Abstract
Normal mammalian development requires a maternal and paternal contribution, which is attributed to imprinted genes, or genes that are expressed from a single parental allele. Approximately 100 imprinted genes have been reported in mammals thus far. Imprinted genes are controlled by cis-acting regulatory elements, termed imprinting control regions (ICRs), which have parental-specific epigenetic modifications, including DNA methylation. ICRs are methylated by de novo DNA methyltransferases during germline development; these parental-specific modifications must be maintained following fertilization when the genome is extensively reprogrammed. Many imprinted genes reside in ∼1-megabase clusters, with two major mechanisms of imprinting regulation currently recognized, CTCF-dependent insulators and long noncoding RNAs. Unclustered imprinted genes are generally regulated by germline-derived differential promoter methylation. Here, we describe the identification and functions of imprinted genes, cis-acting control sequences, trans-acting factors, and imprinting mechanisms in clusters. Finally, we define questions that require more extensive research.
Long noncoding RNAs and CTCF-dependent insulators regulate imprinted gene clusters. Differential promoter methylation regulates unclustered imprinted genes.
In mammals, a small number of genes are marked with their parental origin with the result that only a single parental allele is expressed. These genes, which are termed “imprinted,” are dependent on the epigenetic machinery for their initial designation of parental identity as well as establishment and maintenance of their parent-of-origin-specific gene expression (Fig. 1). Although organisms other than mammals harbor imprinted genes, such as Arabidopsis (Kinoshita et al. 2008), we will confine our discussion to what is known about the identification and regulation of imprinted genes in mammals. Moreover, it is important to note that chromosomal imprinting in mammals was first described from studies on the X chromosome, where paternal-specific inactivation of one of the X chromosomes in all cells of female marsupials and in the extraembryonic tissues of the mouse occur (Cooper et al. 1971).
Figure 1.
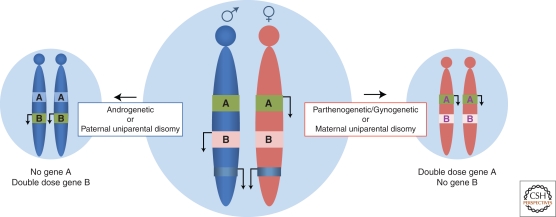
Imprinted genes are expressed in a parental-origin-specific manner. In the center, a cell is depicted with a chromosome inherited maternally (red) or paternally (blue), and two imprinted genes. Gene A is transcribed from the maternal allele (green box and arrow indicating transcription) and repressed on the paternal allele (light blue box). In contrast, gene B is transcribed from the paternal allele (green box and arrow) and repressed on the maternal allele (light pink box). Uniparental embryos and embryos that are uniparental for a given chromosome have been used to show that imprinted genes are critical for development as well as to identify imprinted genes (see text).
FIRST SUGGESTIONS OF IMPRINTING IN MAMMALS
The elegant pronuclear transplantation experiments performed by the Solter and Surani laboratories in the 1980s were among the first to suggest that the mammalian (specifically the mouse) genome possessed imprinted genes (McGrath and Solter 1983, 1984; Surani and Barton 1983; Surani et al. 1984). These experiments showed that mammalian development required both maternal and paternal contributions to proceed normally. Maternal uniparental embryos (gynogenotes or parthogenotes) developed into tissues predominantly of embryonic origin, with a failure of the extraembryonic lineages, whereas paternal uniparental embryos (androgenotes) developed into conceptuses derived of extraembryonic lineages. The investigators reasoned that absence or overexpression of imprinted genes exclusively expressed from either the maternal or paternal genome caused the developmental failure. Experiments by Kono and colleagues have largely shown these ideas to be correct by generating viable mice that are exclusively maternal in origin (Kono et al. 2004; Kawahara et al. 2007). Briefly, these investigators engineered offspring by combining nuclei from nongrowing and fully-grown oocytes that also had mutations at two different imprinted loci. The resulting bimaternal offspring, which were produced at a slightly lower frequency than similarly manipulated wild-type embryos, had normalized the imprinted gene dosage to what is observed in wild-type offspring, suggesting that imprinted gene expression is the main barrier to parthenogenetic development in mammals (Kawahara et al. 2007).
Genetic experiments by Cattanach, Searle, Beechey, and colleagues pointed to specific regions of the genome that harbored imprinted genes (Searle and Beechey 1978, 1990; Cattanach 1982, 1986; Cattanach and Kirk 1985). Offspring with uniparental disomies for specific chromosomal regions were generated by mating mice that were heterozygous for Robertsonian or balanced translocations. That is, rather than having an entire uniparental genome, these experimental manipulations produced offspring that had uniparental origin of only part of the genome—usually both homologs of an individual, or fraction of a, chromosome. If defects resulted, they surmised a requirement for both parental chromosomal regions. This allowed the dissection of the genome approximately one chromosome at a time and the cytogenetic mapping of regions that were imprinted. In some cases, uniparental disomies caused obvious parent-of-origin-specific defects such as growth abnormalities or death, demonstrating a major role of imprinted genes for normal development. Behavioral abnormalities were also detected (Cattanach and Kirk 1985). It is important to note, however, that imprinted genes whose altered dosage exert more subtle or tissue-specific effects would not necessarily be identified in these tests. Nonetheless, more that 90% of the imprinted genes that have been identified in the mouse to date, map to the regions originally identified through these genetic studies.
At approximately the same time as the mouse genetic experiments were performed, genetic disorders in humans were suggested to arise from parental-specific gene expression. One early set of observations came from patients with Prader–Willi syndrome (PWS). PWS, which is characterized by hypotonia in infancy and subsequent hyperphagia, hypogonadism, etc. (MIM176270), was shown to be associated with chromosomal deletions that were paternally derived (Knoll et al. 1989; Nicholls and Knepper 2001). Subsequent experiments identified a number of paternally expressed genes that are likely involved in the diverse phenotypes of PWS, including SNPRN, NECDIN, and HBII-85 snoRNAs (Horsthemke and Wagstaff 2008).
