Abstract
The coordinated and physiological behavior of living cells in an organism critically depends on their ability to interact with surrounding cells and with the extracellular space. For this, cells have to interpret incoming stimuli, correctly process the signals, and produce meaningful responses. A major part of such signaling mechanisms is the translation of incoming stimuli into intracellularly understandable signals, usually represented by second messengers or second-messenger systems. Two key second messengers, namely the calcium ion and signaling lipids, albeit extremely different in nature, play an important and often synergistic role in such signaling cascades. In this report, we will shed some light on an entire family of protein kinases, the protein kinases C, that are perfectly designed to exactly decode these two second messengers in all of their properties and convey the signaling content to downstream processes within the cell.
The kinase domains of protein kinase C lack substrate specificity. Instead, specificity is partly determined by the binding of various lipids and calcium ions to the regulatory domains.
Once generated, second messengers relay their information content in a plethora of properties, including time, quantity (i.e., concentration), space (i.e., subcellular distribution), and interestingly into any combination of these three characteristics. Nevertheless, such information is meaningless for the cell unless it has a toolkit of read-out systems that can actually interpret such second-messenger properties and relate them further downstream into complex signaling networks, or directly to effector systems. An important system is the family of protein kinase Cs (PKCs) that can read-out lipid signals alone, or combine the ability to read-out simultaneous lipid and Ca2+ signals. A common denominator of all PKCs is the property to convey signals downstream by phosphorylation of additional signaling partners or effector proteins. We will briefly introduce the PKC subfamilies with particular emphasis on their signaling ability, discuss the important sensing domains, and their properties, before concentrating on sensing details of the subfamily of conventional PKCs and their role in signal integration in greater depth.
THE PKC FAMILY
In a mammalian cell, members of the PKC family represent ∼2% of the entire kinome (Mellor and Parker 1998; Manning et al. 2002) and display an almost ubiquitous expression throughout the human body. The PKC family comprises 10 family members that are grouped into three subfamilies based on their particular domain composition and arrangement (see Fig. 1). Despite their widespread expression, the assignment of particular isoforms to specific functions has been difficult because PKCs display a signaling network of their own. The activity of novel PKCs (nPKCs) is, for example, modulated by the activity of atypical PKCs (aPKCs) (Parekh et al. 2000). In recent years, the application of knockout (KO) mouse models has proven seminal in unraveling some of the functions that can now be assigned to a particular PKC isoform, for example (Leitges 2007). However, results from KO approaches have proven difficult to interpret. For example, although the nPKC isoform, PKCδ, is known to be expressed ubiquitously, the KO mouse, deficient of PKCδ in all cells, showed no obvious phenotype (Leitges et al. 2001; Miyamoto et al. 2002). Only extensive analysis revealed mild phenotypes in specialized cell types, such as cells from the bone marrow (Leitges et al. 2002). The extensive networking among PKC members might thus render the interpretation of such mouse models rather difficult.
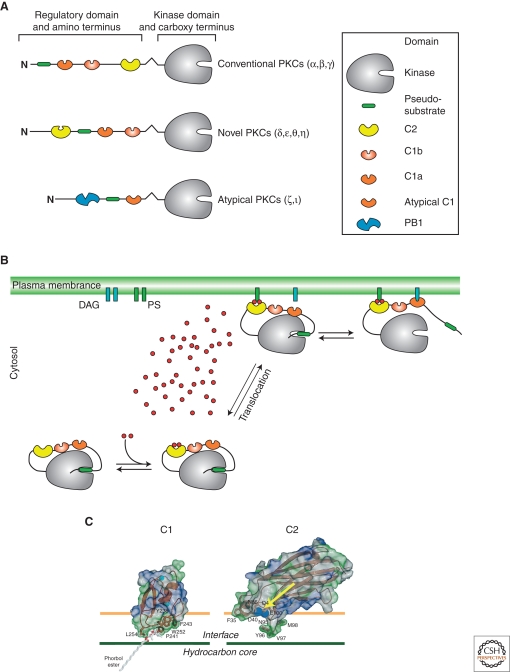
Figure 1.
Basic properties of protein kinases C. (A) The general domain structure of the three subfamilies of the PKCs. The inset explains the symbols used. (B) After maturation and priming steps, activation of the kinase activity involves sequential binding of Ca2+ to the C2 domain, translocation to the plasma membrane, and binding of the C1a domain to DAG. (C) Interaction of the C1 domain with lipids of the plasma membrane (left) is much more intimate than for the Ca2+ (blue spheres marked with an arrow) loaded C2 domain (right). (C, Adapted from Hurley and Misra 2000; reprinted with permission from Annual Reviews, Inc. © 2000.)
Very often enzymes gain substrate specificity via interaction of their activity domain (or nearby domains) and particular substrates. Unfortunately, for PKCs the situation appears to be more complex because their kinase domain, despite smaller differences, appears to be rather unspecific toward substrates (Kennelly and Krebs 1991; Pearce et al. 2010). It is generally believed that the PKC consensus phosphorylation sequence varies little between PKC isoforms. Large overlapping phosphorylation can be found for the same target between different families of AGC kinases (Zhang et al. 2006). From these findings, the following question is rather obvious: How do PKCs gain specificity if the kinase domains’ contribution to specific phosphorylation appears rather low? This question becomes even more central when one considers that the 10 PKC family members share overlap in terms of substrate specificity, but phosphorylate specific targets. Although unique functions have been assigned to particular PKCs, this information does not illuminate the mechanisms underlying their specificity.
An answer to the question of specificity can partly be found in the regulatory domain(s) of PKCs. Although, depending on the particular isoform, the regulatory domain(s) bind lipids and in some cases also Ca2+ ions, how exactly PKCs achieve specificity is still rather unclear. Among other domains, PKCs contain C1 domain(s) (C1a and C1b) that might contribute specificity, but they bind lipids rather than substrates. It should be noted here though, that some evidence for putative direct C1–protein interaction was reported recently (Colon-Gonzalez and Kazanietz 2006). In addition, conventional PKCs (cPKCs) contain a Ca2+-binding C2 domain, but Ca2+ ions of course do not serve as substrates. Thus, we suggest that the lack of substrate specificity attributed to the different PKC kinase domains is actually an important feature for PKCs, because they ought not to express specificity through classical enzyme–substrate interactions but rather through the mechanism of targeting. This means that PKCs follow the idea of local signaling domains in an overwhelmingly dynamic way. Typically, PKCs are only activated within a local signaling domain where specific lipids (such as DAGs) are produced to allow binding to C1 domain(s), or where Ca2+ signaling occurs to trigger a complex choreography of binding steps (see below, and for a recent review see Rosse et al. 2010).
ACTIVATION OF PKCs
To understand substrate-specific interactions, we need to have a close look into the mechanisms that lead to activation of PKCs. The steps leading to activation can be divided into two major episodes that are shared by almost all PKC members: (1) maturation and priming and (2) acute activation of the enzymatic activity. In this review, we will not focus on the first episode(s), because a recent review by Alexandra Newton has explained such steps in detail (Newton 2010).
In brief, once the translation of the PKC protein has been accomplished, highly ordered steps of phosphorylations on the protein itself are needed to gain activation competence. The current understanding is that both maturation and priming might be regulated by an entire group of cofactors, including, but not restricted to, partners stabilizing certain spatial arrangements of the immature PKC protein, such as chaperones (Gould et al. 2009). These stable intermediates are essential for proper phosphorylation to occur. Additional necessary phosphorylation steps involve the activity of the kinase PDK-1 that specifically modifies residues in the activation loop. These phosphorylation events serve a twofold function: (1) stabilizing the structure of the kinase domain and (2) specific priming of the enzymatic activity of the PKC molecule. Such PDK-1-mediated modifications are characteristic for all PKC family members. The particular importance of PDK-1 for PKC was shown by Balendran and coworkers, who reported that PDK-1 knockdown down-regulates PKC levels most likely by increased proteolysis of immature PKCs (Balendran et al. 2000). Nevertheless, all these maturation and priming steps only represent foreplay leading to the real action—the induction of the kinase activity itself.
