Abstract
Spermatogenesis is a complex and ordered differentiation process in which the spermatogonial stem cell population gives rise to primary spermatocytes that undergo two successive meiotic divisions followed by a major biochemical and structural reorganization of the haploid cells to generate mature elongate spermatids. The transcriptional regulatory programs that orchestrate this process have been intensively studied in model organisms such as Drosophila melanogaster and mouse. Genetic and biochemical approaches have identified the factors involved and revealed mechanisms of action that are unique to male germ cells. In a well-studied example, cofactors and pathways distinct from those used in somatic tissues mediate the action of CREM in male germ cells. But perhaps the most striking feature concerns the paralogs of somatically expressed transcription factors and of components of the general transcription machinery that act in distinct regulatory mechanisms in both Drosophila and murine spermatogenesis.
Distinct regulatory mechanisms in Drosophila and murine spermatogenesis have been identified involving paralogs of somatically expressed transcription factors and components of the general transcription machinery (e.g., TAF proteins).
Spermatogenesis is a remarkable differentiation process that takes place continuously in adult organisms. In mammals, spermatogenesis occurs in the seminiferous epithelium under a complex endocrine control, whereas in Drosophila melanogaster it occurs relatively autonomously in the testes. In both mammals and Drosophila melanogaster (hereafter, Drosophila) the spermatogonia stem cell population undergoes self-renewal and gives rise to primary spermatocytes that differentiate through two successive meiotic divisions into haploid round spermatids (Fuller 1998; de Rooij 2001). These processes are controlled by complex regulatory programs directing the expression of specific sets of genes at different developmental stages. In mouse, a first set of germ cell–specific genes are expressed in pachytene spermatocytes followed by a second wave of postmeiotic transcription in round spermatids where most genes required for morphological and biochemical reprogramming are expressed (Sassone-Corsi 2002). In Drosophila, the majority of transcription including that of genes required at late stages takes place prior to meiosis. During the postmeiotic phase, the round spermatids undergo a major morphological and biochemical transformation involving the loss of most somatic histones, the condensation of the genome into a tightly packed protamine-DNA complex, flagellar formation, and finally cytoplasmic exclusion. Transcription is repressed during spermatid elongation as the histone octamer-based chromatin organization is replaced by the incorporation of transition proteins and protamines. In mouse and in humans, however, a small fraction of the genome retains a nucleosomal organization (Gatewood et al. 1990). There is much speculation and interest as to whether inheritance of these histones confers epigenetic information that plays a role in directing gene expression following fertilization (van der Heijden et al. 2008; Albert and Peters 2009).
Study of transcription regulation in spermatogenesis is hampered by the lack of an in vitro differentiation model. Most insight has therefore come from model organisms such as Drosophila and mouse in which the power of genetics has proved essential for identifying the factors and mechanisms involved. This article will discuss several of these mechanisms that are unique to germ cells in both of these model organisms.
SPECIALIZED TRANSCRIPTION MECHANISMS IN MALE GERM CELLS OF DROSOPHILA MELANOGASTER
The Gene Expression Program of Drosophila Primary Spermatocytes
Drosophila primary spermatocytes transcribe genes they themselves require; additionally they transcribe the vast majority of genes required at later stages in spermatids. Although there is significant postmeiotic transcription in mammalian round spermatids (Schultz et al. 2003; Potrzebowski et al. 2008), until recently it was believed to be essentially absent from Drosophila spermatogenesis (Barreau et al. 2008). For example, protamines that repackage chromatin in spermatids are only detected late in elongation, however, protamine transcripts accumulate in primary spermatocytes (Jayaramaiah Raja and Renkawitz-Pohl 2005). Like other organs, testes express a little over 50% of the annotated Drosophila protein coding genes, however, unlike other organs, many of these genes are testis-specific or highly testis enriched (Parisi et al. 2004; Chintapalli et al. 2007). Indeed, transcription of about half the genes for known sperm proteins are testis-specific or highly enriched (Dorus et al. 2006). This large set of testis-expressed genes are activated in primary spermatocytes by the concerted actions of two protein complexes.
