Abstract
The 22q13.3 deletion syndrome results from loss of terminal segments of varying sizes at 22qter. Few genotype–phenotype correlations have been found but all patients have mental retardation and severe delay, or absence of, expressive speech. We carried out clinical and molecular characterization of 13 patients. Developmental delay and speech abnormalities were common to all and comparable in frequency and severity to previously reported cases. Array-based comparative genomic hybridization showed the deletions to vary from 95 kb to 8.5 Mb. We also carried out high-resolution 244K array comparative genomic hybridization in 10 of 13 patients, that defined the proximal and distal breakpoints of each deletion and helped determine the size, extent, and gene content within the deletion. Two patients had a smaller 95 kb terminal deletion with breakpoints within the SHANK3 gene while three other patients had a similar 5.5 Mb deletion implying the recurrent nature of these deletions. The two largest deletions were found in patients with ring chromosome 22. No correlation could be made with deletion size and phenotype although complete/partial SHANK3 was deleted in all patients. There are very few reports on array comparative genomic hybridization analysis on patients with the 22q13.3 deletion syndrome, and we aim to accurately characterize these patients both clinically and at the molecular level, to pave the way for further genotype–phenotype correlations.
Keywords: array CGH, autism spectrum disorders, developmental delay, SHANK3, speech delay, 22q13.3 deletion syndrome
INTRODUCTION
The 22q13.3 deletion syndrome, also known as the Phelan–McDermid syndrome (OMIM # 606232), results from deletions of variable length involving the terminal long arm of chromosome 22, either through a simple deletion or secondary to an unbalanced structural rearrangement. To date, there have been over 100 cases reported in the literature; however, the cryptic nature of this deletion in a significant fraction of cases often precludes a true estimate of its prevalence making it likely to be under diagnosed. Deletion sizes from 100 kb to greater than 9 Mb have been reported to give rise to this syndrome. Over the years, common features have emerged which include developmental delay and delayed or absent speech [Phelan et al., 2005]. While these seem to be the predominant features, autistic features have also been found in nearly 50% of the patients [Cusmano-Ozog et al., 2007]. The clinical phenotype resulting from the submicroscopic terminal deletion has attracted increased attention lately, due to developmental delay and significant speech impairment typical of autism spectrum disorders observed in these children.
Efforts to identify the crucial gene(s) within this terminal segment led to the identification of SHANK3 [SH3 and Ankyrin domain containing protein; formerly known as proline-rich synapse associated protein 2 (PROSAP2)] as the critical gene within this interval. [Sheng et al., 2000; Sala et al., 2001; Grant, 2003] Disruption of SHANK3 resulting in features of 22q13.3 syndrome was first reported in a child with a de novo balanced translocation t(12; 22)(q24.1; q13.3) [Bonaglia et al., 2001]. Subsequently, Anderlid et al. [2002] described the disruption of SHANK3 resulting from a 100-kb deletion in a patient with the phenotype of 22q13.3 deletion syndrome. Wilson et al. [2003] published a report of 56 patients with deletion sizes ranging from 130 kb to 9 Mb and showed that haploinsufficiency of SHANK3 was responsible for the major neurological symptoms of this syndrome. This was followed by identification of a recurrent breakpoint within the SHANK3 gene in two unrelated patients who had a similar SHANK3 disruption but exhibited different degrees of severity in their phenotypes [Bonaglia et al., 2006]. Recently, point mutations resulting in haploinsufficiency for SHANK3 have been identified in a small fraction of patients with idiopathic autism [Durand et al., 2007; Moessner et al., 2007]. The smallest deletion reported involves the last 100 kb of chromosome 22q13.3, which includes three genes, SHANK3, ACR, and RABL2B. Interestingly, a small fraction of patients with this syndrome harbored submicroscopic rearrangements at 22qter that interrupted or led to a complete loss of these three genes only. The largest terminal 22q deletions included more than 50 known genes.