For all PKC family members, activation of the kinase domain involves dissociation of an autoinhibitory domain from the kinase activity center. These domains contain amino acid sequences mimicking substrates for the kinase domain, but lack the ability to be phosphorylated. Although for cPKCs this pseudosubstrate is almost amino-terminal, for nPKCs and aPKCs, it is buried deeper in the regulatory part of the PKC molecule (see Fig. 1).
The members of the nPKC subfamily contain C2 domains, but for these C2 domains the required acidic Ca2+-binding pocket is absent, thus their Ca2+-binding affinity is too low to be relevant physiologically (reviewed in Hurley and Misra 2000). Therefore, nPKCs solely rely on lipid binding of their C1 domains. Both C1 domains, C1a and C1b, display different binding affinities for DAGs and/or phorbol esters. The C1a domain is usually associated with DAG binding, whereas the C1b domain conveys phorbol ester binding (Colon-Gonzalez and Kazanietz 2006). Interestingly, even among nPKCs, differences between this distribution of properties has been reported. Although the C1 domains of PKCε share an apparent equal binding to DAG and phorbol esters, the C1a domain of PKCδ apparently prefers DAG to phorbol esters (Stahelin et al. 2005a). In all instances, binding of the C1 domain(s) to their appropriate lipid partners releases the pseudosubstrate from the kinase-binding pocket leading to kinase activation.
The aPKCs represent a borderline PKC subfamily when considering the activation mechanism. Activation of aPKCs is not mediated by binding to DAG, phosphatidylserines, or phorbol esters because their C1 domain is altered (atypical C1 domain). Binding to these classical PKC activators in this atypical C1 domain is suppressed threefold: (1) The C1 domains structure is de-formed into a “flat surface” that is not able to penetrate into the lipid layer necessary for hydrophobic interactions, (2) the C1 domain cannot unzip to create the necessary lipid-binding pocket, and (3) hydrophilic amino acids disrupt the hydrophobicity of the binding pocket (Mott et al. 1996; Zhou et al. 2002). For a comparison between the varieties of C1 domains, please refer to a recent review (Colon-Gonzalez and Kazanietz 2006). Nevertheless, recently, binding of the atypical C1 domain to other lipid partners such as phosphatidic acid, ceramide, and PIP3 has been implicated (Hirai and Chida 2003). Such interaction with the lipid releases autoinhibition, but still requires activation steps in the form of phosphorylation events at the PKC molecule itself, where PDK-1 again appears to prime the kinase and autophosphorylation finally results in increased kinase activity (Hirai and Chida 2003). In addition to such lipid-dependent phosphorylation of the atypical PKCs, direct protein–protein interactions through their PB1 domain can modulate their activity (see Fig. 1) (Puls et al. 1997; Qiu et al. 2000).
The Dynasty of Conventional PKCs: Decoding Ca2+ and Lipids
The cPKCs display the most complex and dynamic activation patterns among all PKC family members. Although maturation and priming of cPKCs follows a very similar scheme as described above and reviewed in Newton (2010), cPKCs reside in the cytosol during resting periods waiting to be triggered by rises in intracellular Ca2+ concentration. In this section, we will have a closer look at various aspects of how cPKCs decode Ca2+ and lipid signals, whereas the following section highlights feedback and cPKCs’ influence on Ca2+ signaling.
More than anywhere else in the web of PKCs, the activation of cPKCs depends on its distribution; “location, location, location” is critical for specificity of action. As visualized in a seminal report by Oancea and Meyer (1998), cPKCs are positioned at the crossing of two important cellular signaling routes: receptor-mediated activation of PLCs (and consequently production of DAG) and liberation of Ca2+ from internal stores or influx from the extracellular space.
To perform these tasks, cPKCs contain a toolset of two domain structures (namely C1 and C2) that enable the molecules to sense DAG/phorbol ester production and Ca2+ increases, respectively (Newton and Johnson 1998). In this section, we summarize our current knowledge about the complex interaction between cPKCs and Ca2+ with particular respect to how the Ca2+ signaling toolkit (Berridge et al. 2000) can be read-out by cPKCs and referred downstream in the signaling cascade.
As already mentioned for the other PKC subfamilies, we have to consider the particular way cPKCs are activated to understand how cPKCs are indeed able to interpret both Ca2+ and DAG signals in living cells. Figure 1 gives a summary of our current notion on the final activation steps after maturation and priming of the molecule itself. Under resting conditions (i.e., basal Ca2+ concentration of ∼100 nm), cPKCs distribute in the cytosol with basically no membrane binding. It should be noted that in the absence of Ca2+ membrane binding of cPKCs does not occur, even when DAG is present in the plasma membrane. There might be two reasons for this: (1) Negative charges in the Ca2+-binding pocket might actually repel the molecule from the plasma membrane; in particular, when considering the large amount of negatively charged head groups of phospholipids in the inner leaflet of the plasma membrane and (2) in the Ca2+-free form, the conformation of the C1/C2 tandem domain might prevent the C1 domain(s) from encountering DAG in the plasma membrane (Stahelin et al. 2005b).
Interaction Between C2 Domains and Phospholipids
Although binding between DAG and the C1 domain(s) appears reasonably straightforward, there are at least three different mechanisms for the Ca2+-dependent C2 membrane interaction (see also Hurley and Misra 2000).
Conformational Changes Induced by Ligation of Ca2+: Binding of Ca2+ ions at the three Ca2+-binding regions (CBR1-3) induces a conformational change in the C2 domains in such a way that phospholipid binding will become possible. There are at least two lines of evidence that support that notion. Growing crystals of C2 domains is only possible in the state where C2 is occupied by a single Ca2+; binding of the second one renders the crystal unstable (Sutton and Sprang 1998). Although the C2 domains of PKCα and PKCγ only bind 2 Ca2+, PKCβ is able to bind 3 Ca2+ in a cooperative manner, also suggesting major conformational changes in the C2 domains during successive Ca2+ binding (Kohout et al. 2002). In some studies, such changes extend well beyond the C2 domain, giving rise to further conformational changes in the C1 and pseudosubstrate domains (Bolsover et al. 2003).
Bridging between C2 Domain and Phospholipid in Plasma Membrane: In comparison to the electrostatic role of Ca2+ detailed below, the notion of Ca2+ bridging assumes that both partners, the CBRs of the C2 and the head groups of the phospholipids, coordinate the arrangement of the bound Ca2+ ions (Dessen et al. 1999; Verdaguer et al. 1999). Similar Ca2+ bridging had been described for another Ca2+-sensing and membrane-binding protein family, the annexins (Swairjo et al. 1995).
Electrostatic Interaction between Ca2+-C2 and Plasma Membrane: As described above, the binding pocket for Ca2+, flanked by the CBRs, presents the plasma membrane with an array of negative charges that not only discourage membrane phospholipid interactions, but almost results in repelling actions between the C2 domain and phospholipids (Rizo and Südhof 1998; Ubach et al. 1998). Chelating Ca2+ ions by the CBRs switches this behavior entirely, because in the Ca2+-bound form, the surface charge has now changed from negative to positive and the Ca2+-C2 domain now becomes very much attracted to phospholipids. A seminal study by Kohout and coworkers has shown the enormous rapidity with which cPKCs associate with membrane after a stepwise Ca2+ increase (Kohout et al. 2002; Reither et al. 2006).
Seeing Is Believing: Imaging cPKC Dynamics in Living Cells
The advent of sophisticated imaging techniques together with the approach of fusing cPKCs with fluorescent proteins (FPs) allowed the direct visualization and analysis of cPKC dynamics in living cells (Meyer and Oancea 2000; see also Fig. 2A). Here, the group around Tobias Meyer has performed groundbreaking work in not only demonstrating rapid cPKC membrane translocation, but also characterizing such behavior in detail using mutational approaches (Oancea and Meyer 1998). Applying line-scan analysis of cPKC translocation together with line-scan FRAP approaches, Michael Schaefer and coworkers showed that rapid translocation of cPKCs to the plasma membrane does not necessarily require specific transport processes or specific receptors at the plasma membrane, free diffusion was sufficient to explain the observed translocation kinetics (Schaefer et al. 2001). We recently demonstrated that Ca2+-dependent membrane association also occurs in mutated PKCα with a dysfunctional DAG-binding domain (Reither et al. 2006). By rendering the C1 domain DAG insensitive, we were able to show that Ca2+ association to the C2 domain is both sufficient and necessary for rapid membrane association (Reither et al. 2006).
