Abstract
Since its first visualization in 1898, the Golgi has been a topic of intense morphological research. A typical mammalian Golgi consists of a pile of stapled cisternae, the Golgi stack, which is a key station for modification of newly synthesized proteins and lipids. Distinct stacks are interconnected by tubules to form the Golgi ribbon. At the entrance site of the Golgi, the cis-Golgi, vesicular tubular clusters (VTCs) form the intermediate between the endoplasmic reticulum and the Golgi stack. At the exit site of the Golgi, the trans-Golgi, the trans-Golgi network (TGN) is the major site of sorting proteins to distinct cellular locations. Golgi functioning can only be understood in light of its complex architecture, as was revealed by a range of distinct electron microscopy (EM) approaches. In this article, a general concept of mammalian Golgi architecture, including VTCs and the TGN, is described.
Electron microscopy reveals that Golgi stacks are connected by tubules to form a “ribbon.” Vesicular tubular clusters link the cis-face with the ER, whereas the presence of clathrin distinguishes the trans-most cisternae.
In 1898 Camillo Golgi was the first to visualize, describe, and ultimately name the Golgi complex. Using a histochemical impregnation method causing the reduction and deposition of silver, he defined the Golgi in neuronal cells as a reticular apparatus stained by the “black reaction” (Golgi 1898). In the 1950s, the first ultrastructural images of the Golgi were revealed using the then newly developed electron microscope (EM) (Dalton 1954; Farquhar and Rinehart 1954; Sjostrand and Hanzon 1954; Dalton and Felix 1956), reviewed by Farquhar and Palade (1981). In 1961, the thiamine pyrophosphatase reaction developed by Novikoff and Goldfischer allowed cytochemical labeling of Golgi membranes, which revealed the ubiquitous cellular distribution of this organelle (Novikoff and Goldfischer 1961). In the many years of ultrastructural research that have followed, the visualization of the Golgi has gone hand-in-hand with the developing EM techniques.
The intriguing structural complexity of the Golgi has made it one of the most photographed organelles in the cell. However, a full understanding of Golgi architecture is hard to deduce from the ultrathin (70–100 nm) sections used in standard transmission EM preparations. Rambourg and Clermont (1974) were the first to investigate the Golgi in three dimensions (3D), using stereoscopy (Rambourg 1974). In this approach a “thick” (150–200 nm), EM section is photographed at two distinct angles, after which the pairs of photographs are viewed with a stereoscope. Over the years, stereoscopy was applied to a variety of cells and has greatly contributed to our current understanding of Golgi architecture (Lindsey and Ellisman 1985; Rambourg and Clermont 1990; Clermont et al. 1994; Clermont et al. 1995). An alternative approach to study 3D structure is serial sectioning, by which a series of adjacent (serial) thin sections are collected. The Golgi can be followed throughout these sections and be constructed into a 3D model (Beams and Kessel 1968; Dylewski et al. 1984; Rambourg and Clermont 1990). In the nineties, 3D-EM was boosted by the introduction of high-voltage, dual axis 3D electron tomography (Ladinsky et al. 1999; Koster and Klumperman 2003; Marsh 2005; Marsh 2007; Noske et al. 2008), which allows the analysis of sections of up to 3–4 µm with a 4–6 nm resolution in the z-axis. The sections are photographed in a tilt series of different angles, which are reconstructed into a 3D tomogram that allows one to “look beyond” a given structure and reveals how it relates to other cellular compartments.
Membranes with a similar appearance can differ in protein content and function. These differences are revealed by protein localization techniques. Therefore, in addition to the “classical” EM techniques providing ultrastructural details, EM methods that determine protein localization within the context of the cellular morphology have been crucial to further our understanding on the functional organization of the Golgi. For example, by enzyme-activity-based cytochemical staining the cis-to-trans-polarity in the distribution of Golgi glycosylation enzymes was discovered, reviewed by Farquhar and Palade (1981), which was key to understanding the functional organization of the Golgi stack in protein and lipid glycosylation. With the development of immunoEM methods, using antibodies, the need for enzyme activity for protein localization was overcome. This paved the way for the localization of a wide variety of proteins, such as the cytoplasmic coat complexes associated with the Golgi (Rabouille and Klumperman 2005).
A logical next step in EM-based imaging of the Golgi would be to combine protein localization with 3D imaging, but this is technically challenging. A number of protocols enabling protein localization in 3D have recently been described (Trucco et al. 2004; Grabenbauer et al. 2005; Gaietta et al. 2006; Zeuschner et al. 2006; Meiblitzer-Ruppitsch et al. 2008), but these have only been applied in a limited manner to Golgi studies. Another approach that holds great potential for Golgi research is correlative microscopy (CLEM). Live cell imaging of fluorescent proteins has revolutionized cell biology by the real time visualization of dynamic events. However, live cell imaging does not reveal membrane complexity. By CLEM, live cells are first viewed by light microscopy and then prepared for EM (Mironov et al. 2008; van Rijnsoever et al. 2008). When coupled with the recent introduction of super resolution light microscopy techniques for real time imaging, the combination with EM for direct correlation with ultrastructural resolution has great potential (Hell 2009; Lippincott-Schwartz and Manley 2009).
The 100th anniversary of the discovery of the Golgi, in 1998, triggered a wave of reviews on this organelle, including those focusing on Golgi architecture (Rambourg 1997; Farquhar and Palade 1998). More recent reviews that describe Golgi structure in great detail are provided by Marsh (2005) and Hua (2009). In this article, the most recent insights in mammalian Golgi architecture as revealed by distinct EM approaches are integrated into a general concept.
MAMMALIAN GOLGI ARCHITECTURE: GENERAL OVERVIEW
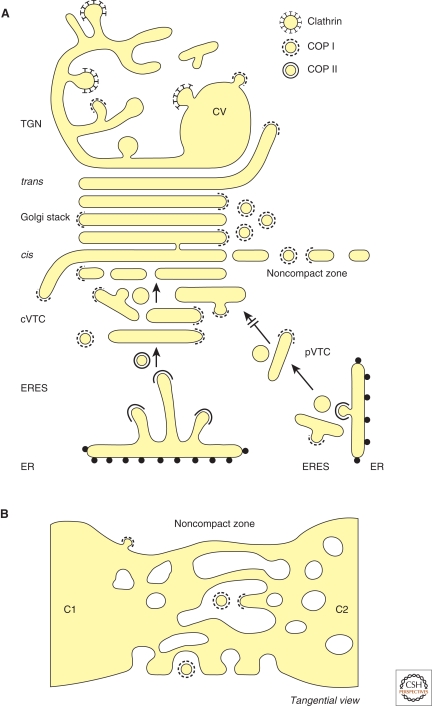
Though different methods of sample preparation and EM techniques highlight different morphological features, a consensus view has emerged over many decades describing the general organization of the mammalian Golgi and its connected membranes (Figs. 1 and 2). A typical mammalian Golgi consists of a pile of stapled, disk-like membranes, the cisternae, which together form the Golgi stack. Based on the presence of distinct sets of resident proteins, the Golgi stack can be divided into three regions: cis, medial and trans. The cisternae move and mature from the cis- into the trans-direction, which in parallel to other pathways of intraGolgi transport, provides a major mode of anterograde membrane flow (for a detailed discussion on intraGolgi transport, see Glick and Luini 2011). A mammalian cell can contain multiple Golgi stacks that are laterally interconnected by a reticulated network of branching and rejoining tubules, referred to as the noncompact zone (Fig. 1B). The compound structure of different stacks interconnected by tubules is called the Golgi ribbon. In addition, the Golgi stack is closely associated with two elaborate membrane networks located at the cis- and trans-sides, which represent the entrance and exit faces of the stack, respectively (Fig. 1A). At the cis-side, a collection of vesicular tubular clusters (VTCs) mediates transport between the ER and the Golgi stack. At the trans-side, the trans-Golgi network (TGN) receives proteins that have passed through the Golgi stack and distributes them to distinct cellular locations. The Golgi ribbon is typically located in the perinuclear area around the microtubule organizing center (MTOC) and maintenance of Golgi structure is intimately related to both the microtubule and actin cytoskeleton (Thyberg and Moskalewski 1985; Weidman et al. 1993; Thyberg and Moskalewski 1999; Egea et al. 2006; Kondylis et al. 2007; Brownhill et al. 2009; Goud and Gleeson 2010). In the paragraphs below the distinct components of Golgi architecture and its associated membranes are described in greater detail.
Figure 1.
Schematic representation of the Golgi and associated membrane networks. (A) Overview of the Golgi stack in relation to ERES, central (c) and peripheral (p) VTCs and TGN and the occurrence of COPII, COPI, and clathrin coats. Membranes are drawn as if cross-sectioned in side view. Because of space limitations the COPI vesicles are indicated at only one side of the Golgi stack and the noncompact zone is only drawn at the cis-cisterna. CV = condensing vacuole. (B) Tangential view of two cisternae (C1 and C2) from adjacent Golgi stacks that are bridged by the tubular network of the noncompact zone.
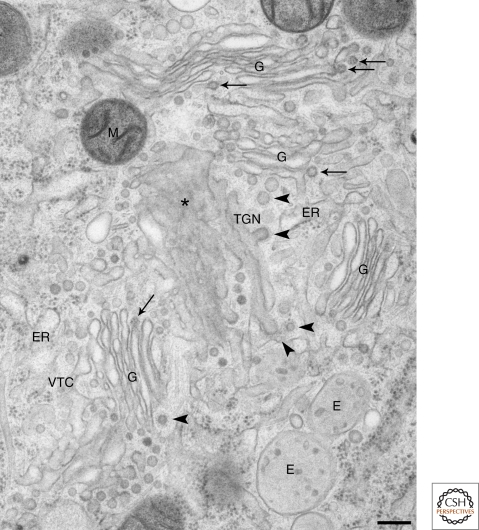
Figure 2.