Array-based comparative genomic hybridization (aCGH) helps to identify genomic rearrangements at a resolution that is 5–10 times higher than that of routine chromosome analysis by karyotyping. The diagnosis of the 22q13.3 deletion syndrome is based on cytogenetic, molecular cytogenetic, and/or molecular demonstration of loss or disruption of the 22q13.3 chromosomal region. Historically, most of the reported deletions in the 22q13.3 region were identified and characterized based on routine karyotyping as well as FISH using the ARSA probe. To our knowledge, no further descriptions in the literature delineating the breakpoints in these deletions have been made. We report the clinical and molecular data for a series of 13 patients with deletion 22q13.3 with sizes varying from 95 kb to ~8.5 Mb. High-resolution aCGH was carried out on 10 of these patients to delineate the size, extent, gene content, and approximate boundaries of the deletions. A comparison of their clinical features with those published in the literature and relevant commonalities in the emerging phenotype are demonstrated.
SUBJECTS AND METHODS
Patients
Clinical aCGH was performed on 8,500 samples that were referred to the Kleberg Cytogenetic Laboratory, Houston, TX, USA, from September 2005 to September 2007. From these, 21 cases were identified with varying sizes of deletion in the 22q13.3 region. A total of 12 patients were included in this study, for which clinical information was available. The 13th patient was also included who was diagnosed with a deletion using FISH. Informed consent approved by the Institutional Review Board for Human Subject Research at Baylor College of Medicine was obtained from their families.
DNA Samples
DNA was extracted from whole blood using the Puregene DNA extraction kit (Gentra, Minneapolis, MN) according to the manufacturer’s instructions.
Chromosome and FISH Analyses
Chromosome and FISH analyses were performed on peripheral blood lymphocytes using standard procedures. FISH analyses with the bacterial artificial chromosome (BAC) clones were performed using standard procedures. Briefly, the BAC clone of interest was grown in TB media with 20 μg/ml chloramphenicol. DNA was extracted from BAC clones (Eppendorf Plasmid Mini Prep kit, Hamburg, Germany) and directly labeled with SpectrumOrange™ dUTP by nick translation (Vysis, Downer Grove, IL) according to the manufacturer’s instructions. Digital FISH images were captured by a Power Macintosh G3 System and MacProbe version 4.4 (Applied Imaging, San Jose, CA).
Microarray Procedures and Data Analysis
Four consecutive versions of arrays, each with a greater genetic coverage and higher genome resolution, representing the chromosome microarray (CMA) evolution in our clinical diagnostic laboratory, were applied during the study period. The CMA V5 BAC and V6 BAC platforms were BAC arrays that have been reported in our previous studies [Shao et al., 2008]. The CMA BAC V6 consisted of 1,475 BAC clones and interrogated over 150 genomic disorder loci, with expanded coverage in pericentromeric and subtelomeric regions, and greater backbone coverage of the genome with inclusion of one clone per band at 650 cytogenetic banding resolutions. Blood samples from Patients 1, 5, 6, 9, 10, and 12 were run on the CMA V5 BAC. The CMA BAC V6 platform was used for Patients 3, 4, and 7.
The CMA V6 OLIGO platform was designed based on the V6 BAC and consisted of 42,640 oligonucleotides, with the average of 20–40 oligonucleotides corresponding to each BAC clone genomic locus [Ou et al., 2008]. Strict oligonucleotide selection criteria and removal of repetitive sequences were employed to ensure high sensitivity and specificity with greater dynamic range. The V6 OLIGO platform was designed by Baylor Medical Genetics Laboratories and manufactured by Agilent Technologies, Inc. (Santa Clara, CA) (http://www.bcm.edu/cma/table.htm). Samples from Patients 8 and 11 were run on V6 BAC.
The oligonucleotide array V7.2 OLIGO is a custom-designed array with about 105,000 oligonucleotides of 60 bp manufactured by Agilent Technologies, Inc. This array selected the best performing oligos from the electronic library from Agilent as probes for virtually all the known microdeletion or microduplication syndromes and the pericentromeric and subtelomeric regions. In addition to these targeted regions, the entire genome (between disease regions) is covered with an average resolution of 30 kb while excluding repetitive sequences through a combination of bioinformatics and computation. The V7.2 OLIGO platform was used for Patient 2. Samples from Patients 3, 5, and 12 that were previously run on older version arrays were re-analyzed on the V7.2 OLIGO.
The procedures for probe labeling and hybridization of the BAC arrays were reported previously [Lu et al., 2007]. The procedures for DNA digestion, labeling, and hybridization for the oligo arrays were performed according to the manufacturer’s instructions, with some modifications [Ou et al., 2008]. The slides were scanned into image files using an Agilent G2565 laser scanner. Microarray image files of oligoarrays were quantified using Agilent Feature extraction software (v9.0), and text file outputs from the quantitation analysis were imported into our in-house analysis package for copy number analysis, as described previously [Ou et al., 2008].