IDENTIFICATION OF IMPRINTED GENES
The first endogenous imprinted genes were identified less than a decade after the original nuclear transplantation and genetic experiments. Diverse strategies were employed to identify these imprinted loci. Some relied on molecularly characterizing regions that likely contained imprinted genes (Barlow et al. 1991; Ferguson-Smith et al. 1991), whereas others incorporated genome-wide studies (Kaneko-Ishino et al. 1995; Wang et al. 2008), and still others were identified as a consequence of gene targeting experiments (DeChiara et al. 1991). Igf2r (insulin-like growth factor type 2 receptor gene) was the first of three imprinted genes that were reported in 1991. The identification of Igf2r was based on a targeted strategy to determine which gene was responsible for the lethality of Hairpin-tail deletion mice that inherited a large deletion of chromosome 17 specifically from their mothers (Johnson 1974; Barlow et al. 1991). Here, positional cloning discovered genes in the deleted region and mice inheriting the deletion from one parent were used to identify the gene(s) showing expression exclusively from the maternal allele. A different strategy was employed to show imprinting of the mouse H19 gene, which encodes a noncoding RNA (ncRNA). In this case, maternal-specific expression of H19 was revealed using single nucleotide polymorphisms (SNPs) that reside in RNA (Bartolomei et al. 1991). By generating F1 hybrid mice between divergent species of Mus, the mouse strain, and therefore the parental allele, from which the RNA product is derived can be tracked. In fact, now that the sequence of multiple mouse strains is known and high densities of strain-specific SNPs are available, whole transcriptome analysis is being used to assess which RNAs are monoallelically expressed (Babak et al. 2008; Wang et al. 2008). With time, this will allow the identification of all imprinted genes. Finally, a few imprinted genes have been identified that result from gene targeting experiments initially designed to test the function of the gene product. In the case of Igf2 (Insulin-like growth factor 2), gene targeting showed that mice inheriting the deletion from the fathers were phenotypically identical to homozygous null mice for the Igf2 gene, whereas mice inheriting the deletion from their mothers were phenotypically wild-type (DeChiara et al. 1991). The conclusion from these breeding studies was that the Igf2 gene is imprinted, with the expressed copy being paternal in origin. In a different approach, using uniparental disomy conceptuses derived from the genetic experiments discussed above and analysis of candidate genes influencing growth, expression of Igf2 was shown to be absent in embryos with maternal uniparental disomy of distal chromosome 7 indicating expression from the paternally inherited chromosome (Ferguson-Smith et al. 1991).
To date, approximately 100 imprinted genes have been described in mammals (for a complete list, see http://www.har.mrc.ac.uk/research/genomic_imprinting/ or http://igc.otago.ac.nz/home.html). Although singletons have been identified, many imprinted genes reside in 1 Mb clusters throughout the genome. These clusters typically contain at least one ncRNA and both maternally and paternally expressed imprinted genes. The imprinting of the multiple genes in the cluster is usually under the control of a discrete DNA element, termed an imprinting control region (ICR; see below for more details). ICRs show parent-of-origin-specific epigenetic modifications that are set up in the germline, such as DNA methylation and posttranslational histone modifications. Where it has been tested, deletion of the ICR results in loss of imprinting of multiple genes in the cluster, emphasizing the importance of this element and its regulatory influence over a multigenic region (Wutz et al. 1997; Thorvaldsen et al. 1998; Yang et al. 1998; Fitzpatrick et al. 2002; Lin et al. 2003; Williamson et al. 2006).
FUNCTIONS OF IMPRINTED GENES
Given their monoallelic expression status and the complexities of their regulation, it is perhaps not surprising that many imprinted genes appear to be dosage sensitive with functional consequences associated with changes in their expression levels. Over the years, phenotypic analyses of humans and mice with altered dosages of individual or multiple imprinted genes have provided insights into their roles. Recurrent themes have emerged indicating that imprinted genes are important for prenatal growth control, the development of particular lineages, for normal brain function and in postnatal energy homeostasis. Some understanding of imprinted gene function in human has come from genotype-phenotype studies in patients with imprinting disorders (Table 1). These include the behavioral and neurodevelopmental disorders Prader–Willi and Angelman syndromes (Horsthemke and Buiting 2006), the imprinted growth disorders Beckwith–Wiedemann and Silver–Russell syndromes (Weksberg et al. 2005; Abu-Amero et al. 2008), the maternal and paternal uniparental disomy 14 syndromes (Ogata et al. 2008), and transient neonatal diabetes (Temple 2007). In general, the features described in patients are consistent with those observed in the corresponding mouse mutants.
Table 1.
Human imprinted disorders
| Syndrome | Clinical features | Etiology | Mouse chromosome |
|---|---|---|---|
| Angelman syndrome (AS) | Mental retardation, speech impairment, ataxia, seizure, microcephaly | 15q11.2-q13 deletion (70%) PatUPD15 (7%), UBE3A mutation (11%), methylation defects (3%), epimutation | 7C |
| Prader–Willi syndrome (PWS) | Neonatal hypotonia, childhood obesity, cognitive impairment, behavioral characteristics, hypogonadism | De novo paternal deletion in 15q11-q13 (70%), MatUPD15 (29%), imprinting defects (1%) | 7C |
| Beckwith–Wiedemann syndrome (BWS) | Pre/postnatal overgrowth, neonatal hypoglycemia, exompholos, macroglossia, hemihypertrophy, increased embryonal tumors | Epimutation of IGF2/H19 DMR1, epimutation of KCNQ1/CDKN1C DMR2 both on 11p15, hypomethylation of DMR2 (50%), hypermethylation DMR1 (2%–7%), PatUPD11, CDKN1C mutation | 7F5 |
| Silver–Russell syndrome (SRS) | Intrauterine/postnatal growth retardation, variable features (inc. 5th finger clinodactyl, learning disabilities) | Paternal DMR1 hypomethylation at 11p15 (>50%), MatUPD7 (5%) Matdup11p15, unknown (30%) | 7F5 |
| Maternal UPD14 (and UPD14 mat-like) syndrome | Low birth weight, short stature, characteristic facies, premature puberty, hypotonia | MatUPD14, paternal microdeletions at 14q32.2, hypomethylated DMRs at DLK1/GTL2 | 12F1 |
| Paternal UPD14 (and UPD14 pat-like) syndrome | Bell-shaped thoracic cage, mental retardation, placentomegaly, polyhydramnios | PatUPD 14, maternal microdeletions at 14q32.2, hypermethylation at DMRs at DLK1/GTL2 | 12F1 |
| Pseudo-hypoparathyroidism 1b | Resistance to parathyroid hormone, hypocalcaemia, hyperphophatemia | Microdeletion upstream of GNAS at 20q, maternal hypomethylation, PatUPD20 | 2H4 |
| Transient neonatal diabetes mellitus | Growth retardation, hyperglycemia with low/undetectable insulin resolved by 6 months old, 40% Type2 diabetes later in life | Paternal UPD6, paternal duplication 6q22-q23, maternal hypomethylation at ZAC1/PLAGL1 DMR | 10A2 |
In mouse, the dosage of imprinted genes has been manipulated by targeted mutagenesis of the active parental allele, through transgenic overexpression studies and by mutation targeted at the ICR resulting in either activation of the repressed allele or inappropriate repression of active alleles. Phenotypic analysis of mouse mutants combined with descriptive studies localizing imprinted gene products in developing and adult cell types in vivo has shown many imprinted genes to be expressed in a range of developing organ systems where defects are manifest in the mutants (for example, see da Rocha et al. 2009). Imprinted genes, in general, are expressed widely and highly during prenatal stages, however, are predominantly down-regulated after birth. The placenta and the brain are sites of expression of many imprinted transcripts consistent with the growth and neurodevelopmental effects seen in human imprinting disorders (Coan et al. 2005; Wilkinson et al. 2007). In particular defective placental development and physiology is a characteristic of perturbed imprinting. For example, absence of expression of a placenta-specific isoform of Igf2 causes impaired nutrient transport to the growing fetus (Sibley et al. 2004). Ascl2 (achaete-scute homolog complex 2) expressed from the maternally inherited chromosome is required for the normal differentiation of spongiotrophoblast cells early within the developing murine placenta (Guillemot et al. 1995). Interestingly, Peg10 and Rtl1 are two imprinted genes required for normal placental function that have evolved from retrotransposons (Ono et al. 2006; Sekita et al. 2008). Although imprinting in the placenta provides a critical role in the control of resources at the interface between mother and fetus, perturbed imprinting in the embryo can influence its growth independent of an influence of the placenta. For example, a double dose of Dlk1 (Delta-like homolog 1) equivalent to expression from both parental chromosomes instead of just the paternally inherited one, results in a growth enhanced fetus even when the placenta expresses normal levels of the gene (da Rocha et al. 2009). Despite a clear role in prenatal growth, the experimental challenges of functionally separating imprinted genes expressed in embryonic and extraembryonic compartments have made it hard to determine the relative contributions of placental and embryonic expression of imprinted genes to the in utero acquisition of resources and growth control of the fetus.
