). BMC Med. 7, 81. 10.1186/1741-7015-7-81 (doi:10.1186/1741-7015-7-81) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moser M. R., Bender T. R., Margolis H. S., Noble G. R., Kendal A. P., Ritter D. G. 1979. An outbreak of influenza aboard a commercial airliner. Am. J. Epidemiol. 110, 1–6 [DOI] [PubMed] [Google Scholar]
- 12.Fabian P., Mcdevitt J. J., Dehaan W. H., Fung R. O. P., Cowling B. J., Chan K. H., Leung G. M., Milton D. K. 2008. Influenza virus in human exhaled breath: an observational study. PLoS ONE 3, e2691. 10.1371/journal.pone.0002691 (doi:10.1371/journal.pone.0002691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee W.-M., Grindle K., Pappas T., Marshall D. J., Moser M. J., Beaty E. L., Shult P. A., Prudent J. R., Gern J. E. 2007. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J. Clin. Microbiol. 45, 2626–2634 10.1128/jcm.02501-06 (doi:10.1128/jcm.02501-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward C. L., Dempsey M. H., Ring C. J. A., Kempson R. E., Zhang L., Gor D., Snowden B. W., Tisdale M. 2004. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J. Clin. Virol. 29, 179–188 10.1016/S1386-6532(03)00122-7 (doi:10.1016/S1386-6532(03)00122-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Elden L. J. R., Nijhuis M., Schipper P., Schuurman R., van Loon A. M. 2001. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol. 39, 196–200 10.1128/jcm.39.1.196-200.2001 (doi:10.1128/jcm.39.1.196-200.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munster V. J., et al. 2009. Practical considerations for high-throughput influenza A virus surveillance studies of wild birds by use of molecular diagnostic tests. J. Clin. Microbiol. 47, 666–673 10.1128/jcm.01625-08 (doi:10.1128/jcm.01625-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Vries E., et al. 2010. Evaluation of a rapid molecular algorithm for detection of pandemic influenza A (H1N1) 2009 virus and screening for a key oseltamivir resistance (H275Y) substitution in neuraminidase. J. Clin. Virol. 47, 34–37 10.1016/j.jcv.2009.09.030 (doi:10.1016/j.jcv.2009.09.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J.-R., Lo J., Ho Y.-L., Wu H.-S., Liu T. 2011. Pandemic H1N1 and seasonal H3N2 influenza infection in the human population show different distributions of viral loads, which substantially affect the performance of rapid influenza tests. Virus Res. 155, 163–167 10.1016/j.virusres.2010.09.015 (doi:10.1016/j.virusres.2010.09.015) [DOI] [PubMed] [Google Scholar]
- 19.Tompkins J. A., White J. A., Bozer Y. A., Tanchoco J. M. A. 2010. Facilities planning. New York, NY: John Wiley & Sons, Inc [Google Scholar]
- 20.Mangili A., Gendreau M. A. 2005. Transmission of infectious diseases during commercial air travel. Lancet 365, 989–996 10.1016/S0140-6736(05)71089-8 (doi:10.1016/S0140-6736(05)71089-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinds W. C. 1999. Aerosol technology, p. 49 New York, NY: Wiley [Google Scholar]
- 22.Poon L. L., Chan K. H., Smith G. J., Leung C. S., Guan Y., Yuen K. Y., Peiris J. S. 2009. Molecular detection of a novel human influenza (H1N1) of pandemic potential by conventional and real-time quantitative RT–PCR assays. Clin. Chem. 55, 1555–1558 10.1373/clinchem.2009.130229 (doi:10.1373/clinchem.2009.130229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ATCC 2009. 8/20/2009 Converting TCID[50] to plaque forming units (PFU). See http://www.atcc.org/CulturesandProducts/TechnicalSupport/FrequentlyAskedQuestions/tabid/469/Default.aspx (retrieved 1 July 2010)
- 24.Yang S., Lee G. W. M., Chen C.-M., Wu C.-C., Yu K.-P. 2007. The size and concentration of droplets generated in human subjects. J. Aerosol. Med. 20, 484–494 10.1089/jam.2007.0610 (doi:10.1089/jam.2007.0610) [DOI] [PubMed] [Google Scholar]
- 25.US EPA 1994. Methods for derivation of inhalation reference concentrations and application of inhalation dosimetry. EPA/600/8-90/066F, US Environmental Protection Agency, Office of Research and Development, Office of Health and Environmental Assessment, Washington, DC [Google Scholar]
- 26.Alford R. H., Kasel J. A., Gerone P. J., Knight V. 1966. Human influenza resulting from aerosol inhalation. Proc. Soc. Exp. Biol. Med. 122, 800–804 [DOI] [PubMed] [Google Scholar]
- 27.Oberdorster G. 1993. Lung dosimetry—pulmonary clearance of inhaled particles. Aerosol Sci. Technol. 18, 279–289 10.1080/02786829308959605 (doi:10.1080/02786829308959605) [DOI] [Google Scholar]
- 28.US CDC 2010. Seasonal influenza (flu). Past weekly surveillance reports See http://www.cdc.gov/flu/weekly/pastreports.htm (retrieved 23 November 2010) [Google Scholar]
- 29.Bean B., Moore B. M., Sterner B., Peterson L. R., Gerding D. N., Balfour H. H., Jr 1982. Survival of influenza viruses on environmental surfaces. J. Infect. Dis. 146, 47–51 [DOI] [PubMed] [Google Scholar]