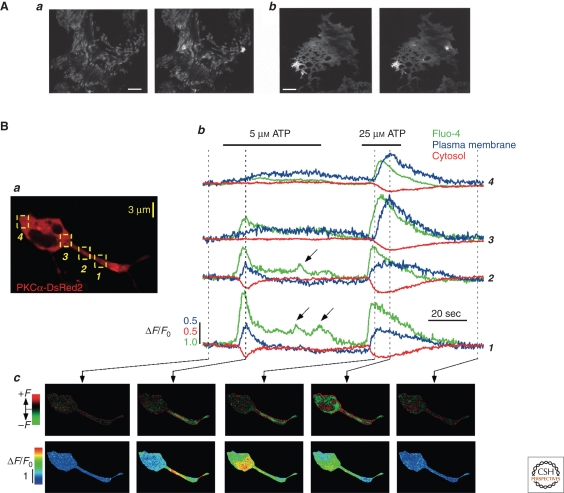
Figure 2.
Spatially restricted cPKC translocations are driven by localized Ca2+ signals. (A) Astrocytes were imaged by TIRF microscopy and stimulated by 100 μm glutamate. Spatially restricted plasma membrane accumulation of GFP-PKCγ (a) and GFP-C2 (b) can be observed. Scale bar, 5 μm. (Adapted from Codazzi et al. 2001; reprinted with permission from Elsevier © 2001.) (B) HEK cells expressing PKCα-DsRed2 were loaded with fluo4 and stimulated with threshold (5 μm ATP) and suprathreshold (25 μm ATP) agonist concentrations. Spatially restricted Ca2+ signals (Bc, lower row, 5 μm) resulted in localized PKC translocation to the plasma membrane (Bc, upper row, 5 μm) while global Ca2+ (Bc, lower row, 25 μm) induced global translocation (Bc, upper row, 25 μm). Numbers in Bb correspond to regions of interest in Ba. (Adapted from Reither et al. 2006; reprinted with permission from The Rockefeller University Press © 2006.)
Most of the studies reporting cPKC dynamics in living cells concentrate on analyzing global changes of the cPKC distribution (e.g., Oancea and Meyer 1998; for more details see Violin and Newton 2003). But if cPKCs are indeed versatile decoding machines for Ca2+ then the question arises as to whether they are able to sense the entire Ca2+ signaling toolkit, from the global, homogeneous Ca2+ signals, to the rapidly propagating Ca2+ wave, and finally spatially restricted Ca2+ signals, including elementary Ca2+ transients, such as Ca2+ puffs (Bootman et al. 1997a,b; Lipp et al. 1997; Koizumi et al. 1999).
First indications for cPKCs' ability to translocate to the plasma membrane in a spatially restricted manner was provided by Tobias Meyers group (Codazzi et al. 2001) in a study using PKCγ fusion proteins with FP tags. They analyzed translocation behavior of PKCγ in astrocytes using total internal reflection microscopy (TIRF) to restrict fluorescence detection to the basal plasma membrane (Fig. 2A). In the same report, the authors showed that similarly restricted translocations could also be found when expressing C2-FP constructs alone, suggesting that they were looking at the effects of local Ca2+ signals (Fig. 2Ab) (Codazzi et al. 2001). Nevertheless, their very low temporal resolution (around 6 s per image) together with the lack of a simultaneous Ca2+ recording prevented a direct link between these two events.
Recently, we were able for the first time to provide compelling evidence that PKCα was able to decode Ca2+ signals generated by various stimulation strengths. Threshold stimulation of nonexcitable cells with Ca2+-mobilizing agonists leads to a breakdown of the global signal into spatially restricted Ca2+ transients (Bootman et al. 1997b; Berridge et al. 2000). Figure 2B depicts such data, indicating that threshold stimulation (5 μm ATP) resulted in a spatially restricted Ca2+ signal (Fig. 2Bc, lower row, left) whereas supra-threshold stimulation caused global Ca2+ increases (Fig. 2Bc, lower row, right). Importantly, translocation of the PKCα fusion protein very closely resembled the spatiotemporal properties of the underlying Ca2+ transients (Fig. 2Bc, upper row). Such data suggested that activation (i.e., membrane association) and deactivation (i.e., membrane dissociation) might actually be extremely dynamic and short lived, a notion we will discuss below in greater detail.
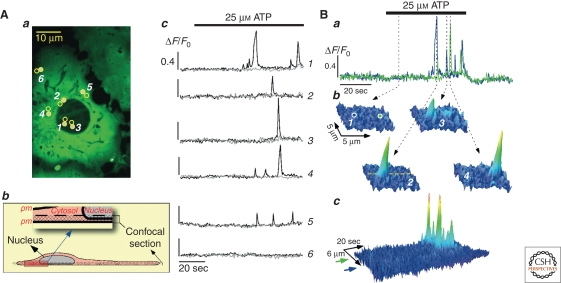
We and others have reported on the breakdown of global Ca2+ transients into elementary Ca2+ signals, referred to as Ca2+ sparks and Ca2+ puffs for ryanodine receptor and InsP3 receptor signals, respectively (reviewed in Lipp and Niggli 1996; Berridge et al. 1999). It is well acknowledged that such elementary Ca2+ transients serve as building blocks for more complex Ca2+ transients, including Ca2+ waves but whether these spatially and temporally restricted signals may have a local signaling role on their own is still unclear. For a universally applied Ca2+ read-out system, cPKCs should thus also be able to sense these Ca2+ transients and translate them into highly localized translocation events. By using rapid 2D confocal imaging of PKCα-FP-expressing cells and threshold stimulations, we were able to show elementary Ca2+ release-mediated local translocation events (LTEs) (see Fig. 3) (Reither et al. 2006). Thus, a possible signaling role can be directly assigned to elementary Ca2+ release sites close to the plasma membrane (≤1.5 μm): they might serve as a tool for cells to induce spatially restricted signaling cascades by local recruitment of cPKCs and ensuing downstream events. Such versatility in reading-out of local Ca2+ signals has as of yet only directly been shown for cPKCs, although similar abilities have been suggested for another very important Ca2+ read-out sensor: calmodulin (Deisseroth et al. 1998; Wheeler et al. 2008).
Figure 3.
Local translocation events (LTEs) in PKCα-expressing cells. (A) LTEs were highly spatially restricted and occurred randomly in the cell. (Aa,c) Open circles represent LTE locations whereas gray circles display neighboring locations. (Ab) Arrangement of confocal recording. (B) Spatiotemporal properties of LTEs. The pseudo-line-scan images in Bc were constructed along the dashed line indicated in Bb2. (B, Adapted from Reither et al. 2006; reprinted with permission from The Rockefeller University Press © 2006.)
Terminating cPKC Signaling
Equally important to initiating cPKC signaling by rises in intracellular Ca2+ is the termination of such potent signaling molecules. For this, it would be beneficial to be able to follow cPKC phosphorylation in living cells.
Two such approaches have been established so far, referred to as CKAR (Violin et al. 2003; Gallegos et al. 2006) and KCP (Schleifenbaum et al. 2004; Brumbaugh et al. 2006). Both have used the fluorescence resonance energy transfer (FRET) approach with different phosphorylation sensors. Roger Tsien and Alexandra Newton have incorporated a construct comprised of a consensus sequence and a specific binding domain recognizing the phosphorylated PKC consensus sequence (Violin et al. 2003). Alternatively, Carsten Schultz's group have used conformational bending based on multiple phosphorylations in a kind of “hinge” region of the sensor (Schleifenbaum et al. 2004). In both cases, PKC-dependent phosphorylation of the sensor results in a conformational rearrangement of CFP and YFP at both ends of the sensors and thus changing the FRET ratio that can be measured.