Golgi region of a human dendritic (D1) cell. Cells were prepared by high-pressure-freezing fixation and freeze-substitution for examination by EM. Multiple Golgi stacks (G) are surrounded by numerous vesicles on which the characteristic dense, regular, coatomer protein complex-I (COPI) coat is clearly visible (arrows). Similar coats are also seen on the rims of Golgi cisternae (arrows); these COP coats are markedly different from the more spiky and wider clathrin coat that is abundant at the trans-side of the Golgi (arrowheads). The asterisk indicates a tangential section of a Golgi cisterna. M = mitochondrion, E = endosomes. Bar, 200 nm. This figure was modified from Rabouille and Klumperman (2005) and reprinted with permission from Nature Review Molecular Cell Biology © 2005.
THE CIS-FACE OF THE GOLGI
All newly synthesized proteins that are destined for secretion or intracellular compartments enter the secretory pathway by co- or posttranslational translocation across the endoplasmic reticulum (ER) membrane. Transport of proteins out of the ER occurs by membrane-bounded carriers, which bud from the ER at specialized ER exit sites (ERES) (Figs. 1A and 3), also known as transitional elements (TEs) or transitional ER (tER). ERES are recognizable by the absence of ribosomes and the presence of Ω-shaped membrane profiles, which represent the forming vesicles, and longer membrane evaginations, which may be up to 500 nm in length (Palade 1975; Sesso 1994; Bannykh et al. 1996; Fan et al. 2003; Mironov et al. 2003). In the EM, ERES are most readily found in the area facing the cis-Golgi (Fig. 3B). However, depending on the cargo load (Farhan et al. 2008); numerous smaller ERES can arise in the peripheral cytoplasm, at some distance from the Golgi (Lotti et al. 1992; Klumperman et al. 1998b).
Figure 3.
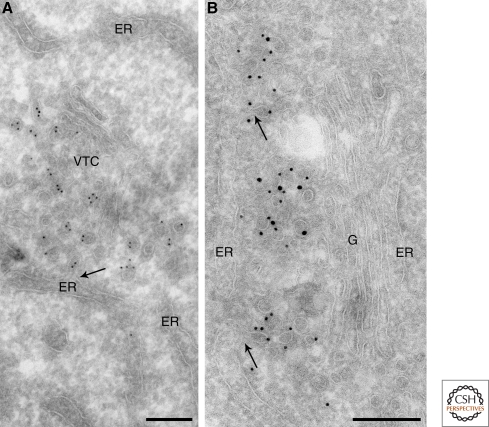
Interface between ER exit sites, VTCs and the Golgi stack. Human hepatoma HepG2 cells were fixed by a mixture of formaldehyde and glutaraldehyde and prepared for ultrathin cryosectioning and immunoEM. (A) Immunogold labeling for the VTC marker ERGIC53 (10 nm gold) shows a budding profile (arrow) on an ER cisterna. The ERES faces a characteristic tubulovesicular VTC. (B) Double immunogold labeling for COPII (10 nm gold) and the ER-Golgi SNARE hsec22b (15 nm gold) outlines ERES located in the central Golgi region. Note that the membrane buds emerging from the ER (arrows) are labeled for COPII. G = Golgi stack. Bars, 200 nm. Figure 3A is modified from Hay et al. (1998) and reprinted with permission from Journal of Cell Biology © 1998. Figure 3B is modified from Klumperman et al. (1998) and reprinted with permission from Journal of Cell Science © 1998.
Many of the ERES budding profiles display a thin, regular coating at their cytoplasmic leaflet, representing the COPII coat. Assembly of the COPII coat requires activation of Sar1-GDP by the integral ER protein sec12, leading to subsequent recruitment of Sec23/Sec24p and Sec31/Sec13p. The COPII coat then drives the deformation of ER membranes into budding vesicles containing cargo for export from the ER (Barlowe et al. 1994; Barlowe 2003). The size and shape of free COPII-coated vesicles, after detachment from the ER, ranges from ∼60 nm vesicles to 200 nm tubules, often with a halter-like appearance; that is, two balls connected by a thinner stem (Sesso 1994; Bannykh et al. 1996; Zeuschner et al. 2006). The COPII vesicles uncoat and fuse with each other or with an independent cluster of vesicular and tubular shaped membranes, the so-called VTCs (Bannykh et al. 1996), which are also commonly referred to as ER-Golgi intermediate compartment, ERGIC (Farquhar and Hauri 1997).
Vesicular Tubular Clusters
As is indicated by their name, VTCs comprise a mixture of vesicles and tubules. Typically, VTCs are located adjacent to an ERES (Bannykh et al. 1996). The tubules are sometimes branched, forming typical tri-angular membrane profiles. In addition, VTCs can display a few flat disc-like membranes (cisternae) of modest diameter, which are mostly found further away from the ER (Sesso et al. 1994; Klumperman et al. 1998b) (Fig. 3A). Continuities between VTCs and ERES are very rarely seen, mostly under nonphysiological circumstances (Bannykh et al. 1998), defining the VTCs as an independent compartment. On VTC membranes, a second type of coat assembles, the COPI coat, which can induce the formation of COPI-coated vesicles that retrieve ER-resident proteins and membranes to the ER (Lewis and Pelham 1990; Letourneur et al. 1994). These coats consist of the small GTPase ARF and a preassembled coatomer complex of seven subunits (Rothman 1994). Since COPII coats exclusively associate with budding profiles forming on ER membranes, whereas COPI coats are absent from these ER-associated buds, the presence of COPI can be used to distinguish VTC membranes from ERES (Martinez-Menarguez et al. 1999).
Cells can have multiple VTCs associated with both the central and peripheral ERES (Lotti et al. 1992; Klumperman et al. 1998b). Generally, the central VTC is the largest and faces the cis-side of the Golgi, whereas the peripheral ERES have no adjacent Golgi (Fig. 1A) (Balch et al. 1994; Klumperman 2000). Size measurements of ER-associated VTCs and those on their way to the Golgi suggest that small clusters of tubulovesicular membranes detach from the peripheral VTCs and head toward the Golgi (Balch et al. 1994; Bannykh et al. 1996; Klumperman 2000). This transport is microtubule-dependent and followed by fusion of the peripherally derived membranes with central VTC membranes or the cis-Golgi cisterna (Presley et al. 1997; Scales et al. 1997). The central VTC flattens out into small-sized cisternal membranes (Mogelsvang et al. 2004) that deposit in parallel to the cis-most Golgi cisternae. They then fuse either directly with the preexisting cis-cisterna or laterally, to make a new cis-cisterna (Fig. 1) (Hauri and Schweizer 1992). Based on a computational model, it was hypothesized that the extent by which VTC membranes can fuse with the cis-Golgi cisterna is determined by the rate by which the maturing cisterna loses its fusion competence, for example by recycling SNARE proteins to the ER (Kuhnle et al. 2010). In electron micrographs, providing a snapshot of these fusion events, continuities between the central VTC and the cis-Golgi are regularly observed and also referred to as the cis-Golgi network (Lindsey and Ellisman 1985; Huttner and Tooze 1989; Rambourg and Clermont 1990; Pelham 1991; Rambourg 1997).
By morphology alone, the boundary between the end of a VTC and the beginning of the Golgi is hard to set (Mellman and Simons 1992). However, marker proteins specific for VTCs, like ERGIC53, show a remarkably strict segregation between VTC and Golgi membranes (Fig. 4A). Notably, this close relationship between VTCs and Golgi was first shown when Golgi’s histochemical staining method was adapted for EM. The electron-dense staining obtained by reducing osmium was specifically found in the cis-most cisterna of the Golgi as well as in the VTCs (Rambourg et al. 1974; Farquhar and Palade 1981).
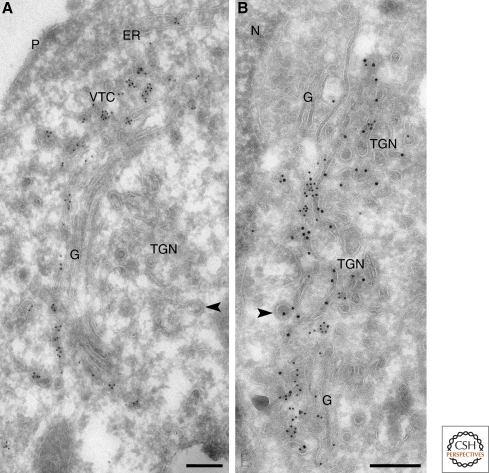
Figure 4.
Interface between the Golgi stack, VTCs and TGN. Human hepatoma HepG2 cells were fixed using a mixture of formaldehyde and glutaraldehyde and prepared for ultrathin cryosectioning and immunoEM. (A) Immunogold labeling for the VTC marker ERGIC53 (10 nm gold) illustrates the boundary between VTCs and the Golgi stack (G). ERGIC53 label is restricted to the VTC membranes and cis-most cisterna of the Golgi stack. The TGN is devoid of ERGIC-53 label. The arrowhead points to a clathrin-coated TGN membrane. (B) Immunogold labeling for the TGN markers TGN46 (10 nm gold) and cation-independent mannose 6-phosphate receptor (CI-MPR; 15 nm gold) outlines the cisternal and tubular membranes of the TGN. Arrowheads point to clathrin-coated TGN membranes. N = nucleus, P = plasma membrane. Bars, 200 nm. Figure 4A is modified from Klumperman et al. (1998) and reprinted with permission from Journal of Cell Science © 1998.