244K aCGH Analysis
Whole Human Genome Oligo Microarray Kits 244K (Agilent Technologies, Inc.) were used to analyze DNA from 10/13 patients to further delineate the identified genomic losses. These included Patients 1 and 2 and 4–11. The procedures for DNA digestion, labeling, and hybridization were performed according to the manufacturer’s instructions with some modifications [Probst et al., 2007].
RESULTS
Molecular Analysis of Deletion
Molecular testing that detected the deletions in the patients was undertaken as follows:
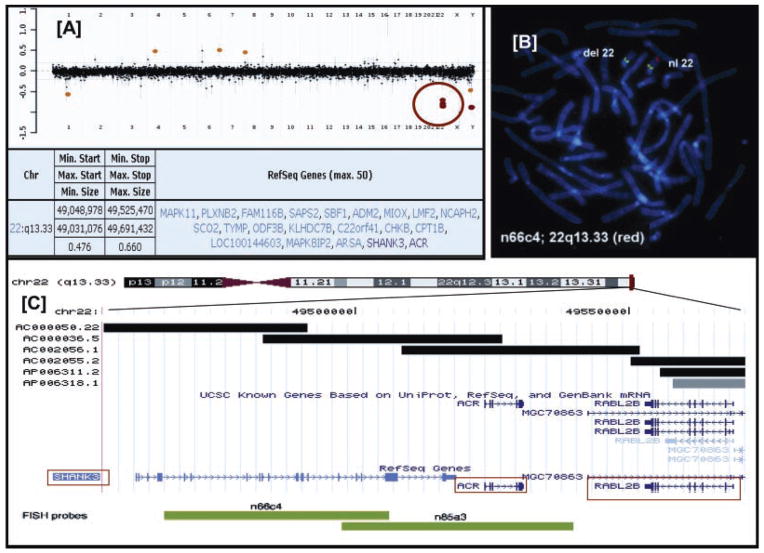
Clinical aCGH was carried out in 12 of 13 patients. An example of Patient 3 is shown in Figure 1A. Deletions were confirmed by partial karyotype (Patients 6 and 9) and by FISH in the remaining cases. All deletions were terminal as confirmed by FISH utilizing subtelomeric probes. Patients 11 and 12 were noted to have ring chromosome 22 by previous chromosomal analysis.
Higher resolution aCGH (244K) was carried out in 10 of 13 (77%) cases to determine the sizes and extent of the deletion (Fig. 2). The smallest terminal deletion was approximately 95 kb (Patients 1 and 2) and the largest deletion was found in Patient 12 (8.55 Mb—data not shown). While Patients 1 and 2 had partial deletions of SHANK3 with proximal breakpoints located within the gene, Patients 3–12 had complete deletions of SHANK3.
FISH testing in Patient 13 was initially performed to diagnose DiGeorge syndrome. Interestingly, it showed a deletion of the ARSA probe, which was used as a control probe for chromosome 22q, while the DiGeorge probe hybridized normally. Further testing showed the deletion to involve SHANK3 using the n66c4 probe as shown in Figure 1B. The location of the probes is shown in Figure 1C.
FIG. 1.
A: Representation of the clinical array CGH data in Patient 3 indicating a loss in copy number (red circle). Below is shown the size and extent of deletions at 22q13 and the genes involved in the region. B: FISH analysis using a cosmid clone (n66c4) as a target probe labeled in red indicating a deletion involving the SHANK3 gene while RP11-316L10 is labeled in green and serves as a control probe. C: Physical map of 22q13.3 region representing SHANK3 and its neighboring genes ACR and RABL2B, the latter toward the telomere. FISH probes (COSMIDS) used in the hybridizations are indicated in the green bar below.
FIG. 2.

Molecular data mapping the deletions in 10 patients on the 244K oligoarray which include the estimated proximal breakpoint and the distal breakpoint and the size of the deletion. Note: the deletion disrupts the SHANK3 gene in Patients 1 and 2.