In the brain, imprinted genes have been implicated in processes modulating metabolic axes, behavior, learning, and maternal care. For example, it was recently shown that the imprinted gene, Gsα, expressed from the maternally inherited chromosome in the hypothalamus, controls melanocortin-mediated energy expenditure (Chen et al. 2009). Gsα is a G protein regulating receptor-mediated cAMP production. Mice with brain-specific deletion of the maternally inherited copy of Gsα, have glucose intolerance, insulin resistance, and obesity. In other examples, Peg1 and Peg3 are two paternally expressed imprinted genes strongly transcribed in the brain and which, when mutated, result in reduced maternal care by mutant mothers (Lefebvre et al. 1998; Li et al. 1999). These findings implicate imprinted genes in the brain in the postnatal control of nutritional resources either directly through the central control of energy homeostasis or indirectly through maternal–offspring interactions.
PWS and Angelman syndrome (AS) are two phenotypically distinct disorders mapping to the same imprinted domain on human chromosome 15q11-q13, the location of a cluster of genes expressed and imprinted in the brain (Horsthemke and Wagstaff 2008). Patients with PWS show hypotonia and early neonatal failure to thrive. Later they have mild to moderate mental retardation, behavioral problems, and hyperphagia in early childhood leading to obesity. Paternally expressed transcripts from the PWS/AS locus including the protein-coding SNRPN gene and its associated noncoding C/D snoRNAs (small nucleolar RNAs), are not expressed in PWS and are implicated in the syndrome though their precise contribution to the etiology of the disorder is not known. Angelman Syndrome, in contrast, is associated with absence of the UBE3A transcript, which is expressed from the maternally inherited chromosome and located at the 3′ end of the imprinted cluster on 15q11-q13. Individuals with AS have ataxia, severe mental retardation, seizures, and absence of speech. Although much has been learnt about the mechanisms of imprinting through the study of the SNRPN/UBE3A imprinted cluster of genes, little is known about how the absence or altered dosage of these genes influences neuronal function and causes the phenotypes that characterize the syndromes.
Despite the emphasis on placenta and brain functions for imprinted genes and an apparently related function in the control of nutrient acquisition, it is clear that imprinted genes also influence the development of other organ systems. For example, the imprinted cyclin-dependent kinase inhibitor, Cdkn1c is strongly expressed in several somatic tissues during development such as muscle, lung, kidney, and the eye. Normal expression of Cdkn1c is associated with cessation of the cell cycle and several studies have shown defects in organ size when Cdkn1c levels are perturbed (Jia et al. 2007b). The paternally expressed Dlk1 gene encodes a protein that is related to ligands functioning in Notch signaling, one of the key signaling pathways regulating the development of multiple lineages. Dlk1 mutants have growth retardation, skeletal abnormalities, adiposity defects and abnormalities of hematopoietic lineages (Moon et al. 2002; Raghunandan et al. 2008).
Findings that defective imprinting of multiple genes results in related phenotypic anomalies mostly involving pre- and postnatal growth control, suggests that perhaps imprinted genes might function in common pathways. Some evidence for this exists. For example, genes whose transcription was perturbed in mice mutant for the paternally expressed transcription factor Plagl1/Zac1 gene, included a large number of other imprinted genes. The work suggested that Plagl1/Zac1 might be a trans-acting regulator of a network of imprinted genes involved in prenatal growth control including Igf2, H19, Cdkn1c, and Dlk1 (Varrault et al. 2006). More detailed analysis of the in vivo targets of Plag1/Zac1 and the functional interactions that might exist between imprinted genes in the proposed network will determine whether this hypothesis is true.
PROPERTIES OF THE IMPRINTING MECHANISM
How does the transcription machinery of the nucleus distinguish between maternally and paternally inherited chromosome homologs and express only one of the two alleles of an imprinted gene—and furthermore, always the same one? How are the two homologs “marked” such that they know their parental origin? The process regulating genomic imprinting has four important properties: (1) The “mark” must be able to influence transcription; (2) it must be heritable in somatic lineages such that a memory of parental origin is faithfully propagated into daughter cells during cell division; (3) the “mark” is likely to be placed on the paternally and maternally inherited chromosomes at a time when they are not in the same nucleus (i.e., during gametogenesis or perhaps immediately after fertilization); (4) there must be a mechanism of erasure of the “mark” in order that paternally inherited chromosomes in the female germline can establish a new “mark” indicative of their maternal origin and vice versa; that maternally inherited chromosomes contributing to spermatogenesis in the developing male would lose their maternal identity and become “marked” as paternal in origin.
DNA methylation is the epigenetic modification that fulfills these criteria. In mammalian cells, it is well established that 5-methyl-cytosine at CpG dinucleotides can influence transcription. CpG dinucleotides are not evenly distributed throughout the genome but are often found as CG-rich sequences called CpG-islands. CpG-islands, which are usually unmethylated, are predominantly associated with the promoters of housekeeping genes (i.e., genes that are ubiquitously expressed and seldom repressed [Bird 2002]). CpG-islands are also found at the promoters of imprinted genes and most, though not all, CG-island promoters of imprinted genes are differentially methylated, where the repressed allele is methylated and the active allele unmethylated. These differentially methylated regions are called DMRs.