THE GOLGI STACK
The morphologically most striking part of the Golgi, the Golgi stack (sometimes referred to as the core region or compact zone of the Golgi), consists of flat, cisternal membranes. The typical structure of the Golgi stack arises because distinct cisternae align on top of each other, forming a pile of closely opposed membranes (Fig. 2). The diameter of the cisternae varies per cell and per condition, but between distinct organisms ranges from 0.7 to 1.1 µm (Rabouille et al. 1995; Pelletier et al. 2002). Most mammalian cells contain multiple Golgi stacks that are laterally interconnected with tubules, designated the noncompact zone of the Golgi. A collection of distinct stacks that are interconnected by the tubular noncompact zones is referred to as the Golgi ribbon (Figs. 1 and 5) (Rambourg et al. 1979; Rambourg 1997). When viewed by EM, an individual cell can show multiple Golgi stacks, representing a single Golgi ribbon winding in and out of the section (Rambourg et al. 1979; Marsh et al. 2001a) (Fig. 2). The number of cisternae within a stack varies between 4 and 11 in mammalian cells (Rambourg 1997) and is characteristic for each cell type. The trans-most cisterna of the Golgi is unique in that it displays a third type of coat i.e., in addition to COPII and COPI, the clathrin coat. This trans-most cisterna also gives rise to the tubular membranes of the TGN (Figs. 1 and 4B). Between and surrounding the cisternae a dense, ribosome-free protein network is present, the “Golgi matrix,” that might contain structural proteins (e.g., coiled coil proteins) supporting the stack structure (Franke et al. 1972; Cluett and Brown 1992; Slusarewicz et al. 1994; Sinka et al. 2008).
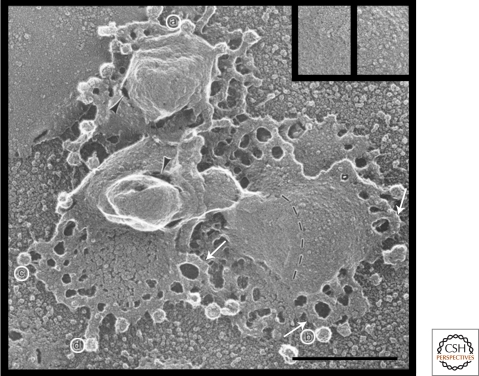
Figure 5.
Replica’s of isolated Golgi cisternae showing tubular interconnections and coated budding profiles. Images were prepared from a Golgi enriched membrane fraction of Chinese hamster ovary cells (CHO). (A) A bud protruding from a nonfenestrated edge of the central part of a cisterna. (B) A coated bud protruding from the side of a tubular loop (arrow). (C) A fully formed coated vesicle with fibrous attachments to the cisterna. (D) A short tubule with a coated bud at the tip. Arrows point to tubules that seem to bend back and fuse with the edge of the same cisterna that they emerge from, thus forming an irregular fenestration. Although not formally identified in these preparations, current evidence indicates that the coats shown here are of the COPI type. Bar, 0.5 µm. This figure was taken from Weidman et al. (1993) and reprinted with permission from Cell © 1993.
The central part of a given Golgi cisterna is usually quite narrow (10–20 nm), whereas the edges, generally indicated as rims, are more dilated. Especially at these dilated rims, the cisternae are perforated with membrane-bounded holes of up to 100 nm diameter (Fig. 5), which typically appear as gaps in 2D EM sections and are called fenestrae. All cisternae are fenestrated, but the fenestrae become smaller in the cis-to-medial direction and then again increase in size and number toward the trans-side of the Golgi (Ladinsky et al. 1999). Some fenestrae are of substantial size (>100 nm) and contain within their lumen a collection of small vesicles, which likely form from the edges of the enclosing cisternae (Rambourg and Clermont 1986). These are referred to as “wells” (Hermo et al. 1991). Especially at the cis-to-medial side of the Golgi, consecutive cisternae can align their wells, which then form indentations of the Golgi stack with a large activity of vesicle budding resulting in the presence of numerous vesicles (Ladinsky et al. 1999). The presence of fenestrae increases the surface-to-volume ratio of the cisternae and introduces more curvature to the membranes. This might be important for the production of vesicles and the segregation of proteins and lipids (Glick and Nakano 2009).
Within a given stack, connections between heterologous cisternae can be formed (Fig. 1). These were first reported by Rambourg and colleagues and are especially well visible in spermatids and Sertoli cells (Rambourg 1997). More recent studies have shown the presence of intercisternal tubular connections in additional cell types, including normal rat kidney (NRK) and pancreatic β cells (Marsh et al. 2004; Trucco et al. 2004). The diameter of these tubules is about 20 nm (Marsh et al. 2004), which is of the same magnitude as the width of the cisternal lumen. Although quantitation of these types of structures is not trivial, it seems that they are rare to absent in control cells with basal secretory activity (Marsh et al. 2001a), but increase in frequency in cells with high secretory activity, such as professional secreting cells (Rambourg 1997) or in cells which are experimentally triggered to protein synthesis (Marsh et al. 2004; Bouchet-Marquis et al. 2008; Vivero-Salmeron et al. 2008). It has been shown that the arrival of cargo at the Golgi triggers the recruitment of Group IVA Ca(2+)-dependent, cytosolic PLA(2) to Golgi membranes, which induces formation of the intercisternal tubules (San Pietro et al. 2009). Thus far, such connections have only been seen in mammalian cells. Their possible function in intraGolgi transport is discussed in Glick and Luini 2011.
The Golgi stack can show multiple sites of close contact with the ER. ER membranes can flatten out to cisternae that closely follow and partially enwrap a trans- or the trans-most cisterna. These flattened ER cisternae lack ribosomes at the side facing the Golgi. Examples of this type of interaction were first described by Novikoff and colleagues (Novikoff et al. 1971) and since then were reported in various cell types and by different EM approaches (Hand and Oliver 1977; Pavelka and Ellinger 1983; Craig and Staehelin 1988; Thorne-Tjomsland et al. 1991; Marsh et al. 2001a; Meiblitzer-Ruppitsch et al. 2008). It is believed that these connections might facilitate protein and lipid exchange between membranes and/or are important for maintaining structure (Levine and Rabouille 2005; Hanada et al. 2007; Peretti et al. 2008; Glick and Nakano 2009; Rocha et al. 2009). By 3D-electron tomography, a second type of ER-Golgi association was visualized consisting of an ER cisterna almost entirely devoid of ribosomes that traversed the Golgi stack through consecutively aligned openings in the cisternae (Marsh et al. 2001a). The functional significance of this observation still needs to be established.
Golgi stacks are conserved throughout eukaryotic evolution, although some exceptions are known, such as the budding yeast Saccharomyces cerevisae (Rabouille and Kondylis 2007), in which the Golgi consists of individual cisternae that are scattered throughout the cytoplasm. These cisternae occasionally associate with each other, but do not form a stack (Preuss et al. 1992; Castillon et al. 2009; Papanikou and Glick 2009), yet can still be sub-divided into cis, medial, trans, and TGN (Brigance et al. 2000). Lower animal cells and organisms (e.g., protozoa, the budding yeast Pichia pastoris, and the fruitfly Drosophila melanogaster) can form Golgi stacks, but these remain separate and are not interconnected by tubules (Prydz et al. 2008; Kondylis and Rabouille 2009). In D. melanogaster the Golgi stacks are typically found in close association with ERES, forming units designated tER-Golgi units. Strikingly, some cells at specific stages of D. melanogaster development do not show a Golgi stack, but instead clusters of vesicles and tubules. Secretion by these cells occurs with high efficiency (Kondylis and Rabouille 2009). Finally, it was found that eukaryotic lineages lacking a morphologically identifiable Golgi do contain conserved Golgi proteins. This suggests that the Golgi can be altered beyond recognition, but that there exist no “Golgi-lacking” eukaryotes (Dacks et al. 2003).
THE NONCOMPACT ZONE OF THE GOLGI
From the fenestrated, peripheral regions of the cisternae of the Golgi stack, lateral networks of ∼30 nm diameter tubules emerge. Some of these tubular membranes bend back to fuse with the same cisterna that they emerge from, forming another fenestration (Fig. 5, arrows). Other tubules emanating from cis- and medial-cisternae project backward to the VTCs (Fig. 1) (Rambourg 1997; Ladinsky et al. 1999; Glick and Nakano 2009). Many tubules, however, extend laterally, sometimes over a distance of several micrometers, and fuse with tubules from adjacent Golgi stacks, forming the tubular network that bridges adjacent stacks (Figs. 1 and 5). These tubular networks are classically referred to as the noncompact zones of the Golgi. The tubules can connect cisternae located at the same positions in the respective stacks, but also cisternae located at different levels (Rambourg 1997).
Volume measurements in insulin producing β cells of mice gave an estimated volume of 3.7–5.8 µm3 for the total volume of the Golgi, of which the Golgi stack comprised 3.1–3.6 µm3 (Noske et al. 2008). Thus, the noncompact zone of the Golgi substantially contributes to total Golgi volume. Recent studies indicate that the formation of a Golgi ribbon is particularly important for protein glycosylation. The Golgi ReAssembly Stacking Proteins GRASP65 and GRASP55 function in the formation and/or maintenance of the tubules connecting the Golgi stacks, presumably by forming protein tethers between adjacent membranes (Barr et al. 1998; Shorter et al. 1999; Rabouille and Kondylis 2007; Feinstein and Linstedt 2008; Vinke et al. 2011). Depletion of GRASP65 and/or the “golgin” protein GM130 (Gillingham and Munro 2003; Barinaga-Rementeria Ramirez and Lowe 2009) (for further details, see Munro 2011) disturbs the Golgi ribbon. This results in the redistribution of Golgi enzymes over the stacks and affects proper protein glycosylation, but does not cause a block in secretion (Puthenveedu et al. 2006; Marra et al. 2007b). It remains to be established, however, whether GM130 acts directly in homotypic fusion of neighboring cisternae (Puthenveedu et al. 2006) or mediates the tethering of ER-derived vesicles with the cis-Golgi (Marra et al. 2007a). Interestingly, unlinking the Golgi ribbon is part of a putative G2/M checkpoint (Sutterlin et al. 2002; Yoshimura et al. 2005; Rabouille and Kondylis 2007).