In the original report using the CKAR probe, the investigators were able to show that oscillations in Ca2+ are directly translated into oscillations of substrate phosphorylation (Violin et al. 2003); a novel and important finding. When analyzing their results in greater detail, especially the temporal relationship between Ca2+ increases and phosphorylation to switch on and off, they found a time delay between these two events of only 10 s, which was not far off the temporal resolution of their recording. From these findings, one has to conclude that the switching of the cPKCs from off to on, and vice versa, is almost instantaneous, even considering that the phosphorylation probe only senses the steady-state between phosphorylation events (via PKCs) and dephosphorylation by phosphatases. But how are cPKC switched off?
In biochemical studies using cPKC-C2 domains and analyzing their membrane association and dissociation kinetics, Newton’s and Kohout’s groups have found very fast kinetics for both processs (Nalefski and Newton 2001; Kohout et al. 2002). Although the kon rate for membrane association was ∼1010 M−1sec−1, membrane dissociation koff was still very fast (∼150 sec−1) (Nalefski and Newton 2001). Because the Ca2+ dissociation was in the same order of magnitude (Kohout et al. 2002), Ca2+ unbinding and membrane dissociation are quasi-simultaneous. But, cPKCs also interact with the plasma membrane through C1 domain(s) that are believed to display much slower kinetics for membrane interactions. A recent biochemical study has indeed shown that the isolated C1 domain of PKCβ exhibits much slower kinetics. Here kon and koff were measured to be 109 M−1sec−1 and 0.1 sec−1, respectively (Dries and Newton 2008). It has to be mentioned here that Dries and Newton found two distinct, kinetically different membrane-binding modes of C1 domains: a weak but fast binding/unbinding to phosphatidylserine and a high affinity but slow interaction with DAG/PMA (Dries and Newton 2008).
Thus, up to now we can draw the following picture (cf. Fig. 1): Membrane association in cPKCs appears to be solely driven by C2-mediated initial membrane contacts with anionic phospholipids, whereas retention of the cPKC at the membrane is dominated by DAG–C1 interactions that apparently not only provide the energy for pseudosubstrate dissociation and kinase activation but also massively increase the likelihood of kinase–substrate interactions by restricting the “search dimensions” from the 3D cytosol to the flat inner surface of the plasma membrane (i.e., 2D) (Kholodenko et al. 2000).
Although all those findings describe basic biochemical properties of cPKCs, the question arises whether any of such properties are reflected in the behavior of cPKCs in the living cell? For this, again, imaging techniques have provided compelling evidence for the notion put forward above. Flash photolysis of caged-Ca2+ compounds in living cells while simultaneously monitoring Ca2+ and PKCα translocation has supported the extremely fast membrane association of the kinase (Reither et al. 2006). Substantial membrane accumulation could be monitored less than 250 msec after photolytic Ca2+ increase.
When considering the kinetics of DAG production measured during agonist stimulation, which has been reported to show no, or only mild, oscillations (Codazzi et al. 2001; Bartlett et al. 2005), DAG production is rather unlikely to cause phosphorylation cycles of cPKCs as reported previously (Violin et al. 2003). As suggested earlier, such oscillations coincide with Ca2+ oscillations, which in turn should also be responsible for switching off kinase activity. But what exactly is the rate-limiting step for returning kinase activity to basal levels? Ca2+ levels alone cannot be the sole driving force behind that, because during stimulation with a Ca2+-mobilizing agonist, DAG is also being produced. It appears feasible to assume that despite rapid decreases in Ca2+ at the plasma membrane, it will be DAG-C1 dissociation that will dominate membrane dissociation and eventually switching off the kinase activity. By analyzing membrane-dissociation kinetics in living cells, we have provided recent evidence that the C1–membrane interaction (presumably via DAG) is a major determinant of this process (Reither et al. 2006). Similar to stop-flow analysis in a spectrometer we have used photolysis of a caged-Ca2+ buffer (diazo-2) to decrease Ca2+ in a quasi-stepwise manner. In living cells, we were able to identify that membrane dissociation of PKCα was indeed slow and characterized by a bi-exponential kinetic. When we rendered the C1a domain DAG insensitive, the same experimental approach revealed fast membrane dissociation described by a single, rapid exponential decay of membrane fluorescence (Reither et al. 2006). The resulting speed of membrane dissociation was increased by more than an order of magnitude, providing compelling evidence for our notion put forward above. The fact that, in the wild-type PKCα, membrane dissociation was bi-exponential also strongly suggests that even during normal cellular stimulation there are two pools of membrane-bound cPKCs: (1) a loosely bound population, where binding is only mediated by weaker PKC–anionic phospholipid interaction mediated by Ca2+-C2 and C1a domains, and (2) a tightly bound cPKC population, which interact via C1a-DAG binding and most likely represent the activated kinase population.
It is therefore feasible to assume that activation and inactivation of cPKCs are primarily driven by the DAG-C1a-binding kinetics, which appear rate limiting in both processes (Nalefski and Newton 2001; Dries and Newton 2008), but that the C2–membrane interaction is the major determinant for the localization of cPKCs to the plasma membrane. Thus, the spatiotemporal properties of the underlying Ca2+ signals determine the strength, timing, and localization of membrane association of cPKCs, but the strength of the agonist-mediated DAG production appears to be the determinant for the strength of cPKC activation. In this respect, the answer to the question of whether cPKC translocation driven by elementary Ca2+ signals (LTEs) does indeed signal downstream is still unknown and will only be answered when we have phosphorylation sensors available with a signal-to-noise ratio that allows sensing their FRET changes in cellular microdomains.
Up to now we have only discussed one part of the Ca2+–cPKC interaction, namely how Ca2+ influences cPKC activity, but we have to consider a dialog between Ca2+ and cPKCs because in return cPKCs’ activity can also modulate Ca2+ signaling, which will be the topic of the following section.
CONVENTIONAL PKCs AND BEYOND: cPKCs AND THEIR REGULATORY INFLUENCE ON Ca2+ HANDLING
In this section, we focus on the immediate functional feedback with respect to the modulation of Ca2+ regulators. The long-term effects of cPKC-dependent regulation of gene expression will not be considered here. In a Ca2+ centric view of cellular signaling, we would like to distinguish direct regulators of Ca2+ such as Ca2+ channels as positive modules together with the counteracting pumps from second level regulators (regulator of regulators). In the following, the effect of cPKCs on the first- and second-level regulators will be discussed.
First Level of Regulation
At the first level of regulation, we consider RyRs, InsP3Rs, and other Ca2+ channels as main positive regulators, whereas SERCA, PMCA, and NCX are main effectors of Ca2+ clearance (Uhlen and Fritz 2010). Consistent with the fact that cPKCs main “operation” field is the plasma membrane; most of the known effectors are membrane associated or in the vicinity of it.
cPKC–PMCA
The plasma membrane ATPase (PMCA) has a PKCγ phosphorylation site at its carboxyl terminus that interferes with the inhibitory binding of calmodulin (Wang et al. 1991). In addition, PKCα is relevant for the up-regulation of PMCA activity (Penniston and Enyedi 1998).
cPKC–TRPs
As already highlighted in a previous contribution of this series (Gees et al. 2010), Ca2+ entry via TRPs is tightly regulated by cPKCs. A comprehensive overview of further regulatory action was provided recently (Venkatachalam and Montell 2007). Don Gill's group showed a down-regulation of the TRPC family by PKC-dependent phosphorylation of the channel protein itself (Venkatachalam et al. 2003). Several members of the TRPVs are up-regulated by cPKCs. Nevertheless, one has to consider that the situation is more complex because the activity of TRPCs and TRPVs is also modulated by PLC-dependent hydrolysis of PIP2. Because PLCs are regulated by PKCs (see below), this relationship displays yet another level of PKC-dependent regulation of TRPs. The other subfamilies of TRP channels seem to only weakly depend on PKC activity, concluding that within the superfamily of TRPs the main effect of cPKC activity might be transduced by TRPC and TRPV.
cPKC–NCX
Na+/Ca2+ exchangers together with SERCA pumps have the main negative effect on the cytosolic Ca2+ concentration (Berridge 2009). There is strong experimental evidence for NCX's activity, up-regulation by cPKC-dependent phosphorylation (Schulze et al. 2003). cPKCs seem to be integrated in a regulatory complex of various enzymes, such as additional kinases and phosphatases (Ruknudin et al. 2007).