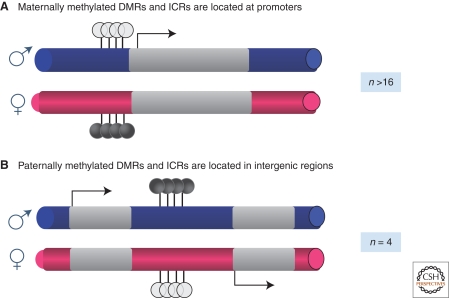
DNA methylation fulfills the second criterion of somatic heritability because DNA methyltransferase 1 (DNMT1) has a maintenance mode of action that recognizes newly replicated DNA comprising an old methylated strand and a new unmethylated strand (hemi-methylated DNA) and places methylation on the newly replicated CpG; hence the critically important heritability of DNA methylation is maintained (Goll and Bestor 2005). In terms of establishment, it is now well-known that DNA is methylated in different places on the maternally and paternally inherited chromosomes during gametogenesis by the de novo methyltransferase DNMT3A (see below). The establishment of these germline DMRs is essential for imprinting after fertilization and initiates the heritable memory that forms the identity “mark” of the two parental chromosomes. Interestingly, in the female germline, methylation marks always act at the promoters of imprinted genes resulting in their heritable repression from the maternally inherited chromosome. In contrast, methylation in the paternal germline is not placed at promoters but rather is deposited within intergenic regions (Fig. 2). Targeted deletion studies of maternally methylated promoter germline DMRs and paternally methylated intergenic germline DMRs in mouse have indicated that these are the imprinting control regions that can regulate multiple imprinted genes at some distance from the ICR (Wutz et al. 1997; Thorvaldsen et al. 1998; Yang et al. 1998; Fitzpatrick et al. 2002; Lin et al. 2003; Williamson et al. 2006). ICRs can therefore be long-range cis-acting control sequences whose function is modulated by their epigenetic state. Finally, DNA methylation imprints are erased in the male and female germlines during a process of germ cell-specific genome-wide reprogramming that occurs in the developing embryo as the cells destined to become germ cells are finishing their migration along the genital ridges toward the developing gonads. This is discussed in more detail below.
Figure 2.
DNA methylation is essential for establishment and maintenance of imprinting. Shown are the two types of DNA methylation present at imprinted loci. (A) Greater than 16 imprinted loci are associated with maternal-specific methylation. For these loci, DNA methylation (filled lollipops) is associated with the promoter of a repressed gene, whereas hypomethylation (gray lollipops) is associated with the promoter of the expressed gene (arrow). (B) Four imprinted loci are associated with DNA methylation in intergenic regions. In one case (H19 and Igf2), the DNA methylation regulates a CTCF-dependent insulator element (see text).
How do paternally methylated ICRs located in an intergenic regions influence the monoallelic activity and repression of multiple imprinted genes? The best-studied example is that of the Igf2-H19 locus. The paternally expressed Igf2 gene is located 90 kb away from the maternally expressed H19 gene (Bartolomei et al. 1991; DeChiara et al. 1991). The two genes, in general, share common sites of expression and share enhancer sequences located downstream of H19. The ICR resides 2–4 kb upstream from the transcriptional start site of H19 (Tremblay et al. 1997). This ICR contains multiple binding sites for the zinc-finger insulator protein, CTCF, which binds exclusively to the unmethylated maternal chromosome (Bell and Felsenfeld 2000; Hark et al. 2000). On the maternal chromosome, the presence of CTCF blocks the enhancers from interacting with Igf2 promoters rendering the gene inactive. The enhancers on the maternal chromosome drive activity from H19 instead. On the paternally inherited chromosome where the ICR is methylated, CTCF cannot bind, and the unblocked enhancers are able to drive activity from the Igf2 promoters. Such methylation-sensitive parental-chromosome-specific promoter–enhancer interaction has been validated by chromatin conformation studies where the expected loops of enhancer–promoter interactions have been identified, modulated by differential DNA methylation and differential CTCF binding on the two parental chromosomes (Murrell et al. 2004; Kurukuti et al. 2006; Engel et al. 2008). However, one should not assume that this CTCF-mediated control of ICRs, which regulates imprinting at the Igf2-H19 locus, acts at all intergenic ICRs. Indeed, there is considerable evidence that this mechanism is not operating at the paternally methylated ICR regulating the Dlk1-Dio3 imprinted domain (da Rocha et al. 2008).
Maternally methylated ICRs, which are located at gene promoters, exert a different effect to mediate imprinting over distances compared to that described for H19 and Igf2. The most extensively studied example of this mode of imprinting control comes from the analysis of the Igf2r cluster of imprinted genes (Koerner et al. 2009). Here, the ICR is located at a CpG island within an intron of the Igf2r gene (Stöger et al. 1993). The Igf2r gene is expressed from the maternally inherited chromosome (Barlow et al. 1991), which has intronic methylation. In contrast, on the paternally inherited chromosome, the CpG-island in the intron is unmethylated and can act as a promoter for a large nonprotein-coding RNA (ncRNA), which is designated Airn, and is transcribed in an antisense direction to Igf2r (Wutz et al. 1997; Lyle et al. 2000). This antisense transcription crosses the Igf2r promoter and is required for its repression on the paternal chromosome (Sleutels et al. 2002). Interestingly, two genes located downstream from Igf2r are also expressed on the maternal chromosome and repressed on the paternally inherited one, even though the direction of antisense transcription is away from those two genes. One hypothesis for which there is some supporting evidence, proposes that the Airn ncRNA interacts in cis with the promoter of at least one of these genes and recruits repressive histone modifications that inactivate the promoter on the paternal chromosome (Nagano et al. 2008). A mechanism in which transcription of a long ncRNA regulated by maternal germline DNA methylation can control multiple genes in an imprinted cluster has also been shown for the Kcnq1 cluster (Fitzpatrick et al. 2002), the Snrpn cluster (Horsthemke and Wagstaff 2008), and for the Gnas cluster (Williamson et al. 2006; Chotalia et al. 2009). Importantly, studies of the mechanisms whereby large ncRNAs act in cis to control the activity and repression of imprinted genes have revealed a useful paradigm for understanding the functional significance of genome-wide expression of ncRNAs in mammalian cells.
GERMLINE ESTABLISHMENT OF IMPRINTS
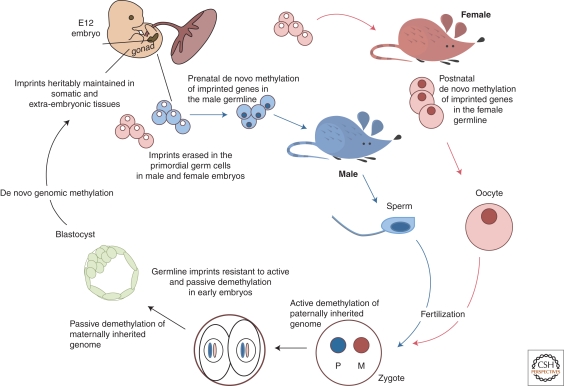
As mentioned above, an important question in imprinting is how parental alleles are marked with their parental origin. If it is assumed that ICRs are key for allelic identity of imprinted genes in cis and that differential epigenetic modifications to the ICRs mediate parental identity, then the question becomes when and how is this ICR marking achieved. It has long been hypothesized that parental-specific marks are assigned in the germline as this is the time when the genomes are in distinct compartments and can be epigenetically modified according to the sex of the transmitting gametes (Fig. 3). Although multiple epigenetic modifications could designate parental origin, DNA methylation has been most widely investigated because of its (1) differential presence on the ICRs as assayed in mature gametes, (2) ease of study on the few cells present in the germline, and (3) proven heritability by the maintenance DNA methyltransferase, DNMT1.
Figure 3.