The Meiotic Arrest Genes Control and Coordinate Transcription in Primary Spermatocyte
There is remarkably little interdependence of cellular events within spermatid differentiation, as defects in one aspect of spermatid formation typically do not cause differentiation arrest; rather they result in abnormal progression through later stages. For example, spermatocytes that fail in the meiotic divisions continue to spermatid differentiation as 4N cells (Gonzalez et al. 1988; Alphey et al. 1992). An exception to this rule is supplied by the “meiotic arrest” mutants in which testes accumulate arrested mature primary spermatocytes (Lin et al. 1996; Table 1).
Table 1.
Classification of the meiotic arrest genes in Drosophila
| Gene name | Homology/domains | Reference(s) | |
|---|---|---|---|
| aly-class | aly (always early) | DIRP | White-Cooper et al. 2000 |
| achi+vis (achintya and vismay) | TALE class homeodomain | Ayyar et al. 2003; Wang and Mann 2003 | |
| comr (cookie monster) | Winged helix | Jiang and White-Cooper 2003 | |
| topi (matotopetli) | 11 Zn fingers | Perezgasga et al. 2004 | |
| tomb (tombola) | CXC | Jiang et al. 2007 | |
| can-class | can (cannonball) | TAF5 WD40 repeats | Hiller et al. 2001 |
| mia (meiosis I arrest) | TAF6 histone fold | Hiller et al. 2004 | |
| mip40 (Myb interacting protein 40kD) | lin-37 | Beall et al. 2007 | |
| nht (no hitter) | TAF4 histone fold | Hiller et al. 2004 | |
| rye (ryan express) | TAF12 histone fold | Hiller et al. 2004 | |
| sa (spermatocyte arrest) | TAF8 histone fold | Hiller et al. 2004 |
The gene names are shown along with distinguishing structural domains and similarity to other factors.
The meiotic arrest genes function in primary spermatocytes to ensure the coordinate progression of meiosis and spermatid differentiation. Reasoning that failure to initiate meiotic divisions in meiotic arrest spermatocytes was likely to result from failure to produce or activate key cell cycle proteins, the expression of the meiotic regulatory proteins CyclinB and Twine was examined. Accumulation of Twine protein was disrupted in all the mutants, and CyclinB failed to accumulate in aly mutants. In aly mutants, the spermatocytes failed to express the mRNAs for these two genes, whereas can (and mia and sa) mutants expressed twine mRNA but not protein. Because meiosis and spermatid differentiation are independent, the expression of eight known spermiogenesis genes in meiotic arrest mutant testes was examined and found to be severely abrogated (in can, mia, and sa mutants) or totally undetectable (aly mutant) (White-Cooper et al. 1998). Thus, the meiotic arrest loci were defined as being critical for transcription of certain spermiogenic genes in primary spermatocytes, and were subdivided into aly-class and can-class on the basis of their effects on particular target genes.
To date eleven meiotic arrest mutant loci have been described, on the basis of this classification five are aly-class, whereas six are can-class (see Table 1). Transcription of all eight known spermiogenesis genes tested in the original 1998 study was dramatically reduced in the meiotic arrest mutants. To assess what proportion of spermiogenesis genes are controlled by the meiotic arrest transcriptional module, a large scale RNA in situ hybridization screen has been performed (www.fly-ted.org) (Barreau et al. 2008). Typically, genes that are transcribed in primary spermatocytes and whose transcripts persist in spermatids consistent with a postmeiotic role are not expressed in mutant testes. As a conservative estimate, based also on unpublished microarray data, over 1500, and probably more than 2000, genes require meiotic arrest gene function for their transcription in Drosophila primary spermatocytes. Therefore, the meiotic arrest genes are critical regulators of gene expression in these cells.
tMAC: A Testis-Specific DREAM Complex
Four of the five known aly-class genes are expressed exclusively or almost exclusively in primary spermatocytes, consistent with their critical function in these cells (Jiang and White-Cooper 2003; Perezgasga et al. 2004; Jiang et al. 2007; White-Cooper 2009). One aly-class locus is complex, with both copies of a recent gene duplication (achi and vis) being mutated to give the male sterile phenotype; vis is strongly testis-enriched in adults; its duplication partner, achi is expressed ubiquitously at low levels (Ayyar et al. 2003; Wang and Mann 2003). aly contains a DIRP domain (domain in Rb-related pathway), whereas topi, comr, tomb, and achi+vis proteins all have predicted DNA binding domains, of the Zn-finger, winged helix, CXC, and homeodomain classes, respectively. Thus, these proteins probably act to directly control transcription by interacting with DNA.