Second Level of Regulation
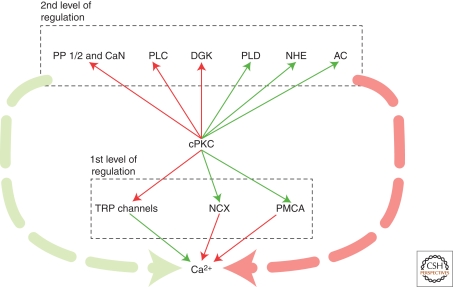
Beside these direct regulators, there are several protein families, which are regulated by cPKCs and have a mediate effect on Ca2+ handling. This second level of regulation also integrates more PKC-independent actions. Therefore, the direct effect of cPKCs is often difficult to separate, especially because the various protein families are in a continuous dynamic state of intimate interaction. This renders predictions rather difficult. Nevertheless, cPKCs can be recognized as a signaling hub translating the increase in cytosolic Ca2+ in activity changes of a plethora of substrate proteins. Here we want to highlight the ubiquitously involved proteins that feed back Ca2+-dependent PKC activity onto Ca2+ handling (see Fig. 4).
Figure 4.
The activity of cPKCs is not only modulated by Ca2+ but in turn affects the activity of a plethora of control mechanisms that influence intracellular Ca2+ handling. Green arrows indicate positive effects, whereas the red color is indicative of negative effects.
cPKC–PLC
PLCs, especially the members of the β subfamily, are known to be phosphorylated by PKCα (Strassheim and Williams 2000; Xu et al. 2004). The phosphorylation of PLCβ1 at S887 has an inhibitory effect. Regulatory effects of PKCα on other isoforms such as PLCδ1, even though it might not be a direct effect, are reported as negative (Fujii et al. 2009). The regulation of PLCs interferes with signaling events dependent on PIP2, DAG, and IP3.
cPKC–PLD
The regulation of phospholipases D (PLD) by cPKCs opens another branch to the signaling effects of phosphatidic acid (PA). Several lines of evidence for a stimulatory effect of PKCα on PLDs have been reported (Hornia et al. 1999; Han et al. 2002; Chen and Exton 2004; Lee et al. 2009).
cPKC–DGK
Interestingly, there is a second route to influence PA signaling via diacylglycerol kinases (DGK). For DGKδ and DGKζ down-regulation by PKCα was reported (Luo et al. 2003; Imai et al. 2004).
cPKC–AC
The family of adenylate cyclases (AC) consists of nine membrane located isoforms. The effect of PKCs on ACs has already been well described by Halls and Cooper within this series of reviews (Halls and Cooper 2011). Despite the fact that not all of the ACs are up-regulated by cPKC phosphorylation, we would still consider a positive effect on the generation of cAMP as the main outcome. Experimental evidence for direct activation by PKCα was found for AC2 and AC5 (Kawabe et al. 1994; Zimmermann and Taussig 1996). The cPKC-driven increase of cAMP levels brings in a whole subnetwork of Ca2+ regulating events downstream from protein kinases A (Reiken et al. 2003; Schulze et al. 2003) and small GTPases via guanine exchange factor EPAC (Schmidt et al. 2001; Pereira et al. 2007).
cPKC–NHE
Another global effect by changing the intracellular pH on signaling could be mediated by the cPKC-dependent up-regulation of the sodium/proton exchangers (NHE). Even though there is only sparse evidence for a direct phosphorylation, supportive evidence has been presented for PKCβ-dependent up-regulation via another interactor (Takahashi et al. 1999; Itoh et al. 2005). The positive effect is also supported by experiments using cPKC-specific inhibitors to down-regulate NHEs (Pederson et al. 2002).
cPKC–Protein Phosphatases
Kinase activity is counteracted by phosphatases (see above). Interestingly, kinases and phosphatases inhibit each other resulting in a stabilizing competition (Srivastava et al. 2002; duBell and Rogers 2004). The effect of cPKCs is mediated by phosphorylation and activation of the inhibitory subunits of CPI-17 (Kolosova et al. 2004; Zemlickova et al. 2004). In addition, there is a report suggesting a competing effect of inhibitory PKA phosphorylation I-1 (Braz et al. 2004). Furthermore, evidence has been presented for down-regulation of calcineurin (Tung 1986; Hashimoto and Soderling 1989). We consider a transient down-regulation by cPKC activity because protein phosphatases remove their own inhibitory phosphates resulting in a positive feedback loop for global phosphatase activity. This will influence the whole kinase-phosphatase balance even though we assume that there might be different outcomes at confined local complex assemblies.
Taking all these feedback loops between cPKCs and global Ca2+ handling together appears to suggest, that cPKCs do impose mainly negative effect via different immediate and mediate regulatory branches. This is already supported by data from different groups (Young et al. 2002; Venkatachalam et al. 2003; Fontainhas et al. 2005; Sakwe et al. 2005).
INTEGRATION OF IMMEDIATE AND INTERMEDIATE cPKC-Ca2+ HANDLING REGULATORY LOOPS
As suggested already in an earlier contribution to this series (Dupont et al. 2010), theoretical models help to provide insights into the regulatory mechanisms of signaling networks. To test the effect of cPKCs on Ca2+ handling in detail and under the condition of a larger set of Ca2+ regulating entities we used the commercially available model DynaCellNet1.0 (www.deplecto.de). This model comprises 45 nodes downstream from G protein coupled receptors that are interconnected by 305 functional relations. The advantage of this modeling approach is that—despite the fact that models reduce complexity—it provides a system still with the characteristics of a complex system. The modeled Ca2+ transients are calculated in dependence of the changing states of the 44 other nodes. DynaCellNet1.0 provides semiquantitative predictions and is mainly a tool to test and generate hypotheses. To investigate the full range of cPKC-dependent regulation we arbitrarily fixed the cPKC activity at different levels, from very low (0) to massive (140). This setup will not directly be transferable into experiments, because fixing levels of enzyme activity is not possible, but it provides an overview of different states of cPKC activity beyond knockdown and overexpression.
The calculated Ca2+ responses (Fig. 5) result from a stimulation of ATP sensitive purinergic receptors (Wettschureck and Offermanns 2005). Beside others, all of the above discussed protein families are integrated. The response under “free-running, physiological” conditions, with no restrictions on cPKCs activity, is shown as control (Fig. 5A–D, black graphs). Despite a slight increase in peak amplitude the main effect of a reduction of cPKC activity levels is an attenuated clearance of cytosolic Ca2+ (Fig. 5A). A similar effect was observed by Ulianich and coworkers (2006). As expected further activation of cPKCs result into an accelerated clearance and a decrease in peak amplitude. Compared to published data, the effect is not very pronounced (Young et al. 2002; Venkatachalam et al. 2003; Sakwe et al. 2005), but the results shown in these reports are based on phorbol ester treatments, which might not exclusively be a result of PKC activity and/or will result in “unphysiological” activity levels and phosphorylation events (Kazanietz 2002). When we model a phorbol ester treatment the model shows a much more pronounced decrease in the amplitude of the Ca2+ transient. An interesting aspect of cPKC regulation is highlighted by the model at extremely high levels of cPKC activity (Fig. 5D). Apart from a massive down-regulation by the first level of regulation, the positive intermediate effects from the second level of regulation become more prominent. The increase of oscillatory behavior results from an “echo” of high cPKC activity in the system. The straightforward notion would be to assume a stimulating effect via cAMP-PKA-EPAC together with a reincrease in protein phosphatase activity via PLD and PA.