Cycle of Imprinting. Imprints are acquired in a sex-specific manner in the germline: Maternally and paternally DNA methylated ICRs gain DNA methylation in oocytes and sperm, respectively. Imprints are maintained despite reprogramming and global changes in DNA methylation after fertilization. Paternal imprints are resistant to the active demethylation of the paternal genome in the zygote, and maternal methylation of ICRs is retained despite passive maternal demethylation in the preimplantation embryo. De novo DNA methylation of the genome begins at the end of the preimplantation stage of development. Imprints are maintained in somatic cells throughout the lifetime of the organism. In the germline, DNA methylation is erased during migration of PGCs into the genital ridge. Imprints are then reestablished appropriately during gametogenesis for transmission to the next generation.
Analysis of methylation patterns in primordial germ cells (PCGs) shows that during embryonic migration imprinted genes have the methylation patterns that are characteristic of somatic cells (Hajkova et al. 2002). During their final migration into the genital ridges, the level of DNA methylation in PGCs is reduced. The mechanisms that control DNA demethylation in PGCs and erasure of imprints are unclear, although a recent study has indicates that these processes are linked to changes in chromatin structure and histone modifications (Hajkova et al. 2008). Following demethylation and differentiation of the PGCs, methylation is imposed on the ICRs in a sex-specific manner (Fig. 3). That is, male and female germ cells show DNA remethylation at different times of development, with ICRs in male germ cells initially methylated in prospermatogonia during the period between mitotic arrest and birth (Davis et al. 1999; Li et al. 2004b) and maternal-specific methylation established after birth during the oocyte growth phase prior to ovulation (Lucifero et al. 2002).
Intriguingly, most ICRs are methylated on the maternal allele, with only four paternally methylated ICRs (H19/Igf2, Dlk1/Dio3, Rasgrf1, Zdbf2) identified thus far. All of the ICRs tested to date, with the exception of Rasgrf1, use the de novo DNA methyltransferase DNMT3A and its stimulatory protein DNMT3L to confer DNA methylation on the ICRs in the respective germ cells (Bourc'his et al. 2001; Hata et al. 2002; Bourc'his and Bestor 2004; Kaneda et al. 2004). Nevertheless, it is still unclear how the epigenetic machinery recognizes ICRs in the presence of all of the sequences in the mammalian genome. One clue comes from the X-ray crystallography studies of the complexed carboxy-terminal domains of DNMT3A and DNMT3L (Jia et al. 2007a). A tetrameric complex consisting of these two enzymes preferentially methylates a pair of CpGs that were 8–10 base pairs apart. Such spacing is found in maternally methylated, but not paternally methylated, imprinted loci, although this CpG spacing is widespread in the genome (Ferguson-Smith and Greally 2007). Additional specificity may arise from the observation that DNMT3L interacts with the amino terminus of histone H3 and this interaction is inhibited by H3 lysine 4 methylation (Ooi et al. 2007).
Recently transcription has been suggested to be important for acquisition of maternal imprints in oocytes. Chotalia and colleagues described transcription, some being oocyte-specific, across differentially methylated regions that they propose are required for the establishment of DNA methylation imprints in the oocyte (Chotalia et al. 2009). In contrast to the well-established role for transcription of ncRNAs in regulating the imprinting of adjacent genes described above, only protein-coding transcripts traversing the germline ICRs appear to be involved in methylation establishment. Although an apparent paradox that transcription is attracting the DNA methylation machinery, the authors suggest that transcription across ICRs is required to establish or maintain open chromatin domains that are permissive for establishment of DNA methylation. Whereas temporal relationships require additional investigation, CpG spacing, posttranslational histone modifications, and transcription in oocytes provide a compelling starting point for the acquisition of maternal-specific DNA methylation imprints.
MAINTENANCE OF GENOMIC IMPRINTS
After imprints are set in the germline, it is imperative that they are maintained so that appropriate expression patterns can be achieved in the developing organism. This is especially difficult right after fertilization in the zygote as this is the time when mammals undergo rapid and extensive reprogramming of the gamete genome into a pluripotent genome (Morgan et al. 2005). As part of this reprogramming, DNA methylation and chromatin modifications are erased and subsequently reset (Fig. 3). Nevertheless, genomic imprints must somehow survive these extensive epigenetic changes. Thus, not only must the imprints survive erasure, they must also be propagated during cleavage divisions. In this case, DNMT1, which has well described role in maintaining DNA methylation at imprinted loci (Li et al. 1993), is present at very low levels in preimplantation embryos. Although the precise mechanism is not known and is, in fact, somewhat controversial, it is probable that DNA methylation is maintained in the preimplantation embryos through a combination of the oocyte-specific form of DNMT1 (DNMT1o) and low levels of the somatic form of DNMT1 (Howell et al. 2001; Cirio et al. 2008; Hirasawa et al. 2008).
Maintenance of parental identity most likely involves the unique combination of cis-acting sequences and trans-acting factors. The cis-acting sequences are presumably ICRs but additional sequences cannot be ruled out. Moreover, with respect to trans-acting factors, it is doubtful that the diverse array of sequences present at imprinted loci would attract a single protein. Consistent with this idea, a number of proteins have been identified in the past few years that contribute to maintenance of imprints after fertilization. Some of these proteins impact DNA methylation maintenance at multiple imprinted loci, whereas others appear to have a more restricted role.
One recently defined factor that is critical for DNA methylation maintenance is ZFP57, a KRAB zinc finger protein. This class of transcription factors represses transcription by recruiting KAP-1/TIF1β corepressor complexes (Friedman et al. 1996; Abrink et al. 2001). Mutations in Zfp57 in the mouse cause both maternal- and zygotic-effect lethality, and result in aberrant DNA methylation and expression patterns of imprinted genes (Li et al. 2008). Lack of both maternal and zygotic ZFP57 resulted in embryonic lethality and complete loss of methylation at numerous imprinted loci, including Snrpn, Peg1, Peg3, Peg5, and Dlk1 DMRs, whereas zygotic disruption alone resulted in partial loss of methylation at these sites and partial lethality. Li and colleagues determined that maternal ZFP57 was additionally required for the establishment of DNA methylation at the Snrpn ICR in oocytes, but surprisingly, zygotic Zfp57 expression compensated for this loss and Snrpn methylation was reestablished after E3.5 (Li et al. 2008). Such a result indicates that despite the lack of DNA methylation at the Snrpn ICR in the oocyte, Snprn harbored a residual imprint that was able to direct de novo methylation; this residual mark was either outside the assayed region or derived from an epigenetic modification other than DNA methylation at the ICR, the latter being more difficult to ascertain in preimplantation embryos. The role of ZFP57 in the maintenance of ICR methylation was independently discovered in cases of transient neonatal diabetes (TND), which is caused by hypomethylation of the promoter of the imprinted gene PLAGL1 (Temple and Shield 2002; Mackay et al. 2008). Autosomal recessive ZFP57 mutations were identified in multiple affected pedigrees; patients with these mutations typically had hypomethylation of other ICRs and DMRs, as well as clinical features not normally associated with TND. These results show conservation in the role of ZFP57 in maintenance of DNA methylation between mice and humans.