COPI COATS IN THE GOLGI STACK
The Golgi area contains numerous 60–100 nm vesicles, especially concentrated in the Golgi wells and at the Golgi rims (Fig. 2, arrows). Depending on the cell type and cellular activity, the number of these vesicles can add up to more than 2000 per Golgi region (Marsh et al. 2001a). Many of these vesicles bear a COP coat. In standard EM sections, COPI and COPII coats cannot be distinguished. However, by immuno-labeling the coats on the rims of the Golgi cisternae as well as on the Golgi associated vesicles were identified as the COPI type, the same coat as found on VTC membranes (Oprins et al. 1993; Griffiths et al. 1995; Orci et al. 1997; Martinez-Menarguez et al. 2001; Rabouille and Klumperman 2005). Many COPI vesicles are tethered to the Golgi cisternae via long, rod-like proteins (Orci et al. 1998; Marsh et al. 2001a), which might represent members of the golgin protein family (Goud and Gleeson 2010).
COPI-coated membrane buds can form at the central, nonfenestrated area of a cisterna, but are much more frequently seen at the fenestrated rims, in which they form directly on the cisternal membrane or at the tips of the tubules that emanate from these regions (Weidman et al. 1993). The majority of the buds associated with Golgi cisternae are coated (Ladinsky et al. 1999). The highest number of COPI-coated buds is found on the cis-cisternae, with declining numbers toward the trans-side and TGN (Oprins et al. 1993; Ladinsky et al. 1999).
The role of COPI vesicles in intraGolgi transport is a topic of intense investigations and debate. It is now generally accepted that COPI coated vesicles mediate retrograde transport of Golgi resident enzymes from maturing cisternae throughout the Golgi stack and of ER-resident proteins to the ER (Emr et al. 2009). In addition, a subpopulation of COPI coated vesicles might be involved in anterograde transport of cargo through the Golgi stack (Orci et al. 1997; Malsam et al. 2005). The small size of the COPI vesicles and the distinct possibility that morphologically identical COPI vesicles represent a mixed population with regard to cargo and directionality, hampers their functional characterization. To further address their role in Golgi transport, a distinction between distinct COPI vesicle subpopulations is required. A first study addressing this point indeed showed that different subtypes of COPI vesicles show a differential distribution relative to the Golgi stack (Moelleken et al. 2007).
THE TRANS-FACE OF THE GOLGI
The TGN is the main site of sorting of proteins and lipids for postGolgi destinations (Griffiths and Simons 1986; Mellman and Simons 1992; Traub and Kornfeld 1997; De Matteis and Luini 2008). The anterograde exit routes from the TGN are to the apical plasma membrane, basolateral plasma membrane, recycling endosomes, early endosomes, late endosomes, and specialized compartments, such as secretory granules, reviewed by De Matteis and Luini (2008). In addition, retrograde transport from the TGN to the Golgi stack occurs (Sandvig et al. 2004; Johannes and Popoff 2008). For each destination a distinct type of carrier might be used (Traub and Kornfeld 1997), but it still remains to be established how many types of TGN exits there are.
TGN Architecture
Morphologically, the TGN is defined as a tubular, anastomosing (meaning branching, reticulating) compartment at the trans-side of the Golgi stack that is continuous with the trans-most Golgi cisterna (Fig. 1) (Griffiths and Simons 1986; Rambourg and Clermont 1990; Geuze and Morre 1991). Hence, the TGN consists of a tubular and a cisternal part, with the latter being the trans-most cisterna of the Golgi stack. The conversion of the cisternal membrane into tubules is dependent on both protein- and lipid-based mechanisms (De Matteis and Luini 2008). In addition to the trans-most cisterna, the penultimate trans-cisterna can also project tubules into the trans-region (Fig. 1), which in 2D sections contributes to the overall membrane complexity of the TGN area (Rambourg et al. 1979; Marsh and Howell 2002; Mogelsvang et al. 2004). The cisternal parts of the two trans-cisternae are sparsely fenestrated and often only partially aligned with the previous cisterna, as if they are detaching, or peeling off, from the Golgi stack (Rambourg et al. 1979; Geuze and Morre 1991). Where these trans-cisternae lose alignment with the stack, they form the anastomosing tubules (Griffiths and Simons 1986; Clermont et al. 1995; Rambourg 1997).
In ultrathin 2D-EM sections of cells with a well-developed TGN, such as the human hepatoma HepG2 cells, the tubular TGN has a striking and distinctive morphology, being a collection of branched tubules with many budding profiles and associated vesicles (Fig. 4B). However, this morphology is strongly dependent on the cell type. In some cells, such as secretory cells or cultured NRK cells, the tubular TGN aligns with only part of the Golgi ribbon or can even be completely absent (Rambourg et al. 1979; Ladinsky et al. 1994; Clermont et al. 1995). In addition, the TGN undergoes dynamic changes in size and structure depending on the level of protein synthesis (Hand and Oliver 1984; Griffiths et al. 1989).
The morphological hallmark of the trans-most/TGN cisterna, which distinguishes it from all other Golgi cisternae, is the presence of clathrin (Pearse and Robinson 1990). Similar to COP coats, clathrin deposits as an electron-dense layer on the cytoplasmic leaflets of membranes. The clathrin coat displays a typical spiky pattern and is thicker than the COP coat (circa 18 nm versus 10 nm), which makes it morphologically distinct from COP coats and recognizable without the need for immuno-labeling (Fig. 2) (Orci et al. 1984; Heuser and Kirchhausen 1985; Kirchhausen et al. 1986; Oprins et al. 1993; Ladinsky et al. 1994). Clathrin can coat relatively long distances of the cisternal part of the TGN membrane, which then is often curved away from the stack. In addition, clathrin is associated with the TGN tubules, in which it deforms the membranes into clathrin coated buds (Ladinsky et al. 2002; Mogelsvang et al. 2004). These buds are wider than COP-coated buds and have an average diameter of circa 100 nm (Mogelsvang et al. 2004; Peden et al. 2004). Notably, in addition to clathrin the TGN can also display COPI-coated buds, albeit with much lower frequency (Oprins et al. 1993; Griffiths et al. 1995; Martinez-Menarguez et al. 1999).
TGN Exits
The clathrin-coated TGN buds give rise to clathrin-coated vesicles that transport mannose 6-phosphate tagged lysosomal enzymes bound to mannose 6-phosphate receptors (MPRs) to endosomes. Clathrin-coated carriers are the best-characterized exit mechanism for anterograde transport from the TGN. After delivery of their cargo to the endosomal system, the MPRs cycle back to the TGN for a new round of transport (Kornfeld and Mellman 1989; Pearse and Robinson 1990; van Meel and Klumperman 2008; Braulke and Bonifacino 2009). Interestingly, cells containing an extensive lysosomal system in general show extensive tubulation of the TGN membranes (Clermont et al. 1995; Rambourg 1997), which might reflect the well-developed MPR recycling pathway in these cells. Other types of membrane-bounded carriers also travel in both anterograde and retrograde directions between the TGN and distinct endosomal intermediates (see Pfeffer 2011). Hence, the membrane complexity of the TGN is determined by both biosynthetic traffic coming from the Golgi stack and incoming traffic from the endocytic system (Farquhar and Palade 1981; Kornfeld and Mellman 1989; Vetterlein et al. 2002). The cargo and destinations of TGN derived COPI-coated vesicles have not been especially studied, but they likely represent a retrograde pathway (Cosson and Letourneur 1994; Martinez-Menarguez et al. 2001).
Proteins lacking a specific sorting signal are by default packaged into vesicles of the constitutive pathway to the plasma membrane (Mellman and Simons 1992; Bossard 2007; De Matteis and Luini 2008). Moreover, in polarized (e.g., epithelial or neuronal) cells additional routes emerge from the TGN to target proteins specifically to the apical (or axonal) versus basolateral (or dendritic) plasma membranes (Bennett et al. 1988; Dotti and Simons 1990; Wandinger-Ness et al. 1990). Polischuk and colleagues have used a CLEM approach to morphologically define the carriers involved in the constitutive transport of exogenously expressed vesicular stomatitis virus G (VSV-G) to the plasma membrane of nonpolarized cells. By live cell imaging these carriers were found to detach from tubular TGN membranes (see Malhotra and Campelo 2010) and by CLEM, they were identified as noncoated, pleiomorphic membranes that could contain branches and fenestrations (Polishchuk and Mironov 2004). However, since there are no distinguishing structural features that specify these carriers when forming at the TGN, the ultrastructural details of this TGN exit have remained elusive.
In professional secretory cells, that is, those possessing a regulated secretory pathway, the major TGN exit is to secretory granules (Kelly 1985; Huttner and Tooze 1989; Arvan and Castle 1998). Secretory granules can be of significant size—up to several hundreds of nanometers—and accumulate in the cytoplasm until they are triggered to fuse with the plasma. The forming secretory granules—indicated as condensing vacuoles (CV)—are in most cells only seen at the TGN and, with clathrin, are morphological hallmarks to determine Golgi polarity (Fig. 1). However, in some cells or after strong stimuli, CVs can also be seen in one or more trans-Golgi cisternae (Rambourg et al. 1984; Rambourg 1997; Slot et al. 1997; Arvan and Castle 1998). These, and other, observations have raised the question as to whether exit from the Golgi might also occur from earlier cisternae and not be exclusively confined to the TGN (for further discussion see below) (Rambourg and Clermont 1990).