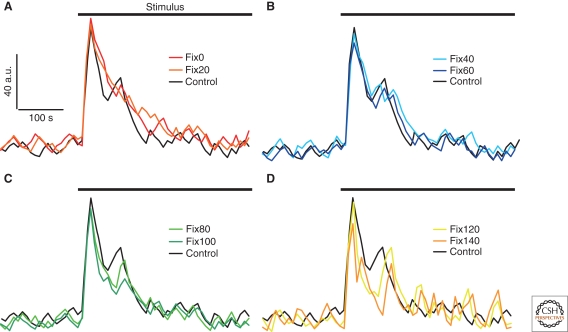
Figure 5.
Modeled Ca2+ transients at different cPKC activity levels. The Ca2+ transient under control condition (A–D; black graph) was calculated with no restrictions on cPKC activity as a result of stimulation of purinergic receptors after 200 sec with ATP. (A) At low levels (fix0 and fix20) of cPKC activity the primary peak is increased in amplitude and Ca2+ removal is slowed down and less noisy. (B) Ca2+ transients modeled with medium cPKC activity levels (fix40 and fix60) are not distinguishable from control conditions. (C) At high levels of cPKC activity (fix80 and fix100) Ca2+ removal is accelerated resulting in a reduced duration and amplitude of the initial transient (fix100). (D) Extremely high levels (fix120 and fix140) of cPKC activity result in a decreased primary peak and an increase of noise in the tail of the transient most likely corresponding to additional Ca2+ oscillations.
CONCLUSIONS
In the context of cellular Ca2+ signaling, cPKCs have the role of an important modulator, acting on the fine-tuning of Ca2+ responses. The question whether the “echo” plays a role might become relevant under patho-physiological conditions were either cPKC expression is up-regulated or one of the alternative routes (cAMP-PKA-EPAC or PLD-PA) has a pronounced effect because of changes in the composition of the different regulatory feedback loops.
Footnotes
Editors: Martin D. Bootman, Michael J. Berridge, James W. Putney, and H. Llewelyn Roderick
Additional Perspectives on Calcium Signaling available at www.cshperspectives.org
REFERENCES
- Balendran A, Hare GR, Kieloch A, Williams MR, Alessi DR 2000. Further evidence that 3-phosphoinositide-dependent protein kinase-1 (PDK1) is required for the stability and phosphorylation of protein kinase C (PKC) isoforms. FEBS Lett 484: 217–223 [DOI] [PubMed] [Google Scholar]
- Bartlett PJ, Young KW, Nahorski SR, Challiss RA 2005. Single cell analysis and temporal profiling of agonist-mediated inositol 1,4,5-trisphosphate, Ca2+, diacylglycerol, and protein kinase C signaling using fluorescent biosensors. J Biol Chem 280: 21837–21846 [DOI] [PubMed] [Google Scholar]
- Berridge MJ 2009. Module 5: Off mechanisms. Cell signalling biology Portland Press, Ltd, London [Google Scholar]
- Berridge M, Lipp P, Bootman M 1999. Calcium signalling. Curr Biol 9: R157–R159 [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, Bootman MD 2000. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol 1: 11–21 [DOI] [PubMed] [Google Scholar]
- Bolsover SR, Gomez-Fernandez JC, Corbalan-Garcia S 2003. Role of the Ca2+/phosphatidylserine binding region of the C2 domain in the translocation of protein kinase Cα to the plasma membrane. J Biol Chem 278: 10282–10290 [DOI] [PubMed] [Google Scholar]
- Bootman M, Niggli E, Berridge M, Lipp P 1997a. Imaging the hierarchical Ca2+ signalling system in HeLa cells. J Physiol 499: 307–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootman MD, Berridge MJ, Lipp P 1997b. Cooking with calcium: The recipes for composing global signals from elementary events. Cell 91: 367–373 [DOI] [PubMed] [Google Scholar]
- Braz JC, Gregory K, Pathak A, Zhao W, Sahin B, Klevitsky R, Kimball TF, Lorenz JN, Nairn AC, Liggett SB, et al. 2004. PKC-α regulates cardiac contractility and propensity toward heart failure. Nat Med 10: 248–254 [DOI] [PubMed] [Google Scholar]
- Brumbaugh J, Schleifenbaum A, Gasch A, Sattler M, Schultz C 2006. A dual parameter FRET probe for measuring PKC and PKA activity in living cells. J Am Chem Soc 128: 24–25 [DOI] [PubMed] [Google Scholar]
- Chen JS, Exton JH 2004. Regulation of phospholipase D2 activity by protein kinase Cα. J Biol Chem 279: 22076–22083 [DOI] [PubMed] [Google Scholar]
- Codazzi F, Teruel MN, Meyer T 2001. Control of astrocyte Ca2+ oscillations and waves by oscillating translocation and activation of protein kinase C. Curr Biol 11: 1089–1097 [DOI] [PubMed] [Google Scholar]
- Colon-Gonzalez F, Kazanietz MG 2006. C1 domains exposed: From diacylglycerol binding to protein–protein interactions. Biochim Biophys Acta 1761: 827–837 [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Heist-Tsien RW 1998. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature 392: 198–202 [DOI] [PubMed] [Google Scholar]
- Dessen A, Tang J, Schmidt H, Stahl M, Clark JD, Seehra J, Somers WS 1999. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell 97: 349–360 [DOI] [PubMed] [Google Scholar]
- Dries DR, Newton AC 2008. Kinetic analysis of the interaction of the C1 domain of protein kinase C with lipid membranes by stopped-flow spectroscopy. J Biol Chem 283: 7885–7893 [DOI] [PubMed] [Google Scholar]
- duBell WH, Rogers TB 2004. Protein phosphatase 1 and an opposing protein kinase regulate steady-state L-type Ca2+ current in mouse cardiac myocytes. J Physiol 556: 79–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont G, Combettes L, Birds GS, Putney JW 2011. Calcium oscillations. Cold Spring Harb Perspect Biol 3: a004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontainhas AM, Obukhov AG, Nowycky MC 2005. Protein kinase Cα modulates depolarizaton-evoked changes of intracellular Ca2+ concentration in a rat pheochromocytoma cell line. Neuroscience 133: 393–403 [DOI] [PubMed] [Google Scholar]
- Fujii M, Yi KS, Kim MJ, Ha SH, Ryu SH, Suh PG, Yagisawa H 2009. Phosphorylation of phospholipase C-δ 1 regulates its enzymatic activity. J Cell Biochem 108: 638–650 [DOI] [PubMed] [Google Scholar]
- Gallegos LL, Kunkel MT, Newton AC 2006. Targeting protein kinase C activity reporter to discrete intracellular regions reveals spatiotemporal differences in agonist-dependent signaling. J Biol Chem 281: 30947–30956 [DOI] [PubMed] [Google Scholar]
- Gees M, Colsoul B, Nilius B 2010. The role of transient receptor potential cation channels in Ca2+ signaling. Cold Spring Harb Perspect Biol 2: a003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould CM, Kannan N, Taylor SS, Newton AC 2009. The chaperones Hsp90 and Cdc37 mediate the maturation and stabilization of protein kinase C through a conserved PXXP motif in the C-terminal tail. J Biol Chem 284: 4921–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halls ML, Cooper DM 2011. Regulation by Ca2+-signaling pathways of adenylyl cyclases. Cold Spring Harb Perspect Biol 3: a004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JM, Kim Y, Lee JS, Lee CS, Lee BD, Ohba M, Kuroki T, Suh PG, Ryu SH 2002. Localization of phospholipase D1 to caveolin-enriched membrane via palmitoylation: Implications for epidermal growth factor signaling. Mol Biol Cell 13: 3976–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Soderling TR 1989. Regulation of calcineurin by phosphorylation. Identification of the regulatory site phosphorylated by Ca2+/calmodulin-dependent protein kinase II and protein kinase C. J Biol Chem 264: 16524–16529 [PubMed] [Google Scholar]
- Hirai T, Chida K 2003. Protein kinase Cζ (PKCζ): Activation mechanisms and cellular functions. J Biochem 133: 1–7 [DOI] [PubMed] [Google Scholar]