Additional proteins have been identified that are involved in the stability of imprints, some of which have restricted roles and others of which have a more global role in the maintenance of DNA methylation. In the former category, RBBP1/ARID4A and RBBP1L1/ARID4B are involved in maintenance of imprinting at the Snprn locus (Wu et al. 2006), whereas RNAi-mediated depletion of MBD3 in the oocyte and preimplantation embryo resulted in biallelic expression of H19 in blastocysts, suggesting a role for components of the NuRD complex in maintenance of H19 imprinting (Reese et al. 2007). A general role in imprinting maintenance is indicated for PGC7/STELLA (Nakamura et al. 2007); Stella−/− eggs have normal ICR methylation but embryos derived from these eggs are hypomethylated at multiple loci with maternally or paternally methylated ICRs. Zygotes also show premature global loss of DNA methylation on the maternal pronucleus, indicating a more widespread role for STELLA. Thus, these results suggest that the maternal genome must be protected from the active demethylation occurring on the paternal genome immediately after fertilization.
Two highly related members of the CATERPILLER protein family have also been suggested to be involved in early imprinting regulation in humans, although it is not clear whether these proteins function specifically in imprinting establishment in the germline or early imprinting maintenance, or both. NLRP7 (NALP7), which has no mouse ortholog, was identified as the causative gene for familial recurrent hydatidiform mole, a rare maternal-effect autosomal recessive disorder in which affected women have recurrent molar pregnancies of diploid biparental origin (Murdoch et al. 2006). These women generate eggs that are capable of fertilization, but the resulting conceptus has extensive hypomethylation at the ICRs of imprinted genes. Mutation in a second family member, NLRP2 (NALP2), has recently been associated with a more restricted loss of ICR methylation (Meyer et al. 2009). A family that has two siblings with Beckwith–Wiedemann syndrome was reported in which the mother was homozygous for an NLRP2 mutation. The siblings showed hypomethylation in the ICR that regulates the imprinting of the KCNQ1 cluster, although one child also had a partial loss of methylation of the PEG1 DMR. Together these experiments suggest that NLRP family members are involved in the regulation of imprinting. Consistent with this, these genes are highly expressed early in mammalian development. Nevertheless, their mechanism of action is uncertain, as some family members are involved in the inflammasome, which detects and reacts to extracellular pathogens.
IMPRINTING IN VARIOUS MAMMALIAN SPECIES
Gene imprinting is found in flowering plants and in mammals and it is likely that imprinting in the two organisms evolved convergently. Interestingly, in plants, the altered dosage of imprinted genes influences the development of the endosperm, a vegetative tissue that can be considered extraembryonic and, like in mammals, supports the development of the embryo. Most imprinted genes identified to date in plants are endosperm genes. Similarly, in plants, imprinting depends on DNA methylation for its maintenance, but as endosperm is a vegetative tissue, there is no requirement for germline erasure like in mammals (Feil and Berger 2007). The dependence on imprinting in extraembryonic tissues to control nutritional resources and influence the growth of the embryo perhaps indicates common evolutionary pressures acting to establish and maintain this process of dosage control in both plants and mammals.
Interestingly, not all genes are imprinted in all mammals. For example, some genes that are imprinted in the mouse are not imprinted in the human. This is true for the placenta-specific imprinted genes of the Kcnq1 cluster (Monk et al. 2006). In contrast, the L3MBTL gene is imprinted in human but not imprinted in mouse (Li et al. 2004a, 2005). To add further complexity, the Igf2r gene, which is imprinted in the mouse, has been shown to exhibit polymorphic imprinting in human—it is imprinted in some individuals but not in others (Xu et al. 1993). These findings suggest that imprinting of particular genes varies between different mammalian species and that imprinting may be adaptable and flexible both within and between species.
It is not known how or why mammalian imprinting evolved although its emergence appears to be associated with the evolution of a placenta (Constancia et al. 2004; Kaneko-Ishino et al. 2006), suggesting that even distantly related placental mammals such as metatherians (marsupials) will have imprinting, whereas oviparous mammals, the prototherians (monotremes), will not. Recently, this idea has been put to the test and data investigating the imprinting status of a few individual mammalian imprinted genes in marsupials and monotremes is consistent with this idea. These studies have shown that some eutherian imprinted genes such as Igf2-H19 are imprinted in marsupials. In the tammar wallaby, like in mouse and human, a DMR located upstream of H19 containing CTCF binding sites suggests conservation of the imprinting control mechanism as well (Smits et al. 2008). Peg10, essential for placentation in mouse, is also imprinted in marsupials and has a differentially methylated promoter (Suzuki et al. 2007). However, other eutherian imprinted genes such as those of the Dlk1 imprinted cluster are either absent or not imprinted in the marsupial (Edwards et al. 2008). This suggests that imprinting has evolved independently at different individual loci in some mammalian clades as the need has arisen. It will be interesting to determine why selective pressures have acted to control gene dosage by imprinting at some loci and not at others in metatherian mammals. No evidence exists for imprinting in oviparous mammals such as the platypus or the echidna (Suzuki et al. 2007; Edwards et al. 2008).
CONCLUDING REMARKS
Whereas much has been learned about the numbers and types of imprinted genes, imprinting mechanisms and epigenetic machinery that impacts imprinted gene expression, much remains to be determined. For example, how many more imprinted genes remain to be identified and to what functional pathways do they contribute? Questions remain about the imprinting mechanisms and in particular, what are the underlying germline features that confer maternal- and paternal-specific imprints differentially in egg and sperm? What are the chromatin-associated complexes that recognize, establish, and maintain genomic imprints? What are the mechanistic relationships between DNA methylation and epigenetic modifications to core histones at imprinted domains, and how do these modifications interact with large ncRNAs to influence gene regulation in cis over large genomic regions containing multiple genes? Mouse models and rare human patients continue to be a useful paradigm for exploring this and results can continue to be applied more generally to other nonimprinted regions contributing to a wider understanding of genome function. The advent of technologies in which epigenetic modifications in germ cells and early embryos can be characterized in vivo during key stages of development, applying emerging molecular technologies to very small populations of cells, will help determine the controlling factors and temporal order of events that define the developmental epigenetic program.
The dynamic and variable nature of imprinted gene activity and repression within and between mammalian species raises questions about the extent to which imprints can be adaptable to normal and abnormal environmental influences. Experiments addressing the functional importance of gene dosage control at these loci have the potential to shed light on this. Such functional and mechanistic studies will contribute to our understanding of the complex developmental pathways that have evolved to make a mammal, will provide deeper insights into the relationships between DNA sequence, chromatin structure, and genome function, and along the way, might help us know how and why this remarkable process evolved in the first place.
ACKNOWLEDGMENTS
We acknowledge our many colleagues whose hard work and numerous ideas have led to the numerous breakthroughs discussed in this article. Work in the Bartolomei laboratory was supported by U.S. Public Health Service grants GM51279 and HD42026 and in the Ferguson-Smith lab by grants from the MRC, BBSRC, and Wellcome Trust.