Clinical Profiling of Patients
The age of patients studied ranged from 3 to 19 years, and there were six males and seven females. Developmental delay and significant speech delay and/or loss of speech were common to all. Additional problems including history of feeding difficulties and recurrent infections were less common (3/13, 23%) and found to coexist in Patients 8, 9, and 12 only. Seizures were reported in only 4/13 (31%) individuals while Patient 4 had an abnormal EEG with no clinically apparent seizure disorder. Table I outlines clinical features commonly seen in patients with the 22q13.3 deletion syndrome and their presence in the patients described here.
TABLE I.
Clinical Features of Patients in This Study, Along With Their Deletion Sizes Arranged From Smallest to Largest Deletion
| Patient | Age/sex | Deletion size | Growth | Feeding problems | DD | Speech delay/loss of speech | Behavioral issues | Autism/ASD | Regression | Seizures | Recurrent infections |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 9y/M | 95 kb | Normal | − | + | + | + | + | + | + | − |
| 2 | 3y/F | 95 kb | Normal | − | + | + | + | N/A | + | − | − |
| 3 | 13y/M | 504 kb | Normal | Unknown | + | + | + | N/A | − | − | − |
| 4 | 9y/M | 1.01 Mb | Normal | − | + | + | + | + | − | Abn EEG | − |
| 5 | 3y/F | 3.03 Mb | Normal | − | + | + | + | + | − | − | +Ear |
| 6 | 11y/F | 3.06 Mb | Accelerated | − | + | + | + | N/A | − | − | − |
| 7 | 19y/M | 5.65 Mb | Accelerated | + | + | + | + | N/A | − | − | − |
| 8 | 5y/F | 5.66 Mb | Normal | + | + | + | + | N/A | − | + | +UTI |
| 9 | 31y/F | 5.79 Mb | Normal | + | + | + | + | − | − | + | +URI |
| 10 | 3y/F | 8.23 Mb | Normal | + | + | + | + | N/A | − | − | − |
| 11 | 9y/M | 8.44 Mb r(22) | Normal | − | + | + | + | N/A | − | − | − |
| 12 | 5y/M | 8.55 Mb r(22) | Short | + | + | + | − | N/A | − | + | + PNA |
| 13 | 15y/F | Terminal deletiona | Short | − | + | + | + | − | − | − | − |
ASD, autism spectrum disorder; Abn EEG, abnormal electroencephalogram; UTI, urinary tract infections; URI, upper respiratory tract infections; PNA, pneumonia; y, years; N/A, patients who did not get a formal neuropsychological evaluation.
Size not determined, terminal deletion detected by deletion of ARSA probe on FISH analysis. Patient not available for re-analysis.
Behavioral problems such as repetitive behaviors, impaired socialization, hyperactivity, and self-stimulation were observed in 12 of 13 (92%) patients. Patient 12 was ventilator-dependent and unable to move; hence his behavior could not be assessed. Five of 13 (38%) patients had a formal neuro-developmental assessment. Patient 5 was diagnosed with pervasive developmental delay with a developmental quotient (DQ) < 50. Patients 1 and 4 were diagnosed with autism using Autism Diagnostic Observation Schedule (ADOS), Autism Diagnostic Interview (ADI), and Gilliam and Childhood Autism Rating Scale, respectively. Patients 9 and 13 had formal testing but did not meet criteria for a diagnosis of autism.
Common physical findings noted in the patients are listed in Table II (Fig. 3). Facial dysmorphism was significant in at least four patients. Hypotonia was present in 4 of 13 (31%) patients. A comparison of the frequencies of the common clinical features observed in patients with the 22q13.3 deletion syndrome as seen in this study and two previously published reports is listed in Table III [Phelan et al., 2005; Cusmano-Ozog et al., 2007]. Brain imaging was performed in 9 of 13 patients as an initial test to evaluate the developmental delay. Patients 3 and 5 showed mild thinning of the corpus callosum and Patients 3, 8, and 10 had mildly delayed myelination. Brain MRI in Patient 1 showed brain asymmetry with a larger size of left lateral ventricle compared to the right and loss of normal left temporal horn morphology. The remainder (4 of 9 or 44%) had a normal brain MRI. These results are similar to reports of brain imaging abnormalities in patients with the 22q13.3 deletion syndrome [Philippe et al., 2008].
TABLE II.