TGN Sorting
There are currently two distinct models to explain protein sorting in the TGN. The prevalent model is that all proteins that travel through the Golgi stack enter the TGN from which they are sorted to their distinct destinations. However, based on the observations that (i) both the trans-most and the penultimate trans-cisternae can project tubules into the TGN area (Marsh et al. 2001a; Marsh et al. 2001b; Mogelsvang et al. 2004), (ii) that not all TGNs display the tubular compartment and that (iii) induction of a 20°C block, hampering TGN exit, resulted in the swelling of the three last trans-Golgi cisternae (Ladinsky et al. 2002) it was postulated that distinct trans-cisternae might receive distinct sets of cargoes that exit the Golgi sequentially (Ladinsky et al. 1994; Ladinsky et al. 1999; Ladinsky et al. 2002; Mogelsvang et al. 2004). Thus, this second model proposes that multiple trans-Golgi cisternae each form functionally distinct types of exit compartments, as opposed to the original model that one membrane compartment (the TGN), facilitates the sorting of mixed cargo types to multiple destinations.
The second model proposes that proteins destined for the endo-lysosomal pathways would exit the Golgi from the clathrin-coated trans-most/TGN cisterna, whereas proteins destined for the constitutive pathway would exit from the preceding trans-cisternae (Mogelsvang et al. 2004). A key question to distinguish between the two models is therefore how distinct cargoes distribute over the distinct trans-cisternae and TGN. There are several observations in the literature indicating that the trans-most/TGN membrane can contain mixed cargo. For example, CVs in the TGN of professional secretory cells can display clathrin- and COPI-coated buds (Fig. 1) (Orci et al. 1984; Martinez-Menarguez et al. 1999), indicating the presence of three types of cargo (secretory, lysosomal, retrograde) within the same membrane domain. The actual presence of lysosomal enzymes in CVs was shown by immunoEM (Kuliawat et al. 1997; Klumperman et al. 1998a). A second example is provided by polarized cells, in which two types of clathrin-coated vesicles emerge from the TGN. One of these, containing the μ1B adaptin isoform of the AP-1 complex (Ohno et al. 1999), targets VSV-G to the basolateral membrane (Folsch et al. 2003). Since clathrin only associates with the trans-most/TGN cisterna this finding implies that VSV-G enters the clathrin-coated TGN membrane. This was confirmed by immunoEM showing the presence of VSV-G in all cisternae (Griffiths et al. 1985; Griffiths et al. 1989). Other examples of mixed cargoes within the same trans-most/TGN cisterna are: secretory lipoprotein particles and lysosomal enzymes (Farquhar et al. 1974; Farquhar and Palade 1981); endocytosed transferrin and the constitutive secreted protein albumin (Stoorvogel et al. 1988); albumin, MPR and lysosomal enzymes (Geuze et al. 1985); the recycling asialoglycoprotein receptor and MPRs (Geuze et al. 1987); the constitutive secreted protein procollagen and clathrin (Bonfanti et al. 1998).
Another prediction of the alternative model for TGN sorting is that the penultimate trans cisterna is consumed into the vesicles that mediate constitutive transport to the plasma membrane (Mogelsvang et al. 2004). This is a difficult point to prove since, as discussed above, constitutive vesicles do not have distinguishing structural features when forming at the TGN and, once formed, travel rapidly away from the Golgi. Fluorescence measurements in live cells, however, have indicated that anterograde moving cargo might exit the Golgi stack at the level of multiple cisternae (Patterson et al. 2008), although it is not clear how this relates to the fact that most vesicles surrounding the Golgi are COPI-coated. In the Golgi region of insulin producing HIT-T15 cells the presence of noncoated vesicles was described (Mogelsvang et al. 2004), but it cannot be excluded that these represent uncoated COPI vesicles and their cargo has remained unidentified.
Thus, the prevalent evidence is in favor of the idea that the trans-Golgi/TGN cisternae is the main sorting station of the Golgi, containing mixed cargo and showing exit sites to different destinations. However, the presence of multiple cargo types and exits from the TGN does not exclude the possibility that a subset of transport vesicles detaches at an earlier stage during passage through the Golgi stack.
VARIATIONS ON A THEME
Within the general concept outlined above, the architecture of the Golgi and its associated compartments is subject to many variables. Golgi structure varies by cell type. For example, in neurons and Sertoli cells, the Golgi covers a large area of the cytoplasm, whereas in, for example, leukocytes or plasma cells the Golgi is compact and spherical (Rambourg 1997). Within a given cell type, the Golgi volume depends on the level of protein synthesis. Inhibition of ER to Golgi traffic can lead to the disappearance of the Golgi (Lee and Linstedt 1999; Glick 2000; Morin-Ganet et al. 2000; Prescott et al. 2001), whereas an increase in synthesis can lead to increased Golgi volume (Clermont et al. 1993; Noske et al. 2008). The Golgi adapts to these changes by Golgi-based signaling systems (Farhan and Rabouille 2011), for example, by monitoring protein chaperones that escape the ER. In the Golgi, these proteins bind to the KDEL receptor for recycling to the ER, which initiates a Golgi-based signaling cascade that activates intraGolgi trafficking (Pulvirenti et al. 2008). Golgi structure is also determined by a diversity of additional, nonrelated processes, such as cell polarity and wound healing (Yadav et al. 2009). Finally yet importantly, the Golgi undergoes profound remodeling during mitosis and apoptosis (Ramirez and Lowe 2009).
CONCLUDING REMARKS
Extensive ultrastructural research has generated a general outline of Golgi architecture in mammalian cells. The challenge for the years to come lies in filling in the fine details. For example, the increasing number of proteins found to be involved in Golgi structure and function should allow further definition of Golgi subdomains. By studying variations in cell type and different cellular conditions, additional features of the Golgi should be revealed. Further unraveling of Golgi structure will greatly benefit from developments in microscopy techniques, such as super resolution microscopy, CLEM, and 3D protein localization studies.
ACKNOWLEDGMENTS
The author thanks Catherine Rabouille, Viola Oorschot, and Maaike Pols for critical reading and input on the manuscript and Rene Scriwanek and Marc van Peski for assistance with the preparation of the figures. J.K. is the recipient of VICI grant 918.56.611 of the Netherlands Organization for Scientific research (NWO).
Footnotes
Editors: Graham Warren and James Rothman
Additional Perspectives on The Golgi available at www.cshperspectives.org
REFERENCES
- Arvan P, Castle D 1998. Sorting and storage during secretory granule biogenesis: Looking backward and looking forward. Biochem J 332 (Pt 3): 593–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, McCaffery JM, Plutner H, Farquhar MG 1994. Vesicular stomatitis virus glycoprotein is sorted and concentrated during export from the endoplasmic reticulum. Cell 76: 841–852 [DOI] [PubMed] [Google Scholar]
- Bannykh SI, Nishimura N, Balch WE 1998. Getting into the Golgi. Trends Cell Biol 8: 21–25 [DOI] [PubMed] [Google Scholar]
- Bannykh SI, Rowe T, Balch WE 1996. The organization of endoplasmic reticulum export complexes. J Cell Biol 135: 19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C 2003. Signals for COPII-dependent export from the ER: what’s the ticket out?. Trends in cell biology 13: 295–300 [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77: 895–907 [DOI] [PubMed] [Google Scholar]
- Barr FA, Nakamura N, Warren G 1998. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J 17: 3258–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beams HW, Kessel RG 1968. The Golgi apparatus: structure and function. Int Rev Cytol 23: 209–276 [DOI] [PubMed] [Google Scholar]
- Bennett MK, Wandinger-Ness A, Simons K 1988. Release of putative exocytic transport vesicles from perforated MDCK cells. EMBO J 7: 4075–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfanti L, Mironov AA Jr, Martinez-Menarguez JA, Martella O, Fusella A, Baldassarre M, Buccione R, Geuze HJ, Mironov AA, Luini A 1998. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell 95: 993–1003 [DOI] [PubMed] [Google Scholar]
- Bossard C, Bresson D, Polishchuk RS, Malhotra V 2007. Dimeric PKD regulates membrane fission to form transport carriers at the TGN. J Cell Biol 179: 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet-Marquis C, Starkuviene V, Grabenbauer M 2008. Golgi apparatus studied in vitreous sections. J Microsc 230: 308–316 [DOI] [PubMed] [Google Scholar]
- Braulke T, Bonifacino JS 2009. Sorting of lysosomal proteins. Biochim Biophys Acta 1793: 605–614 [DOI] [PubMed] [Google Scholar]
- Brigance WT, Barlowe C, Graham TR 2000. Organization of the yeast Golgi complex into at least four functionally distinct compartments. Mol Biol Cell 11: 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownhill K, Wood L, Allan V 2009. Molecular motors and the Golgi complex: staying put and moving through. Semin Cell Dev Biol 20: 784–792 [DOI] [PubMed] [Google Scholar]
- Castillon GA, Watanabe R, Taylor M, Schwabe TM, Riezman H 2009. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic 10: 186–200 [DOI] [PubMed] [Google Scholar]