Footnotes
Editors: Paolo Sassone-Corsi, Margaret T. Fuller, and Robert Braun
Additional Perspectives on Germ Cells available at www.cshperspectives.org
REFERENCES
- Abrink M, Ortiz JA, Mark C, Sanchez C, Looman C, Hellman L, Chambon P, Losson R 2001. Conserved interaction between distinct Krüppel-associated box domains and the transcriptional intermediary factor 1 β. Proc Natl Acad Sci 98: 1422–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Amero S, Monk D, Frost J, Preece M, Stanier P, Moore GE 2008. The genetic aetiology of Silver–Russell syndrome. J Med Genet 45: 193–199 [DOI] [PubMed] [Google Scholar]
- Babak T, Deveale B, Armour C, Raymond C, Cleary MA, van der Kooy D, Johnson JM, Lim LP 2008. Global survey of genomic imprinting by transcriptome sequencing. Curr Biol 18: 1735–1741 [DOI] [PubMed] [Google Scholar]
- Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N 1991. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349: 84–87 [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Zemel S, Tilghman SM 1991. Parental imprinting of the mouse H19 gene. Nature 351: 153–155 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Bird A 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99 [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294: 2536–2539 [DOI] [PubMed] [Google Scholar]
- Cattanach BM 1982. Non-disjunction tests with Robertsonian translocations. Mouse News Lett 66: 62–63 [Google Scholar]
- Cattanach BM 1986. Parental origin effects in mice. J Embrol Exp Morph Suppl 97: 137–150 [PubMed] [Google Scholar]
- Cattanach BM, Kirk M 1985. Differential activity of maternally and paternally derived chromosome reigons in mice. Nature 315: 496–498 [DOI] [PubMed] [Google Scholar]
- Chen M, Wang J, Dickerson KE, Kelleher J, Xie T, Gupta D, Lai EW, Pacak K, Gavrilova O, Weinstein LS 2009. Central nervous system imprinting of the G protein G(s)α and its role in metabolic regulation. Cell Metab 9: 548–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G 2009. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev 23: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirio MC, Martel J, Mann M, Toppings M, Bartolomei M, Trasler J, Chaillet JR 2008. DNA methyltransferase 1o functions during preimplantation development to preclude a profound level of epigenetic variation. Dev Biol 324: 139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coan PM, Burton GJ, Ferguson-Smith AC 2005. Imprinted genes in the placenta—A review. Placenta 26 Suppl A: S10–S20 [DOI] [PubMed] [Google Scholar]
- Constancia M, Kelsey G, Reik W 2004. Resourceful imprinting. Nature 432: 53–57 [DOI] [PubMed] [Google Scholar]
- Cooper DW, VandeBerg JL, Sharman GB, Poole WE 1971. Phosphoglycerate kinase polymorphism in kangaroos provides further evidence for paternal X inactivation. Nature New Biology 230: 155–157 [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC 2008. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet 24: 306–316 [DOI] [PubMed] [Google Scholar]
- da Rocha S, Charalambous M, Lin SP, Gutteridge I, Ito Y, Gray D, Dean W, Ferguson-Smith AC 2009. Gene dosage effects of the imprinted Delta-like homologue 1 (Dlk1/Pref1) in development: Implications for the evolution of imprinting. PLoS Genet 5: e1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Trasler JM, Moss SB, Yang GJ, Bartolomei MS 1999. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics 58: 18–28 [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Robertson EJ, Efstratiadis A 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64: 849–859 [DOI] [PubMed] [Google Scholar]
- Edwards CA, Mungall AJ, Matthews L, Ryder E, Gray DJ, Pask AJ, Shaw G, Graves JA, Rogers J, Dunham I, et al. 2008. The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol 6: e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel N, Raval AK, Thorvaldsen JL, Bartolomei MS 2008. Three-dimensional conformation at the H19/Igf2 locus supports a model of enhancer tracking. Hum Mol Genet 17: 3021–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Berger F 2007. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 23: 192–199 [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Greally JM 2007. Epigenetics: Perceptive enzymes. Nature 449: 148–149 [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Cattanach BM, Barton SC, Beechey CV, Surani MA 1991. Embryological and molecular investigations of parental imprinting on mouse chromosome 7. Nature 351: 667–670 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deleion of KvDMR1. Nat Genet 32: 426–431 [DOI] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ III 1996. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev 10: 2067–2078 [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH 2005. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Guillemot F, Caspary T, Tilghman SM, Copeland NG, Gilbery DJ, Jenkins NA, Anderson DJ, Joyner AL, Rossant J, Nagy A 1995. Genomic imprinting of Mash-2, a mouse gene required for trophoblast development. Nature Genet 9: 235–241 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA 2008. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117: 15–23 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129: 1983–1993 [DOI] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H 2008. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev 22: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthemke B, Buiting K 2006. Imprinting defects on human chromosome 15. Cytogenet Genome Res 113: 292–299 [DOI] [PubMed] [Google Scholar]
- Horsthemke B, Wagstaff J 2008. Mechanisms of imprinting of the Prader–Willi/Angelman region. Am J Med Genet A 146A: 2041–2052 [DOI] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR 2001. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104: 829–838 [DOI] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X 2007a. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449: 248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Lin M, Zhang L, York JP, Zhang P 2007b. The Notch signaling pathway controls the size of the ocular lens by directly suppressing p57Kip2 expression. Mol Cell Biol 27: 7236–7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DR 1974. Hairpin-tail: A case of post-reductional gene action in the mouse egg. Genetics 76: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429: 900–903 [DOI] [PubMed] [Google Scholar]
- Kaneko-Ishino T, Kohda T, Ono R, Ishino F 2006. Complementation hypothesis: The necessity of a monoallelic gene expression mechanism in mammalian development. Cytogenet Genome Res 113: 24–30 [DOI] [PubMed] [Google Scholar]
- Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, Viville S, Barton SC, Ishino F, Surani MA 1995. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat Genet 11: 52–59 [DOI] [PubMed] [Google Scholar]
- Kawahara M, Wu Q, Takahashi N, Morita S, Yamada K, Ito M, Ferguson-Smith AC, Kono T 2007. High-frequency generation of viable mice from engineered bi-maternal embryos. Nat Biotechnol 25: 1045–1050 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Ikeda Y, Ishikawa R 2008. Genomic imprinting: A balance between antagonistic roles of parental chromosomes. Semin Cell Dev Biol 19: 574–579 [DOI] [PubMed] [Google Scholar]
- Knoll JHM, Nicholls RD, Magenis RE, Graham JMJ, Lalande M, Latt SA 1989. Angelman and Prader–Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet 32: 285–290 [DOI] [PubMed] [Google Scholar]
- Koerner MV, Pauler FM, Huang R, Barlow DP 2009. The function of non-coding RNAs in genomic imprinting. Development 136: 1771–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono T, Obata Y, Wu Q, Niwa K, Ono Y, Yamamoto Y, Park ES, Seo JS, Ogawa H 2004. Birth of parthenogenetic mice that can develop to adulthood. Nature 428: 860–864 [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci 103: 10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA 1998. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet 20: 163–169 [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R 1993. Role for DNA methylation in genomic imprinting. Nature 366: 362–365 [DOI] [PubMed] [Google Scholar]