- Hornia A, Lu Z, Sukezane T, Zhong M, Joseph T, Frankel P, Foster DA 1999. Antagonistic effects of protein kinase C α and δ on both transformation and phospholipase D activity mediated by the epidermal growth factor receptor. Mol Cell Biol 19: 7672–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Misra S 2000. Signaling and subcellular targeting by membrane-binding domains. Annu Rev Biophys Biomol Struct 29: 49–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Kai M, Yamada K, Kanoh H, Sakane F 2004. The plasma membrane translocation of diacylglycerol kinase δ1 is negatively regulated by conventional protein kinase C–dependent phosphorylation at Ser-22 and Ser-26 within the pleckstrin homology domain. Biochem J 382: 957–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh S, Ding B, Bains CP, Wang N, Takeishi Y, Jalili T, King GL, Walsh RA, Yan C, Abe J 2005. Role of p90 ribosomal S6 kinase (p90RSK) in reactive oxygen species and protein kinase C β (PKC-β)–mediated cardiac troponin I phosphorylation. J Biol Chem 280: 24135–24142 [DOI] [PubMed] [Google Scholar]
- Kawabe J, Iwami G, Ebina T, Ohno S, Katada T, Ueda Y, Homcy CJ, Ishikawa Y 1994. Differential activation of adenylyl cyclase by protein kinase C isoenzymes. J Biol Chem 269: 16554–16558 [PubMed] [Google Scholar]
- Kazanietz MG 2002. Novel "nonkinase" phorbol ester receptors: The C1 domain connection. Mol Pharmacol 61: 759–767 [DOI] [PubMed] [Google Scholar]
- Kennelly PJ, Krebs EG 1991. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem 266: 15555–15558 [PubMed] [Google Scholar]
- Kholodenko BN, Hoek JB, Westerhoff HV 2000. Why cytoplasmic signalling proteins should be recruited to cell membranes. Trends Cell Biol 10: 173–178 [DOI] [PubMed] [Google Scholar]
- Kohout SC, Corbalan-Garcia S, Torrecillas A, Gomez-Fernandez JC, Falke JJ 2002. C2 domains of protein kinase C isoforms α, β, and γ: Activation parameters and calcium stoichiometries of the membrane-bound state. Biochemistry 41: 11411–11424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi S, Bootman MD, Bobanovic LK, Schell MJ, Berridge MJ, Lipp P 1999. Characterization of elementary Ca2+ release signals in NGF-differentiated PC12 cells and hippocampal neurons. Neuron 22: 125–137 [DOI] [PubMed] [Google Scholar]
- Kolosova IA, Ma SF, Adyshev DM, Wang P, Ohba M, Natarajan V, Garcia JG, Verin AD 2004. Role of CPI-17 in the regulation of endothelial cytoskeleton. Am J Physiol Lung Cell Mol Physiol 287: L970–980 [DOI] [PubMed] [Google Scholar]
- Lee CS, Kim KL, Jang JH, Choi YS, Suh PG, Ryu SH 2009. The roles of phospholipase D in EGFR signaling. Biochim Biophys Acta 1791: 862–868 [DOI] [PubMed] [Google Scholar]
- Leitges M 2007. Functional PKC in vivo analysis using deficient mouse models. Biochem Soc Trans 35: 1018–1020 [DOI] [PubMed] [Google Scholar]
- Leitges M, Sanz L, Martin P, Duran A, Braun U, Garcia JF, Camacho F, Diaz-Meco MT, Rennert PD, Moscat J 2001. Targeted disruption of the ζPKC gene results in the impairment of the NF-κB pathway. Mol Cell 8: 771–780 [DOI] [PubMed] [Google Scholar]
- Leitges M, Gimborn K, Elis W, Kalesnikoff J, Hughes MR, Krystal G, Huber M 2002. Protein kinase C-δ is a negative regulator of antigen-induced mast cell degranulation. Mol Cell Biol 22: 3970–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipp P, Niggli E 1996. A hierarchical concept of cellular and subcellular Ca2+-signalling. Prog Biophys Mol Biol 65: 265–296 [DOI] [PubMed] [Google Scholar]
- Lipp P, Thomas D, Berridge MJ, Bootman MD 1997. Nuclear calcium signalling by individual cytoplasmic calcium puffs. EMBO J 16: 7166–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Prescott SM, Topham MK 2003. Protein kinase C α phosphorylates and negatively regulates diacylglycerol kinase ζ. J Biol Chem 278: 39542–39547 [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S 2002. The protein kinase complement of the human genome. Science 298: 1912–1934 [DOI] [PubMed] [Google Scholar]
- Mellor H, Parker PJ 1998. The extended protein kinase C superfamily. Biochem J 332: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T, Oancea E 2000. Studies of signal transduction events using chimeras to green fluorescent protein. Methods Enzymol 327: 500–513 [DOI] [PubMed] [Google Scholar]
- Miyamoto A, Nakayama K, Imaki H, Hirose S, Jiang Y, Abe M, Tsukiyama T, Nagahama H, Ohno S, Hatakeyama S, et al. 2002. Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cδ. Nature 416: 865–869 [DOI] [PubMed] [Google Scholar]
- Mott HR, Carpenter JW, Zhong S, Ghosh S, Bell RM, Campbell SL 1996. The solution structure of the Raf-1 cysteine-rich domain: A novel ras and phospholipid binding site. Proc Natl Acad Sci 93: 8312–8317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski EA, Newton AC 2001. Membrane binding kinetics of protein kinase C βII mediated by the C2 domain. Biochemistry 40: 13216–13229 [DOI] [PubMed] [Google Scholar]
- Newton AC 2010. Protein kinase C: Poised to signal. Am J Physiol Endocrinol Metab 298: E395–E402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC, Johnson JE 1998. Protein kinase C: A paradigm for regulation of protein function by two membrane-targeting modules. Biochim Biophys Acta 1376: 155–172 [DOI] [PubMed] [Google Scholar]
- Oancea E, Meyer T 1998. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell 95: 307–318 [DOI] [PubMed] [Google Scholar]
- Parekh DB, Ziegler W, Parker PJ 2000. Multiple pathways control protein kinase C phosphorylation. Embo J 19: 496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce LR, Komander D, Alessi DR 2010. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol 11: 9–22 [DOI] [PubMed] [Google Scholar]
- Pederson SF, Varming C, Christensen ST, Hoffmann EK 2002. Mechanisms of activation of NHE by cell shrinkage and by calyculin A in Ehrlich ascites tumor cells. J Membr Biol 189: 67–81 [DOI] [PubMed] [Google Scholar]
- Penniston JT, Enyedi A 1998. Modulation of the plasma membrane Ca2+ pump. J Membr Biol 165: 101–109 [DOI] [PubMed] [Google Scholar]
- Pereira L, Metrich M, Fernandez-Velasco M, Lucas A, Leroy J, Perrier R, Morel E, Fischmeister R, Richard S, Benitah JP, et al. 2007. The cAMP binding protein Epac modulates Ca2+ sparks by a Ca2+/calmodulin kinase signalling pathway in rat cardiac myocytes. J Physiol 583: 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls A, Schmidt S, Grawe F, Stabel S 1997. Interaction of protein kinase C ζ with ZIP, a novel protein kinase C-binding protein. Proc Natl Acad Sci 94: 6191–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu RG, Abo A, Steven Martin G 2000. A human homolog of the C. elegans polarity determinant Par-6 links Rac and Cdc42 to PKCζ signaling and cell transformation. Curr Biol 10: 697–707 [DOI] [PubMed] [Google Scholar]