Physical Exam Findings of the Patients Described in This Study
| Patient | Age/sex | Deletion size | Epicanthal folds | Macrotia | Bulbous nose | Pointed chin | Large hands | Small toe nails | Hypotonia | Lymphedema |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 9y/M | 95 kb | − | − | − | − | − | − | − | − |
| 2 | 3y/F | 95 kb | − | + | + | − | − | − | − | − |
| 3 | 13y/M | 504 kb | − | + | − | − | − | − | Unknown | − |
| 4 | 9y/M | 1.01 Mb | + | + | + | − | + | − | − | − |
| 5 | 3y/F | 3.03 Mb | − | + | + | − | − | − | − | − |
| 6 | 11y/F | 3.06 Mb | − | − | + | − | − | − | + | − |
| 7 | 19y/M | 5.65 Mb | + | + | + | + | + | + | − | + |
| 8 | 5y/F | 5.66 Mb | − | − | + | − | + | − | + | − |
| 9 | 31y/F | 5.79 Mb | + | + | + | + | + | + | − | + |
| 10 | 3y/F | 8.23 Mb | − | + | + | + | + | + | + | − |
| 11 | 9y/M | 8.44 Mb r(22) | + | + | + | − | − | − | − | − |
| 12 | 5y/M | 8.55 Mb r(22) | − | + | − | − | + | − | +Neonatal | + |
| 13 | 15y/F | Terminal deletiona | − | − | − | − | − | − | − | − |
Size of deletion not determined, terminal deletion detected by deletion of ARSA probe on FISH analysis. (Patient not available for re-analysis).
FIG. 3.

Facial features of six 22q13.3 deletion syndrome patients from this cohort. Photographs of Patients 2, 4, 5, 7, 8, and 10 from left to right then bottom left to right, respectively. The common features are listed in Table II. Individual features include: Patient 2: full cheeks; Patient 4: mild ptosis and long eyelashes with thick, bushy eyebrows; Patient 5: upslanting palpebral fissures with flat nasal bridge and long philtrum; Patient 7: bitemporal narrowing, thick bushy eyebrows, and low set ears; Patient 8: flat nasal bridge; Patient 10: bitemporal narrowing, upslanting palpebral fissures, and long eyelashes.
TABLE III.
Comparison of Frequencies of Common Clinical Features as Seen in the Patients in This Study and Previously Published Reports [Phelan et al., 2001; Cusmano-Ozog et al., 2007]
| Phenotype | This study (n = 13) | Cusmano-Ozog et al. (n = 107) | Phelan et al. (n = 37) |
|---|---|---|---|
| Growth—normal to accelerated growth | 11 | 95 | 35 |
| Feeding difficulties | 5 | N/A | N/A |
| Seizures | 4 | 25 | 10 |
| Autistic-like features and other behavioral issues | 12 | 47 | N/A |
| Hypotonia | 4 | 92 | 36 |
| Developmental delay | 13 | 105 | 37 |
| Absent/severely delayed speech | 13 | 103 | 37 |
| Epicanthal folds | 4 | 32 | 15 |
| Macrotia | 9 | 58 | 24 |
| Bulbous nose | 8 | N/A | N/A |
| Large hands | 6 | 35 | 25 |
| Small nails | 3 | 39 | 29 |
DISCUSSION
Patients with the 22q13.3 deletion syndrome have a common phenotype characterized by developmental delay, including gross motor delays and absent or delayed speech, hypotonia, and craniofacial dysmorphic features. Clinical features of the patients in the present study are similar to that of previous reports (Table III) except for the lower prevalence of hypotonia in the present report. As established in previous reports, the present data also suggest that developmental delay is an almost universal feature of the 22q13.3 deletion syndrome. Behavioral problems such as impaired social skills, persistent mouthing, and chewing of nonfood items, inattentiveness, and hyperactivity were found in 12 of 13 (92%) patients. Sleep disturbances were also seen in 6 of 13 (46%) of the patients. A common association of the 22q13.3 deletion syndrome with autism spectrum disorders has begun to emerge, which is also supported in this cohort although only five patients received a formal neuropsychological evaluation. The first reported association between autism and 22q13.3 deletion was published in a 14-year-old girl [Goizet et al., 2000].