- Li J, Bench AJ, Piltz S, Vassiliou G, Baxter EJ, Ferguson-Smith AC, Green AR 2005. L3mbtl, the mouse orthologue of the imprinted L3MBTL, displays a complex pattern of alternative splicing and escapes genomic imprinting. Genomics 86: 489–494 [DOI] [PubMed] [Google Scholar]
- Li J, Bench AJ, Vassiliou GS, Fourouclas N, Ferguson-Smith AC, Green AR 2004a. Imprinting of the human L3MBTL gene, a polycomb family member located in a region of chromosome 20 deleted in human myeloid malignancies. Proc Natl Acad Sci 101: 7341–7346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Lees-Murdock DJ, Xu GL, Walsh CP 2004b. Timing of establishment of paternal methylation imprints in the mouse. Genomics 84: 952–960 [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC 2008. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Keverne EB, Aparicio SA, Ishino F, Barton SC, Surani MA 1999. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science 284: 330–333 [DOI] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC 2003. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet 35: 97–102 [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM 2002. Methylation dynamics of imprinted genes in mouse germ cells. Genomics 79: 530–538 [DOI] [PubMed] [Google Scholar]
- Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP 2000. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet 25: 19–21 [DOI] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, et al. 2008. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet 40: 949–951 [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D 1983. Nuclear transplantation in mouse embryos. J Exp Zool 228: 355–362 [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D 1984. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37: 179–183 [DOI] [PubMed] [Google Scholar]
- Meyer E, Lim D, Pasha S, Tee LJ, Rahman F, Yates JR, Woods CG, Reik W, Maher ER 2009. Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith–Wiedemann syndrome). PLoS Genet 5: e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE 2006. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci 103: 6623–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YS, Smas CM, Lee K, Villena JA, Kim KH, Yun EJ, Sul HS 2002. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol 22: 5585–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W 2005. Epigenetic reprogramming in mammals. Hum Mol Genet 14 Spec No 1: R47–R58 [DOI] [PubMed] [Google Scholar]
- Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, Bagga R, Kircheisen R, Ao A, Ratti B, et al. 2006. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet 38: 300–302 [DOI] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36: 889–893 [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. 2007. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9: 64–71 [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL 2001. Genome organization, function, and imprinting in Prader–Willi and Angelman syndromes. Annu Rev Genomics Hum Genet 2: 153–175 [DOI] [PubMed] [Google Scholar]
- Ogata T, Kagami M, Ferguson-Smith AC 2008. Molecular mechanisms regulating phenotypic outcome in paternal and maternal uniparental disomy for chromosome 14. Epigenetics 3: 181–187 [DOI] [PubMed] [Google Scholar]
- Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, Ogonuki N, Miki H, et al. 2006. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet 38: 101–106 [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunandan R, Ruiz-Hidalgo M, Jia Y, Ettinger R, Rudikoff E, Riggins P, Farnsworth R, Tesfaye A, Laborda J, Bauer SR 2008. Dlk1 influences differentiation and function of B lymphocytes. Stem Cells Dev 17: 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese KJ, Lin S, Verona RI, Schultz RM, Bartolomei MS 2007. Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3. PLoS Genetics 3: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle AG, Beechey CV 1978. Complementation studies with mouse translocations. Cytogenet Cell Genet 20: 282–303 [DOI] [PubMed] [Google Scholar]
- Searle AG, Beechey CV 1990. Genome imprinting phenomena on mouse chromosome 7. Genet Res 56: 237–244 [DOI] [PubMed] [Google Scholar]
- Sekita Y, Wagatsuma H, Nakamura K, Ono R, Kagami M, Wakisaka N, Hino T, Suzuki-Migishima R, Kohda T, Ogura A, et al. 2008. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto–maternal interface of mouse placenta. Nat Genet 40: 243–248 [DOI] [PubMed] [Google Scholar]
- Sibley CP, Coan PM, Ferguson-Smith AC, Dean W, Hughes J, Smith P, Reik W, Burton GJ, Fowden AL, Constancia M 2004. Placental-specific insulin-like growth factor 2 (Igf2) regulates the diffusional exchange characteristics of the mouse placenta. Proc Natl Acad Sci 101: 8204–8208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810–813 [DOI] [PubMed] [Google Scholar]
- Smits G, Mungall AJ, Griffiths-Jones S, Smith P, Beury D, Matthews L, Rogers J, Pask AJ, Shaw G, Vandeberg JL, et al. 2008. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet 40: 971–976 [DOI] [PubMed] [Google Scholar]
- Stöger R, Kubicka P, Liu C-G, Kafri T, Razin A, Cedar H, Barlow DP 1993. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell 73: 61–71 [DOI] [PubMed] [Google Scholar]
- Surani MAH, Barton SC 1983. Development of gynogenetic eggs in the mouse: implications for parthenogenetic embryos. Science 222: 1034–1036 [DOI] [PubMed] [Google Scholar]
- Surani MAH, Barton SC, Norris ML 1984. Development of reconstituted mouse eggs suggest imprinting of the genome during gametogenesis. Nature 308: 548–550 [DOI] [PubMed] [Google Scholar]
- Suzuki S, Ono R, Narita T, Pask AJ, Shaw G, Wang C, Kohda T, Alsop AE, Marshall Graves JA, Kohara Y, et al. 2007. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet 3: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple IK 2007. Imprinting in human disease with special reference to transient neonatal diabetes and Beckwith–Wiedemann syndrome. Endocr Dev 12: 113–123 [DOI] [PubMed] [Google Scholar]
- Temple IK, Shield JP 2002. Transient neonatal diabetes, a disorder of imprinting. J Med Genet 39: 872–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 12: 3693–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Duran KL, Bartolomei MS 1997. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol 17: 4322–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, et al. 2006. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell 11: 711–722 [DOI] [PubMed] [Google Scholar]
- Wang X, Sun Q, McGrath SD, Mardis ER, Soloway PD, Clark AG 2008. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS ONE 3: e3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksberg R, Shuman C, Smith AC 2005. Beckwith–Wiedemann syndrome. Am J Med Genet C Semin Med Genet 137: 12–23 [DOI] [PubMed] [Google Scholar]
- Wilkinson LS, Davies W, Isles AR 2007. Genomic imprinting effects on brain development and function. Nat Rev Neurosci 8: 832–843 [DOI] [PubMed] [Google Scholar]
- Williamson CM, Turner MD, Ball ST, Nottingham WT, Glenister P, Fray M, Tymowska-Lalanne Z, Plagge A, Powles-Glover N, Kelsey G, et al. 2006. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet 38: 350–355 [DOI] [PubMed] [Google Scholar]
- Wu MY, Tsai TF, Beaudet AL 2006. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev 20: 2859–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP 1997. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389: 745–749 [DOI] [PubMed] [Google Scholar]
- Xu Y, Goodyer CG, Deal C, Polychronakos C 1993. Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem Biophys Res Commun 197: 747–754 [DOI] [PubMed] [Google Scholar]
- Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, Jenkins NA, Copeland NG, Brannan CI 1998. A mouse model for Prader–Willi syndrome imprinting-centre mutations. Nature Genet 19: 25–31 [DOI] [PubMed] [Google Scholar]