Most animal genomes have only one DIRP protein and one CXC-domain protein, however, in Drosophila these genes have duplicated and aly and tomb are testis-specific paralogs of the broadly expressed genes mip130 and mip120. Mip130 and Mip120 proteins copurify in a complex (MMB/dREAM) from somatic tissues with Mip40, Myb, CAF1, Rbf, E2F2, Dp, and Lin-52 (Korenjak et al. 2004; Lewis et al. 2004). An orthologous complex has also been purified from Caenorhabditis elegans (DRM) (Harrison et al. 2006) and from humans (DREAM/LINC) (Litovchick et al. 2007; Schmit et al. 2007).
Given the evolutionary conservation of the protein families it is not surprising that Aly and Tomb proteins copurify with Mip40 from testes in the tMAC complex (testis meiotic arrest complex) that also contains Topi, Comr, and CAF1 (Beall et al. 2007). Direct protein interactions have been shown for Topi:Comr, for Tomb:Aly and for Tomb:Comr (Perezgasga et al. 2004; Jiang et al. 2007). Notably, Vis (and Achi) was not found in tMAC, neither were Myb, Rbf, Dp, or E2F2. Because Achi/Vis, Aly, and Comr have been shown to coimmunoprecipitate from testes (Wang and Mann 2003), it is likely that tMAC comes in at least two forms, one with Achi/Vis, but lacking Mip40, the other with Mip40, but lacking Achi/Vis. Curiously, whereas mip40 mutants are meiotic arrest male sterile, they do not have an aly-class phenotype, indicating that Mip40 functions differently from the other components of the complex (Beall et al. 2007).
Analysis of protein localizations in various mutant backgrounds also sheds light on the interdependence of the proteins. Aly and Comr proteins must both be present for either to localize to the nucleus (Jiang and White-Cooper 2003), and for Tomb to be stable, whereas Tomb is essential to allow Aly and Comr to interact with chromatin (Jiang et al. 2007). Topi and Achi/Vis seem to be more peripheral to the core complex, although all testis-specific members of tMAC must be present for the complex to efficiently interact with chromatin (Ayyar et al. 2003; Perezgasga et al. 2004).
The Meiotic Arrest Genes—Transcriptional Activators or Repressors of Repressors?
Theoretically, activation of a set of genes in a developmentally orchestrated manner can be achieved by several different mechanisms. The most straightforward is direct activation by a transcription factor (complex) of all the target genes. More complex are transcription factor cascades, in which factors are sequentially activated as differentiation proceeds. Finally, the genes could have a default setting of ON, and be repressed in all cells in which their transcription is not desired. Activation would then involve repressing this repressor. The finding that transcription of any one meiotic arrest locus is independent of all the others argues against a potential transcription factor cascade mechanism (White-Cooper et al. 2000; Hiller et al. 2004).
Although the human homolog of Achi/Vis, TGIF, is a transcriptional repressor, Achi in Drosophila testes is an activator (Wang et al. 2008). dREAM (Beall et al. 2004; Korenjak et al. 2004; Lewis et al. 2004), and orthologous complexes in other species have predominantly been linked to a transcriptional repression rather than activation (Cui et al. 2006), although some activating functions are reported (Georlette et al. 2007). LINC does activate expression of certain cell cycle genes (Pilkinton et al. 2007), and the activation versus repression role of LINC seems to be related to the DNA binding factors with which it is associated (Osterloh et al. 2007; Schmit et al. 2007). It is likely that dREAM, which contains Myb, Rbf, and Dp is predominantly repressive, whereas tMAC, using Comr, Topi, and Achi/Vis to interact with promoters, is predominantly activatory. Consistent with this, all the tMAC subunits associate with euchromatin in primary spermatocytes, although their binding to specific target promoters has not been reported. The C. elegans SynMuvB pathway genes also include known chromatin modifiers, for example, subunits of the NuRD nucleosome remodeling and histone deacetylase complex (Fay and Yochem 2007). It is possible that tMAC interacts with such a complex in primary spermatocytes, and activates gene expression by regulating chromatin architecture and accessibility in primary spermatocytes (White-Cooper et al. 2000).