Autism spectrum disorders are etiologically heterogeneous and are associated with a recognizable cause in only 10% of patients, most commonly with fragile X syndrome, tuberous sclerosis, and cytogenetically detectable chromosomal anomalies. Mutations in the neuroligin genes (NLGN3 and NLGN4) on the X chromosome [Jamain et al., 2003] and SHANK3 on chromosome 22q13 [Durand et al., 2007] are monogenic causes of autism spectrum disorders. In one study of 400 unrelated subjects with autism spectrum disorder, one de novo mutation and two gene deletions of the SHANK3 gene were discovered, contributing 0.75% causality to this cohort [Moessner et al., 2007]. However, in another recent study by Philippe et al. [2008] autism was suspected in seven of eight patients with the 22q13.3 deletion syndrome. These patients did not fulfill DSM-IV criteria for autism. Their relationship pattern, language development, and nature of repetitive behaviors were distinct from autism. In the patients described here, two of five (40%) patients who underwent neuropsychological evaluations were diagnosed with autism. This is similar to previous studies [Cusmano-Ozog et al., 2007].
Two of the patients reported here were found to have a ring chromosome 22 (Patients 11 and 12). r(22) is assumed to arise from breakage and subsequent fusion of both chromosome arms to generate a ring chromosome with concomitant loss of terminal short and long arm sequences. A review of 17 patients with r(22) showed that clinical features are similar in patients with simple 22q13 deletion and r(22) patients when they are of similar ages and similarly sized deletions [Luciani et al., 2003]. The features common in both conditions are global developmental delay with severe speech delay, hypotonia, macrotia, epicanthal folds, and small toenails. Patient 12 showed a degree of growth failure which is similar to previously published reports of patients with r(22). These patients are known to exhibit growth failure rather than accelerated growth as seen in 22q13.3 deletion syndrome [Hunter et al., 1977; Jeffries et al., 2005].
Historically, FISH and chromosome G-banding have been commonly used to demonstrate the deletions in this syndrome [Doheny et al., 1997]. Koolen et al. [2005] used aCGH on nine patients with subtelomeric deletions of 22q and reported deletion sizes ranging from 3.3 to 8.4 Mb. At that time, they were unable to demonstrate a deletion–phenotype correlation. They used a tiling resolution array with an average clone spacing of 100 kb. However, minimal coverage across the most terminal segment of 22q precluded identification of disruptions within or in the immediate vicinity of the SHANK3 gene, the haploinsufficiency of which has been established to be associated with the neurological phenotype of patients with the 22q13.3 deletion syndrome. It is clear that aCGH analysis offers higher resolution compared to microscopic cytogenetics for molecular characterization of patients with this phenotype.
All the patients described here had terminal deletion of the 22q13 region and in all of them, either all or a part of the SHANK3 gene was found to be deleted. The deletion sizes ranged from 95 kb (Patient 1) to 8.55 Mb (Patient 12). The two patients with the smallest deletion had distinct phenotypes with Patient 1 showing more severe behavioral issues compared to Patient 2. This is an example of the difficulty in making genotype–phenotype correlations in patients with the 22q13.3 deletion syndrome. One hypothesis is that epigenetic factors may play a role in the clinical features of this syndrome. Alternatively, sequence variations in alleles on the nondeleted chromosome may have qualitative and/or quantitative effects on their expression that is unmasked by deletion of their paired allele [Ching et al., 2005]. Three patients (Patients 7–9) had a similar sized ~5.5 Mb deletion. All three patients had feeding problems while two of three had behavioral issues, seizures, and recurrent infections. As more such patients are reported with exact deletion sizes and breakpoints, a genotype–phenotype correlation could emerge, possibly delineating the role of other genes contributing to the 22q13.3 phenotype. A recent report by Wilson et al. [2008] of two patients with interstitial deletion of the 22q13 region with intact SHANK3 indicates that haploinsufficiency for other 22q13 genes could also have major effects on cognitive and language development. More studies are required to explore the interaction of these proximal genes and their role in the neurological phenotype of this syndrome.
Approximately 90 genes are deleted in the largest sized deletion found in Patient 12, whereas the smallest deletion segment contained the three genes, SHANK3, ACR, and RABL2B (Fig. 1C). Some of the genes (besides SHANK3) responsible for clinically significant disorders that are present in this interval include ARSA (metachromatic leukodystrophy), TYMP (mitochondrial neuro-gastrointestinal encephalomyopathy), MLC1 (megalencephalic leukoencephalopathy with subcortical cysts), ALG12 (congenital disorders of glycosylation 1g), ATXN10 (cerebellar ataxia), UPK3A (renal adysplasia), and SULT4A1 (susceptibility to schizophrenia).