- Clermont Y, Rambourg A, Hermo L 1994. Connections between the various elements of the cis- and mid-compartments of the Golgi apparatus of early rat spermatids. Anat Rec 240: 469–480 [DOI] [PubMed] [Google Scholar]
- Clermont Y, Rambourg A, Hermo L 1995. Trans-Golgi network (TGN) of different cell types: three-dimensional structural characteristics and variability. Anat Rec 242: 289–301 [DOI] [PubMed] [Google Scholar]
- Clermont Y, Xia L, Rambourg A, Turner JD, Hermo L 1993. Structure of the Golgi apparatus in stimulated and nonstimulated acinar cells of mammary glands of the rat. Anat Rec 237: 308–317 [DOI] [PubMed] [Google Scholar]
- Cluett EB, Brown WJ 1992. Adhesion of Golgi cisternae by proteinaceous interactions: intercisternal bridges as putative adhesive structures. J Cell Sci 103 (Pt 3): 773–784 [DOI] [PubMed] [Google Scholar]
- Cosson P, Letourneur F 1994. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science (New York) 263: 1629–1631 [DOI] [PubMed] [Google Scholar]
- Craig S, Staehelin LA 1988. High pressure freezing of intact plant tissues. Evaluation and characterization of novel features of the endoplasmic reticulum and associated membrane systems. Eur J Cell Biol 46: 81–93 [PubMed] [Google Scholar]
- Dacks JB, Davis LA, Sjogren AM, Andersson JO, Roger AJ, Doolittle WF 2003. Evidence for Golgi bodies in proposed ‘Golgi-lacking’ lineages. Proc Biol Sci 270 Suppl 2: S168–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton AJ, Felix MD 1954. Cytological and cytochemical characteristics of the Golgi substance of epithelial cells of the epididymis - in situ, in homogenates and after isolation. Am J Anat 94: 171–208 [DOI] [PubMed] [Google Scholar]
- Dalton AJ, Felix MD 1956. A comparative study of the Golgi complex. J Biophys Biochem Cytol 2: 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis MA, Luini A 2008. Exiting the Golgi complex. Nat Rev Mol Cell Biol 9: 273–284 [DOI] [PubMed] [Google Scholar]
- Dotti CG, Simons K 1990. Polarized sorting of viral glycoproteins to the axon and dendrites of hippocampal neurons in culture. Cell 62: 63–72 [DOI] [PubMed] [Google Scholar]
- Dylewski DP, Haralick RM, Keenan TW 1984. Three-dimensional ultrastructure of the Golgi apparatus in bovine mammary epithelial cells during lactation. J Ultrastruct Res 87: 75–85 [DOI] [PubMed] [Google Scholar]
- Egea G, Lazaro-Dieguez F, Vilella M 2006. Actin dynamics at the Golgi complex in mammalian cells. Curr Opin Cell Biol 18: 168–178 [DOI] [PubMed] [Google Scholar]
- Emr S, Glick BS, Linstedt AD, Lippincott-Schwartz J, Luini A, Malhotra V, Marsh BJ, Nakano A, Pfeffer SR, Rabouille C, et al. 2009. Journeys through the Golgi–taking stock in a new era. J Cell Biol 187: 449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JY, Roth J, Zuber C 2003. Ultrastructural analysis of transitional endoplasmic reticulum and pre-Golgi intermediates: a highway for cars and trucks. Histochem Cell Biol 120: 455–463 [DOI] [PubMed] [Google Scholar]
- Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP 2008. Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J 27: 2043–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H, Rabouille C 2011. Signalling to and from the secretory pathway. J Cell Sci 124: 171–180 [DOI] [PubMed] [Google Scholar]
- Farquhar MG, Hauri HP 1997. Protein sorting and vesicular traffic in the Golgi apparatus. In The Golgi Apparatus (ed. Berger EG, Roth J.), pp. 63–129 Birkhauser Verlag, Basel [Google Scholar]
- Farquhar MG, Palade GE 1981. The Golgi apparatus (complex)-(1954-1981)-from artifact to center stage. J Cell Biol 91: 77s–103s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar MG, Palade GE 1998. The Golgi apparatus: 100 years of progress and controversy. Trends Cell Biol 8: 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar MG, Rinehart JF 1954. Cytologic alterations in the anterior pituitary gland following thyroidectomy: an electron microscope study. Endocrinology 65: 857–876 [DOI] [PubMed] [Google Scholar]
- Farquhar MG, Bergeron JJ, Palade GE 1974. Cytochemistry of Golgi fractions prepared from rat liver. J Cell Biol 60: 8–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein TN, Linstedt AD 2008. GRASP55 regulates Golgi ribbon formation. Mol Biol Cell 19: 2696–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsch H, Pypaert M, Maday S, Pelletier L, Mellman I 2003. The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J Cell Biol 163: 351–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke WW, Kartenbeck J, Krien S, VanderWoude WJ, Scheer U, Morre DJ 1972. Inter- and intracisternal elements of the Golgi apparatus. A system of membrane-to-membrane cross-links. Z Zellforsch Mikrosk Anat 132: 365–380 [DOI] [PubMed] [Google Scholar]
- Gaietta GM, Giepmans BN, Deerinck TJ, Smith WB, Ngan L, Llopis J, Adams SR, Tsien RY, Ellisman MH 2006. Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc Natl Acad Sci 103: 17777–17782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuze HJ, Morre DJ 1991. Trans-Golgi reticulum. J Electron Microsc Tech 17: 24–34 [DOI] [PubMed] [Google Scholar]
- Geuze HJ, Slot JW, Schwartz AL 1987. Membranes of sorting organelles display lateral heterogeneity in receptor distribution. J Cell Biol 104: 1715–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuze HJ, Slot JW, Strous GJ, Hasilik A, von Figura K 1985. Possible pathways for lysosomal enzyme delivery. J Cell Biol 101: 2253–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S 2003. Long coiled-coil proteins and membrane traffic. Biochimica et biophysica acta 1641: 71–85 [DOI] [PubMed] [Google Scholar]
- Glick BS 2000. Organization of the Golgi apparatus. Curr Opin Cell Biol 12: 450–456 [DOI] [PubMed] [Google Scholar]
- Glick BS, Nakano A 2009. Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev Biol 25: 113–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Luini A 2011. Models for Golgi traffic: a critical assessment. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golgi C 1898. Sur la structure des cellules nerveuses des ganglions spinaux. Arch Ital Biologie 30: 278–286 [Google Scholar]
- Goud B, Gleeson PA 2010. TGN golgins, Rabs and cytoskeleton: regulating the Golgi trafficking highways. Trends Cell Biol 20: 329–336 [DOI] [PubMed] [Google Scholar]
- Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, Nilsson T 2005. Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods 2: 857–862 [DOI] [PubMed] [Google Scholar]
- Griffiths G, Simons K 1986. The trans Golgi network: sorting at the exit site of the Golgi complex. Science 234: 438–443 [DOI] [PubMed] [Google Scholar]
- Griffiths G, Fuller SD, Back R, Hollinshead M, Pfeiffer S, Simons K 1989. The dynamic nature of the Golgi complex. J Cell Biol 108: 277–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G, Pepperkok R, Locker JK, Kreis TE 1995. Immunocytochemical localization of β-COP to the ER-Golgi boundary and the TGN. J Cell Sci 108 (Pt 8): 2839–2856 [DOI] [PubMed] [Google Scholar]
- Griffiths G, Pfeiffer S, Simons K, Matlin K 1985. Exit of newly synthesized membrane proteins from the trans cisterna of the Golgi complex to the plasma membrane. J Cell Biol 101: 949–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Tomishige N, Kawano M 2007. CERT and intracellular trafficking of ceramide. Biochim Biophys Acta 1771: 644–653 [DOI] [PubMed] [Google Scholar]
- Hand AR, Oliver C 1977. Relationship between the Golgi apparatus, GERL, and secretory granules in acinar cells of the rat exorbital lacrimal gland. J Cell Biol 74: 399–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand AR, Oliver C 1984. Effects of secretory stimulation on the Golgi apparatus and GERL of rat parotid acinar cells. J Histochem Cytochem 32: 403–412 [DOI] [PubMed] [Google Scholar]
- Hauri HP, Schweizer A 1992. The endoplasmic reticulum-Golgi intermediate compartment. Curr Opin Cell Biol 4: 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell SW 2009. Microscopy and its focal switch. Nat Methods 6: 24–32 [DOI] [PubMed] [Google Scholar]
- Hermo L, Green H, Clermont Y 1991. Golgi apparatus of epithelial principal cells of the epididymal initial segment of the rat: structure, relationship with endoplasmic reticulum, and role in the formation of secretory vesicles. Anat Rec 229: 159–176 [DOI] [PubMed] [Google Scholar]
- Heuser J, Kirchhausen T 1985. Deep-etch views of clathrin assemblies. J Ultrastruct Res 92: 1–27 [DOI] [PubMed] [Google Scholar]
- Hua Z, Graham TR 2009. The Golgi Apparatus. In Trafficking Inside Cells: Pathways, Mechanisms and Regulation (ed. Segev N.), pp. 42–61 Landes Bioscience and Springer Science+Business Media, Chicago [Google Scholar]
- Huttner WB, Tooze SA 1989. Biosynthetic protein transport in the secretory pathway. Curr Opin Cell Biol 1: 648–654 [DOI] [PubMed] [Google Scholar]
- Johannes L, Popoff V 2008. Tracing the retrograde route in protein trafficking. Cell 135: 1175–1187 [DOI] [PubMed] [Google Scholar]
- Kelly RB 1985. Pathways of protein secretion in eukaryotes. Science 230: 25–32 [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC, Heuser J 1986. Configuration of clathrin trimers: evidence from electron microscopy. J Ultrastruct Mol Struct Res 94: 199–208 [DOI] [PubMed] [Google Scholar]
- Klumperman J 2000. Transport between ER and Golgi. Curr Opin Cell Biol 12: 445–449 [DOI] [PubMed] [Google Scholar]
- Klumperman J, Kuliawat R, Griffith JM, Geuze HJ, Arvan P 1998a. Mannose 6-phosphate receptors are sorted from immature secretory granules via adaptor protein AP-1, clathrin, and syntaxin 6-positive vesicles. J Cell Biol 141: 359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J, Schweizer A, Clausen H, Tang BL, Hong W, Oorschot V, Hauri HP 1998b. The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J Cell Sci 111 (Pt 22): 3411–3425 [DOI] [PubMed] [Google Scholar]