- Reiken S, Gaburjakova M, Guatimosim S, Gomez AM, D’Armiento J, Burkhoff D, Wang J, Vassort G, Lederer WJ, Marks AR 2003. Protein kinase A phosphorylation of the cardiac calcium release channel (ryanodine receptor) in normal and failing hearts. Role of phosphatases and response to isoproterenol. J Biol Chem 278: 444–453 [DOI] [PubMed] [Google Scholar]
- Reither G, Schaefer M, Lipp P 2006. PKCα: A versatile key for decoding the cellular calcium toolkit. J Cell Biol 174: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC 1998. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem 273: 15879–15882 [DOI] [PubMed] [Google Scholar]
- Rosse C, Linch M, Kermorgant S, Cameron AJ, Boeckeler K, Parker PJ 2010. PKC and the control of localized signal dynamics. Nat Rev Mol Cell Biol 11: 103–112 [DOI] [PubMed] [Google Scholar]
- Ruknudin AM, Wei SK, Haigney MC, Lederer WJ, Schulze DH 2007. Phosphorylation and other conundrums of Na/Ca exchanger, NCX1. Ann NY Acad Sci 1099: 103–118 [DOI] [PubMed] [Google Scholar]
- Sakwe AM, Rask L, Gylfe E 2005. Protein kinase C modulates agonist-sensitive release of Ca2+ from internal stores in HEK293 cells overexpressing the calcium sensing receptor. J Biol Chem 280: 4436–4441 [DOI] [PubMed] [Google Scholar]
- Schaefer M, Albrecht N, Hofmann T, Gudermann T, Schultz G 2001. Diffusion-limited translocation mechanism of protein kinase C isotypes. FASEB J 15: 1634–1636 [DOI] [PubMed] [Google Scholar]
- Schleifenbaum A, Stier G, Gasch A, Sattler M, Schultz C 2004. Genetically encoded FRET probe for PKC activity based on pleckstrin. J Am Chem Soc 126: 11786–11787 [DOI] [PubMed] [Google Scholar]
- Schmidt M, Evellin S, Weernink PA, von Dorp F, Rehmann H, Lomasney JW, Jakobs KH 2001. A new phospholipase-C-calcium signalling pathway mediated by cyclic AMP and a Rap GTPase. Nat Cell Biol 3: 1020–1024 [DOI] [PubMed] [Google Scholar]
- Schulze DH, Muqhal M, Lederer WJ, Ruknudin AM 2003. Sodium/calcium exchanger (NCX1) macromolecular complex. J Biol Chem 278: 28849–28855 [DOI] [PubMed] [Google Scholar]
- Srivastava J, Goris J, Dilworth SM, Parker PJ 2002. Dephosphorylation of PKCδ by protein phosphatase 2Ac and its inhibition by nucleotides. FEBS Lett 516: 265–269 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Digman MA, Medkova M, Ananthanarayanan B, Melowic HR, Rafter JD, Cho W 2005a. Diacylglycerol-induced membrane targeting and activation of protein kinase Cε: Mechanistic differences between protein kinases Cδ and Cε. J Biol Chem 280: 19784–19793 [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Wang J, Blatner NR, Rafter JD, Murray D, Cho W 2005b. The origin of C1A–C2 interdomain interactions in protein kinase Cα. J Biol Chem 280: 36452–36463 [DOI] [PubMed] [Google Scholar]
- Strassheim D, Williams CL 2000. P2Y2 purinergic and M3 muscarinic acetylcholine receptors activate different phospholipase C-β isoforms that are uniquely susceptible to protein kinase C–dependent phosphorylation and inactivation. J Biol Chem 275: 39767–39772 [DOI] [PubMed] [Google Scholar]
- Sutton RB, Sprang SR 1998. Structure of the protein kinase Cβ phospholipid-binding C2 domain complexed with Ca2+. Structure 6: 1395–1405 [DOI] [PubMed] [Google Scholar]
- Swairjo MA, Concha NO, Kaetzel MA, Dedman JR, Seaton BA 1995. Ca2+-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V. Nat Struct Biol 2: 968–974 [DOI] [PubMed] [Google Scholar]
- Takahashi E, Abe J, Gallis B, Aebersold R, Spring DJ, Krebs EG, Berk BC 1999. p90(RSK) is a serum-stimulated Na+/H+ exchanger isoform-1 kinase. Regulatory phosphorylation of serine 703 of Na+/H+ exchanger isoform-1. J Biol Chem 274: 20206–20214 [DOI] [PubMed] [Google Scholar]
- Tung HY 1986. Phosphorylation of the calmodulin-dependent protein phosphatase by protein kinase C. Biochem Biophys Res Commun 138: 783–788 [DOI] [PubMed] [Google Scholar]
- Uhlen P, Fritz N 2010. Biochemistry of calcium oscillations. Biochem Biophys Res Commun 396: 28–32 [DOI] [PubMed] [Google Scholar]
- Ubach J, Zhang X, Shao X, Sudhof TC, Rizo J 1998. Ca2+ binding to synaptotagmin: How many Ca2+ ions bind to the tip of a C2-domain? EMBO J 17: 3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulianich L, Elia MG, Treglia AS, Muscella A, Di Jeso B, Storelli C, Marsigliante S 2006. The sarcoplasmic-endoplasmic reticulum Ca2+ ATPase 2b regulates the Ca2+ transients elicited by P2Y2 activation in PC Cl3 thyroid cells. J Endocrinol 190: 641–649 [DOI] [PubMed] [Google Scholar]
- Venkatachalam K, Montell C 2007. TRP channels. Annu Rev Biochem 76: 387–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K, Zheng F, Gill DL 2003. Regulation of canonical transient receptor potential (TRPC) channel function by diacylglycerol and protein kinase C. J Biol Chem 278: 29031–29040 [DOI] [PubMed] [Google Scholar]
- Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC 1999. Ca2+ bridges the C2 membrane-binding domain of protein kinase Cα directly to phosphatidylserine. EMBO J 18: 6329–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violin JD, Newton AC 2003. Pathway illuminated: Visualizing protein kinase C signaling. IUBMB Life 55: 653–660 [DOI] [PubMed] [Google Scholar]
- Violin JD, Zhang J, Tsien RY, Newton AC 2003. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol 161: 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KK, Wright LC, Machan CL, Allen BG, Conigrave AD, Roufogalis BD 1991. Protein kinase C phosphorylates the carboxyl terminus of the plasma membrane Ca2+-ATPase from human erythrocytes. J Biol Chem 266: 9078–9085 [PubMed] [Google Scholar]
- Wettschureck N, Offermanns S 2005. Mammalian G proteins and their cell type specific functions. Physiol Rev 85: 1159–1204 [DOI] [PubMed] [Google Scholar]
- Wheeler DG, Barrett CF, Groth RD, Safa P, Tsien RW 2008. CaMKII locally encodes L-type channel activity to signal to nuclear CREB in excitation-transcription coupling. J Cell Biol 183: 849–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Wang J, Kodavatiganti R, Zeng Y, Kass IS 2004. Activation of protein kinase C contributes to the isoflurane-induced improvement of functional and metabolic recovery in isolated ischemic rat hearts. Anesth Analg 99: 993–1000 [DOI] [PubMed] [Google Scholar]
- Young SH, Wu SV, Rozengurt E 2002. Ca2+-stimulated Ca2+ oscillations produced by the Ca2+-sensing receptor require negative feedback by protein kinase C. J Biol Chem 277: 46871–46876 [DOI] [PubMed] [Google Scholar]
- Zemlickova E, Johannes FJ, Aitken A, Dubois T 2004. Association of CPI-17 with protein kinase C and casein kinase I. Biochem Biophys Res Commun 316: 39–47 [DOI] [PubMed] [Google Scholar]
- Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD 2006. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol Cell 24: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Horita DA, Waugh DS, Byrd RA, Morrison DK 2002. Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR). J Mol Biol 315: 435–446 [DOI] [PubMed] [Google Scholar]
- Zimmermann G, Taussig R 1996. Protein kinase C alters the responsiveness of adenylyl cyclases to G protein α and βγ subunits. J Biol Chem 271: 27161–27166 [DOI] [PubMed] [Google Scholar]