We hypothesize that there may be as yet uncharacterized genes in the vicinity of SHANK3, SNP, or other variants within the regulatory sequences of SHANK3 itself, or modifier effects of unlinked genes that qualitatively or quantitatively affect its expression. If the deletion breakpoint is located within these regulatory genes, expression of SHANK3 could be altered leading to a clinical phenotype. Point mutations in SHANK3, just as in the case of the neuroligin genes, are rare in patients with autism. Micro-deletions and disruptions of SHANK3, however, provide the most convincing evidence for a role in autism. It is possible that similar microdeletions will identify additional genes that may contribute to the autism phenotype [Jacquemont et al., 2006]. Such additional candidates may include genes that are involved in the growth and development of the pre- and postsynaptic compartments.
In summary, this study illustrates the use of aCGH in delineating deletion breakpoints in the 22q13.3 deletion syndrome and highlights the utility of aCGH as a diagnostic tool for analysis of patients with developmental delay, unexplained hypotonia, and autism spectrum disorder with or without craniofacial dysmorphism. We anticipate that as more patients are diagnosed with the 22q13.3 deletion syndrome using aCGH, a clearer picture of genotype–phenotype correlation may emerge based on the variety of the deleted genes and the differing sizes of the deletions.
Acknowledgments
We are grateful to Dr. James Lupski for reading the manuscript and offering valuable comments and to the patients and their families, whose contribution and support made this possible.
References
- Anderlid BM, Schoumans J, Anneren G, Tapia-Paez I, Dumanski J, Blennow E, Nordenskjold M. FISH-mapping of a 100-kb terminal 22q13 deletion. Hum Genet. 2002;110:439–443. doi: 10.1007/s00439-002-0713-7. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Borgatti R, Felisari G, Gagliardi C, Selicorni A, Zuffardi O. Disruption of the ProSAP2 gene in a t(12;22)-(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. Am J Hum Genet. 2001;69:261–268. doi: 10.1086/321293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Mani E, Aceti G, Anderlid BM, Baroncini A, Pramparo T, Zuffardi O. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet. 2006;43:822–828. doi: 10.1136/jmg.2005.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching TT, Maunakea AK, Jun P, Hong C, Zardo G, Pinkel D, Albertson DG, Fridlyand J, Mao J-H, Shchors K, Weiss WA, Costello JF. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat Genet. 2005;37:645–651. doi: 10.1038/ng1563. [DOI] [PubMed] [Google Scholar]
- Cusmano-Ozog K, Manning MA, Hoyme HE. 22q13.3 deletion syndrome: A recognizable malformation syndrome associated with marked speech and language delay. Am J Med Genet Part C. 2007;145C:393–398. doi: 10.1002/ajmg.c.30155. [DOI] [PubMed] [Google Scholar]
- Doheny KF, McDermid HE, Harum K, George TH, Raymond GV. Cryptic terminal rearrangement of chromosome 22q13.32 detected by FISH in two unrelated patients. J Med Genet. 1997;34:640–644. doi: 10.1136/jmg.34.8.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Roge B, Heron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–32. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goizet C, Excoffier E, Taine L, Taupiac E, El Moneim AA, Arveiler B, Bouvard M, Lacombe D. Case with autistic syndrome and chromosome 22q13.3 deletion detected by FISH. Am J Med Genet. 2000;96:839–844. [PubMed] [Google Scholar]
- Grant SG. Synapse signalling complexes and networks: Machines underlying cognition. BioEssays. 2003;25:1229–1235. doi: 10.1002/bies.10381. [DOI] [PubMed] [Google Scholar]
- Hunter AG, Ray M, Wang HS, Thompson DR. Phenotypic correlations in patients with ring chromosome 22. Clin Genet. 1977;12:239–249. doi: 10.1111/j.1399-0004.1977.tb00933.x. [DOI] [PubMed] [Google Scholar]