- Kondylis V, Rabouille C 2009. The Golgi apparatus: lessons from Drosophila. FEBS letters 583: 3827–3838 [DOI] [PubMed] [Google Scholar]
- Kondylis V, van Nispen tot Pannerden HE, Herpers B, Friggi-Grelin F, Rabouille C 2007. The golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Developmental cell 12: 901–915 [DOI] [PubMed] [Google Scholar]
- Kornfeld S, Mellman I 1989. The biogenesis of lysosomes. Annu Rev Cell Biol 5: 483–525 [DOI] [PubMed] [Google Scholar]
- Koster AJ, Klumperman J 2003. Electron microscopy in cell biology: integrating structure and function. Nat Rev Mol Cell Biol 4 Suppl: SS6–10 [PubMed] [Google Scholar]
- Kuhnle J, Shillcock J, Mouritsen OG, Weiss M 2010. A modeling approach to the self-assembly of the Golgi apparatus. Biophys J 98: 2839–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuliawat R, Klumperman J, Ludwig T, Arvan P 1997. Differential sorting of lysosomal enzymes out of the regulated secretory pathway in pancreatic β-cells. J Cell Biol 137: 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky MS, Kremer JR, Furcinitti PS, McIntosh JR, Howell KE 1994. HVEM tomography of the trans-Golgi network: structural insights and identification of a lace-like vesicle coat. J Cell Biol 127: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA 1999. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J Cell Biol 144: 1135–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky MS, Wu CC, McIntosh S, McIntosh JR, Howell KE 2002. Structure of the Golgi and distribution of reporter molecules at 20 degrees C reveals the complexity of the exit compartments. Mol Biol Cell 13: 2810–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Linstedt AD 1999. Osmotically induced cell volume changes alter anterograde and retrograde transport, Golgi structure, and COPI dissociation. Mol Biol Cell 10: 1445–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur F, Gaynor EC, Hennecke S, Demolliere C, Duden R, Emr SD, Riezman H, Cosson P 1994. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 79: 1199–1207 [DOI] [PubMed] [Google Scholar]
- Levine T, Rabouille C 2005. Endoplasmic reticulum: one continuous network compartmentalized by extrinsic cues. Curr Opin Cell Biol 17: 362–368 [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR 1990. A human homologue of the yeast HDEL receptor. Nature 348: 162–163 [DOI] [PubMed] [Google Scholar]
- Lindsey JD, Ellisman MH 1985. The neuronal endomembrane system. II. The multiple forms of the Golgi apparatus cis element. J Neurosci 5: 3124–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Manley S 2009. Putting super-resolution fluorescence microscopy to work. Nat Methods 6: 21–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti LV, Torrisi MR, Pascale MC, Bonatti S 1992. Immunocytochemical analysis of the transfer of vesicular stomatitis virus G glycoprotein from the intermediate compartment to the Golgi complex. J Cell Biol 118: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra V, Campelo F 2010. PKD regulates membrane fission to generate TGN to cell surface transport carriers. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malsam J, Satoh A, Pelletier L, Warren G 2005. Golgin tethers define subpopulations of COPI vesicles. Science 307: 1095–1098 [DOI] [PubMed] [Google Scholar]
- Marra P, Salvatore L, Mironov A Jr, Di Campli A, Di Tullio G, Trucco A, Beznoussenko G, Mironov A, De Matteis MA 2007a. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol Biol Cell 18: 1595–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra P, Salvatore L, Mironov A Jr, Di Campli A, Di Tullio G, Trucco A, Beznoussenko G, Mironov A, De Matteis MA 2007b. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol Biol Cell 18: 1595–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh BJ 2005. Lessons from tomographic studies of the mammalian Golgi. Biochim Biophys Acta 1744: 273–292 [DOI] [PubMed] [Google Scholar]
- Marsh BJ 2007. Reconstructing mammalian membrane architecture by large area cellular tomography. Methods Cell Biol 79: 193–220 [DOI] [PubMed] [Google Scholar]
- Marsh BJ, Howell KE 2002. The mammalian Golgi–complex debates. Nat Rev Mol Cell Biol 3: 789–795 [DOI] [PubMed] [Google Scholar]
- Marsh BJ, Mastronarde DN, Buttle KF, Howell KE, McIntosh JR 2001a. Organellar relationships in the Golgi region of the pancreatic β cell line, HIT-T15, visualized by high resolution electron tomography. Proc Natl Acad Sci 98: 2399–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh BJ, Mastronarde DN, McIntosh JR, Howell KE 2001b. Structural evidence for multiple transport mechanisms through the Golgi in the pancreatic β-cell line, HIT-T15. Biochem Soc Trans 29: 461–467 [DOI] [PubMed] [Google Scholar]
- Marsh BJ, Volkmann N, McIntosh JR, Howell KE 2004. Direct continuities between cisternae at different levels of the Golgi complex in glucose-stimulated mouse islet β cells. Proc Natl Acad Sci 101: 5565–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Menarguez JA, Geuze HJ, Slot JW, Klumperman J 1999. Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell 98: 81–90 [DOI] [PubMed] [Google Scholar]
- Martinez-Menarguez JA, Prekeris R, Oorschot VM, Scheller R, Slot JW, Geuze HJ, Klumperman J 2001. Peri-Golgi vesicles contain retrograde but not anterograde proteins consistent with the cisternal progression model of intra-Golgi transport. J Cell Biol 155: 1213–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiblitzer-Ruppitsch C, Vetterlein M, Stangl H, Maier S, Neumuller J, Freissmuth M, Pavelka M, Ellinger A 2008. Electron microscopic visualization of fluorescent signals in cellular compartments and organelles by means of DAB-photoconversion. Histochem Cell Biol 130: 407–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Simons K 1992. The Golgi complex: in vitro veritas?. Cell 68: 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AA, Polishchuk RS, Beznoussenko GV 2008. Combined video fluorescence and 3D electron microscopy. Methods Cell Biol 88: 83–95 [DOI] [PubMed] [Google Scholar]
- Mironov AA, Mironov AA Jr, Beznoussenko GV, Trucco A, Lupetti P, Smith JD, Geerts WJ, Koster AJ, Burger KN, Martone ME, et al. 2003. ER-to-Golgi carriers arise through direct en bloc protrusion and multistage maturation of specialized ER exit domains. Dev Cell 5: 583–594 [DOI] [PubMed] [Google Scholar]
- Moelleken J, Malsam J, Betts MJ, Movafeghi A, Reckmann I, Meissner I, Hellwig A, Russell RB, Sollner T, Brugger B, et al. 2007. Differential localization of coatomer complex isoforms within the Golgi apparatus. Proc Natl Acad Sci 104: 4425–4430 [DOI] [PubMed] [Google Scholar]
- Mogelsvang S, Marsh BJ, Ladinsky MS, Howell KE 2004. Predicting function from structure: 3D structure studies of the mammalian Golgi complex. Traffic 5: 338–345 [DOI] [PubMed] [Google Scholar]
- Morin-Ganet MN, Rambourg A, Deitz SB, Franzusoff A, Kepes F 2000. Morphogenesis and dynamics of the yeast Golgi apparatus. Traffic 1: 56–68 [DOI] [PubMed] [Google Scholar]
- Munro S 2011. The Golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noske AB, Costin AJ, Morgan GP, Marsh BJ 2008. Expedited approaches to whole cell electron tomography and organelle mark-up in situ in high-pressure frozen pancreatic islets. J Struct Biol 161: 298–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikoff AB, Goldfischer S 1961. Nucleosidediphosphatase activity in the Golgi apparatus and its usefulness for cytological studies. Proc Natl Acad Sci 47: 802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikoff PM, Novikoff AB, Quintana N, Hauw JJ 1971. Golgi apparatus, GERL, and lysosomes of neurons in rat dorsal root ganglia, studied by thick section and thin section cytochemistry. J Cell Biol 50: 859–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Tomemori T, Nakatsu F, Okazaki Y, Aguilar RC, Foelsch H, Mellman I, Saito T, Shirasawa T, Bonifacino JS 1999. Mu1B, a novel adaptor medium chain expressed in polarized epithelial cells. FEBS Lett 449: 215–220 [DOI] [PubMed] [Google Scholar]
- Oprins A, Duden R, Kreis TE, Geuze HJ, Slot JW 1993. β-COP localizes mainly to the cis-Golgi side in exocrine pancreas. J Cell Biol 121: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Perrelet A, Rothman JE 1998. Vesicles on strings: morphological evidence for processive transport within the Golgi stack. Proc Natl Acad Sci 95: 2279–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Halban P, Amherdt M, Ravazzola M, Vassalli JD, Perrelet A 1984. A clathrin-coated, Golgi-related compartment of the insulin secreting cell accumulates proinsulin in the presence of monensin. Cell 39: 39–47 [DOI] [PubMed] [Google Scholar]
- Orci L, Stamnes M, Ravazzola M, Amherdt M, Perrelet A, Sollner TH, Rothman JE 1997. Bidirectional transport by distinct populations of COPI-coated vesicles. Cell 90: 335–349 [DOI] [PubMed] [Google Scholar]
- Palade G 1975. Intracellular aspects of the process of protein synthesis. Science 189: 867. [DOI] [PubMed] [Google Scholar]
- Papanikou E, Glick BS 2009. The yeast Golgi apparatus: insights and mysteries. FEBS Lett 583: 3746–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Hirschberg K, Polishchuk RS, Gerlich D, Phair RD, Lippincott-Schwartz J 2008. Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system. Cell 133: 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka M, Ellinger A 1983. The trans Golgi face in rat small intestinal absorptive cells. Eur J Cell Biol 29: 253–261 [PubMed] [Google Scholar]
- Pearse BM, Robinson MS 1990. Clathrin, adaptors, and sorting. Annu Rev Cell Biol 6: 151–171 [DOI] [PubMed] [Google Scholar]
- Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J 2004. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J Cell Biol 164: 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR 1991. Recycling of proteins between the endoplasmic reticulum and Golgi complex. Curr Opin Cell Biol 3: 585–591 [DOI] [PubMed] [Google Scholar]
- Pelletier L, Stern CA, Pypaert M, Sheff D, Ngo HM, Roper N, He CY, Hu K, Toomre D, Coppens I, et al. 2002. Golgi biogenesis in Toxoplasma gondii. Nature 418: 548–552 [DOI] [PubMed] [Google Scholar]
- Peretti D, Dahan N, Shimoni E, Hirschberg K, Lev S 2008. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell 19: 3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR 2011. Entry at the trans-face of the Golgi. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polishchuk RS, Mironov AA 2004. Structural aspects of Golgi function. Cell Mol Life Sci 61: 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott AR, Farmaki T, Thomson C, James J, Paccaud JP, Tang BL, Hong W, Quinn M, Ponnambalam S, Lucocq J 2001. Evidence for prebudding arrest of ER export in animal cell mitosis and its role in generating Golgi partitioning intermediates. Traffic (Copenhagen, Denmark) 2: 321–335 [DOI] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J 1997. ER-to-Golgi transport visualized in living cells. Nature 389: 81–85 [DOI] [PubMed] [Google Scholar]
- Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D 1992. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol Biol Cell 3: 789–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prydz K, Dick G, Tveit H 2008. How many ways through the Golgi maze?. Traffic 9: 299–304 [DOI] [PubMed] [Google Scholar]
- Pulvirenti T, Giannotta M, Capestrano M, Capitani M, Pisanu A, Polishchuk RS, San Pietro E, Beznoussenko GV, Mironov AA, Turacchio G, et al. 2008. A traffic-activated Golgi-based signalling circuit coordinates the secretory pathway. Nat Cell Biol 10: 912–922 [DOI] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD 2006. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol 8: 238–248 [DOI] [PubMed] [Google Scholar]
- Rabouille C, Klumperman J 2005. Opinion: The maturing role of COPI vesicles in intra-Golgi transport. Nat Rev Mol Cell Biol 6: 812–817 [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kondylis V 2007. Golgi ribbon unlinking: an organelle-based G2/M checkpoint. Cell Cycle (Georgetown, Tex) 6: 2723–2729 [DOI] [PubMed] [Google Scholar]
- Rabouille C, Misteli T, Watson R, Warren G 1995. Reassembly of Golgi stacks from mitotic Golgi fragments in a cell-free system. J Cell Biol 129: 605–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y 1986. Tridimensional structure of the Golgi apparatus in type A ganglion cells of the rat. Am J Anat 176: 393–409 [DOI] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y 1990. Three-dimensional electron microscopy: structure of the Golgi apparatus. Eur J Cell Biol 51: 189–200 [PubMed] [Google Scholar]
- Rambourg A, Clermont Y 1997. Three-dimensional structure of the Golgi apparatus in mammalian cells. In The Golgi Apparatus (ed. Berger EG, Roth J.), pp. 37–61 Birkhauser Verlag, Basel [Google Scholar]
- Rambourg A, Clermont Y, Hermo L 1979. Three-dimensional architecture of the golgi apparatus in Sertoli cells of the rat. Am J Anat 154: 455–476 [DOI] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, Marraud A 1974. Three-dimensional structure of the osmium-impregnated Golgi apparatus as seen in the high voltage electron microscope. Am J Anat 140: 27–45 [DOI] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, Marraud A 1974. Three-dimensional structure of the osmium-impregnated Golgi apparatus as seen in the high voltage electron micrscope. Am J Anat 140: 27–46 [DOI] [PubMed] [Google Scholar]
- Rambourg A, Segretain D, Clermont Y 1984. Tridimensional architecture of the Golgi apparatus in the atrial muscle cell of the rat. Am J Anat 170: 163–179 [DOI] [PubMed] [Google Scholar]
- Ramirez IB, Lowe M 2009. Golgins and GRASPs: Holding the Golgi together. Seminars in Cell & Developmental Biology 7: 770–779 [DOI] [PubMed] [Google Scholar]
- Ramirez IB, Lowe M 2009. Golgins and GRASPs: holding the Golgi together. Semin Cell Dev Biol 20: 770–779 [DOI] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol 185: 1209–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE 1994. Mechanisms of intracellular protein transport. Nature 372: 55–63 [DOI] [PubMed] [Google Scholar]
- San Pietro E, Capestrano M, Polishchuk EV, DiPentima A, Trucco A, Zizza P, Mariggio S, Pulvirenti T, Sallese M, Tete S, et al. 2009. Group IV phospholipase A(2)α controls the formation of inter-cisternal continuities involved in intra-Golgi transport. PLoS Biol 7: e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K, Spilsberg B, Lauvrak SU, Torgersen ML, Iversen TG, van Deurs B 2004. Pathways followed by protein toxins into cells. Int J Med Microbiol 293: 483–490 [DOI] [PubMed] [Google Scholar]
- Scales SJ, Pepperkok R, Kreis TE 1997. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell 90: 1137–1148 [DOI] [PubMed] [Google Scholar]
- Sesso A, de Faria FP, Iwamura ES, Correa H 1994. A three-dimensional reconstruction study of the rough ER-Golgi interface in serial thin sections of the pancreatic acinar cell of the rat. J Cell Sci 107: 517–528 [PubMed] [Google Scholar]
- Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA 1999. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. The EMBO journal 18: 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinka R, Gillingham AK, Kondylis V, Munro S 2008. Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J Cell Biol 183: 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostrand FS, Hanzon V 1954. Ultrastructure of Golgi apparatus of exocrine cells of mouse pancreas. Exp Cell Res 7: 415–429 [DOI] [PubMed] [Google Scholar]
- Slot JW, Garruti G, Martin S, Oorschot V, Posthuma G, Kraegen EW, Laybutt R, Thibault G, James DE 1997. Glucose transporter (GLUT-4) is targeted to secretory granules in rat atrial cardiomyocytes. J Cell Biol 137: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarewicz P, Nilsson T, Hui N, Watson R, Warren G 1994. Isolation of a matrix that binds medial Golgi enzymes. J Cell Biol 124: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoorvogel W, Geuze HJ, Griffith JM, Strous GJ 1988. The pathways of endocytosed transferrin and secretory protein are connected in the trans-Golgi reticulum. J Cell Biol 106: 1821–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C, Hsu P, Mallabiabarrena A, Malhotra V 2002. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell 109: 359–369 [DOI] [PubMed] [Google Scholar]
- Thorne-Tjomsland G, Clermont Y, Tang XM 1991. Glucose-6-phosphatase activity of endoplasmic reticulum and Golgi apparatus in spermatocytes and spermatids of the rat: an electron microscopic cytochemical study. Biol Cell 71: 33–41 [DOI] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S 1985. Microtubules and the organization of the Golgi complex. Exp Cell Res 159: 1–16 [DOI] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S 1999. Role of microtubules in the organization of the Golgi complex. Exp Cell Res 246: 263–279 [DOI] [PubMed] [Google Scholar]
- Traub LM, Kornfeld S 1997. The trans-Golgi network: a late secretory sorting station. Curr Opin Cell Biol 9: 527–533 [DOI] [PubMed] [Google Scholar]
- Trucco A, Polishchuk RS, Martella O, Di Pentima A, Fusella A, Di Giandomenico D, San Pietro E, Beznoussenko GV, Polishchuk EV, Baldassarre M, et al. 2004. Secretory traffic triggers the formation of tubular continuities across Golgi sub-compartments. Nat Cell Biol 6: 1071–1081 [DOI] [PubMed] [Google Scholar]
- van Meel E, Klumperman J 2008. Imaging and imagination: understanding the endo-lysosomal system. Histochem Cell Biol 129: 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijnsoever C, Oorschot V, Klumperman J 2008. Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nat Methods 5: 973–980 [DOI] [PubMed] [Google Scholar]
- Vetterlein M, Ellinger A, Neumuller J, Pavelka M 2002. Golgi apparatus and TGN during endocytosis. Histochem Cell Biol 117: 143–150 [DOI] [PubMed] [Google Scholar]
- Vinke FP, Grieve AG, Rabouille C 2011. The multiple facets of the Golgi reassembly stacking proteins. Biochem J 433: 423–433 [DOI] [PubMed] [Google Scholar]
- Vivero-Salmeron G, Ballesta J, Martinez-Menarguez JA 2008. Heterotypic tubular connections at the endoplasmic reticulum-Golgi complex interface. Histochem Cell Biol 130: 709–717 [DOI] [PubMed] [Google Scholar]
- Wandinger-Ness A, Bennett MK, Antony C, Simons K 1990. Distinct transport vesicles mediate the delivery of plasma membrane proteins to the apical and basolateral domains of MDCK cells. J Cell Biol 111: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidman P, Roth R, Heuser J 1993. Golgi membrane dynamics imaged by freeze-etch electron microscopy: views of different membrane coatings involved in tubulation versus vesiculation. Cell 75: 123–133 [PubMed] [Google Scholar]
- Yadav S, Puri S, Linstedt AD 2009. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell 20: 1728–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Yoshioka K, Barr FA, Lowe M, Nakayama K, Ohkuma S, Nakamura N 2005. Convergence of cell cycle regulation and growth factor signals on GRASP65. J Biol Chem 280: 23048–23056 [DOI] [PubMed] [Google Scholar]
- Zeuschner D, Geerts WJ, van Donselaar E, Humbel BM, Slot JW, Koster AJ, Klumperman J 2006. Immuno-electron tomography of ER exit sites reveals the existence of free COPII-coated transport carriers. Nat Cell Biol 4: 377–383 [DOI] [PubMed] [Google Scholar]