- Jacquemont ML, Sanlaville D, Redon R, Raoul O, Cormier-Daire V, Lyonnet S, Amiel J, Le Merrer M, Heron D, de Blois MC, Prieur M, Vekemans M, Carter NP, Munnich A, Colleaux L, Philippe A. Array-based comparative genomic hybridization identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:843–849. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T Paris Autism Research International Sibpair Study. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries AR, Curran S, Elmslie F, Sharma A, Wenger S, Hummel M, Powell J. Molecular and phenotypic characterization of ring chromosome 22. Am J Med Genet Part A. 2005;137A:139–147. doi: 10.1002/ajmg.a.30780. [DOI] [PubMed] [Google Scholar]
- Koolen DA, Reardon W, Rosser EM, Lacombe D, Hurst JA, Law CJ, Bongers EM, van Ravenswaaij-Arts CM, Leisink MA, van Kessel AG, Veltman JA, de Vries BB. Molecular characterization of patients with subtelomeric 22q abnormalities using chromosome specific array-based comparative genomic hybridization. Eur J Hum Genet. 2005;13:1019–1024. doi: 10.1038/sj.ejhg.5201456. [DOI] [PubMed] [Google Scholar]
- Lu X, Shaw CA, Patel A, Li J, Cooper ML, Wells WR, Sullivan CM, Sahoo T, Yatsenko SA, Bacino CA, Stankiewics P, Ou Z, Chinault AC, Beaudet AL, Lupski JR, Cheung SW, Ward PA. Clinical implementation of chromosomal microarray analysis: Summary of 2513 postnatal cases. PLoS ONE. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani JJ, de Mas P, Depetris D, Mignon-Ravix C, Bottani A, Prieur M, Jonveaux P, Philippe A, Bourrouillou G, de Martinville B, Delobel B, Vallee L, Croquette MF, Mattei MG. Telomeric 22q13 deletions resulting from rings, simple deletions, and translocations: Cytogenetic, molecular, and clinical analyses of 32 new observations. J Med Genet. 2003;40:690–696. doi: 10.1136/jmg.40.9.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, Scherer SW. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z, Kang SH, Shaw CA, Carmack CE, White LD, Patel A, Beaudet AL, Cheung SW, Chinault AC. Bacterial artificial chromosome—Emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan MC, Rogers RC, Saul RA, Stapleton GA, Sweet K, McDermid H, Shaw SR, Claytor J, Willis J, Kelly DP. 22q13 deletion syndrome. Am J Med Genet. 2001;101:91–99. doi: 10.1002/1096-8628(20010615)101:2<91::aid-ajmg1340>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Phelan MC, Stapleton GA, Rogers RC. Deletion 22q13 syndrome (Phelan-McDermid syndrome) In: Cassidy SB, Allanson JE, editors. The management of genetic syndromes. 2. Hoboken, NJ: John Wiley & Sons Inc; 2005. pp. 171–181. [Google Scholar]
- Philippe A, Boddaert N, Vaivre-Douret L, Robel L, Danon-Boileau L, Malan V, de Blois M, Heron D, Colleaux B, Golse B, Zilbovicius B, Munnich A. Neurobehavioural profile and brain imaging study of the 22q13.3 deletion syndrome in childhood. Pediatrics. 2008;122:e376–e382. doi: 10.1542/peds.2007-2584. [DOI] [PubMed] [Google Scholar]
- Probst FJ, Roeder ER, Enciso VB, Ou Z, Cooper ML, Eng P, Li J, Gu Y, Stratton RF, Chinault AC, Shaw CA, Sutton VR, Cheung SW, Nelson DL. Chromosomal microarray analysis (CMA) detects a large X chromosome deletion including FMR1, FMR2, and IDS in a female patient with mental retardation. Am J Med Genet Part A. 2007;143A:1358–1365. doi: 10.1002/ajmg.a.31781. [DOI] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- Shao L, Shaw C, Lu X-Y, Sahoo T, Bacino C, Lalani S, Stankiewicz P, Yatsenko S, Li Y, Neill S, Pursley A, Chinault C, Patel A, Beaudet A, Lupski J, Cheung S. Identification of chromosome abnormalities in subtelomeric regions by microarray analysis: A study of 5,380 cases. Am J Med Genet Part A. 2008;146A:2242–2251. doi: 10.1002/ajmg.a.32399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113:1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Wong AC, Shaw SR, Tse WY, Stapleton GA, Phelan MC, Hu S, Marshall J, McDermid HE. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HL, Crolla JA, Walker D, Artifoni L, Dallapiccola B, Takano T, Vasudevan P, Huang S, Maloney V, Yobb T, Quarrell O, McDermid HE. Interstitial 22q13 deletions: Genes other than SHANK3 have major effects on cognitive and language development. Eur J Hum Genet. 2008;16:1301–1310. doi: 10.1038/ejhg.2008.107. [DOI] [PubMed] [Google Scholar]