tTAFs as Repressors of a Repressor
An alternative mechanism is observed for the members of the can-class factors that encode paralogs of the subunits of the general transcription factor TFIID. The TFIID complex is formed by the TATA-binding protein (TBP) and 13–14 TBP-associated factors (TAFs) (Tora 2002; Matangkasombut et al. 2004; Cler et al. 2009). The composition and organization of TFIID is highly conserved through evolution. TFIID has a modular structure with a core domain formed by TAF5 and a set of histone fold-containing TAFs, in which a second module comprising TAF1, TAF7, and TBP associates to form TFIID (Fig. 1) (Gangloff et al. 2001; Papai et al. 2009). In addition to the TFIID TAFs that are widely expressed in embryonic and adult tissues, male germ cells express paralogs of TBP and several TAFs that play critical, but distinct roles in spermatogenesis in Drosophila and mouse.
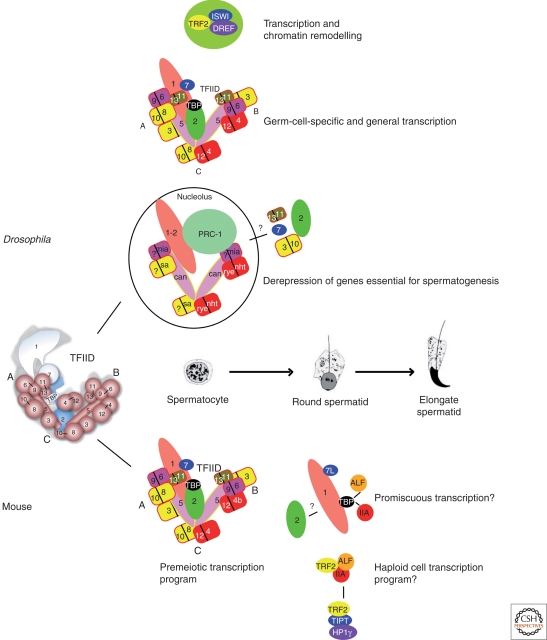
Figure 1.
Schematic description of TFIID complexes that are involved in transcription in Drosophila and murine male germ cells. The structure of TFIID as determined from immunoelectron microscopy (EM) experiments is shown on the left of the figure. The TBP and TAF subunits are represented by colored balls superimposed on the reconstructed EM model. Drosophila spermatocytes express paralogs of several TAFs that are shown organized in the form of a partial TFIID complex. The heterodimerization partners of mia and sa remain to be determined along with the presence of TAFs 2, 7, 11, and 13 in this complex. These spermatocytes likely also contain a TFIID complex comprising somatically expressed TAFs and the TRF2 complex. Murine spermatocytes predominantly contain a normal TFIID complex containing TAF4 and/or TAF4b. In haploid cells, TAF4 is strongly down-regulated leading to loss of full TFIID and accumulation of the TBP-TAF1-TAF7L submodule. TBP can interact with either TFIIA or ALF to mediate elevated levels of mRNA transcription in these cells. TRF2 also interacts with either TFIIA or ALF and may mediate transcription of a subset of essential genes or it may interact with chromatin via TIPT and HP1γ.
The can-class meiotic arrest genes can, mia, nht, rye, and sa, encode testis-specific paralogs of ubiquitously expressed TAFs that form the TFIID core (Hiller et al. 2001, 2004). These tTAFs share the same structural domains (histone-folds, WD40 repeats) and form heterodimers similar to those described for the corresponding core TAFs (see Table 1, and Kolthur-Seetharam et al. 2008). A simple hypothesis is that these factors along with nontestis-specific components, and testis-specific splice isoforms of TAFs (e.g., TAF1-2; Metcalf and Wassarman 2007) assemble to form a testis-specific TFIID complex. Alternatively, as a subset of TAFs are also components of other transcriptional regulatory complexes, for example, in the Polycomb Repression Complex 1 (PRC1) with Polycomb group (PcG) proteins (Saurin et al. 2001) or the GCN5/PCAF histone acetyltransferases in the SAGA complex (Timmers and Tora 2005), the tTAFs could also function through their incorporation in these complexes.
The idea that the tTAFs form a testis-specific TFIID complex that binds directly to the promoters of target genes would predict that this complex be localized to euchromatin in primary spermatocytes. Although some tTAF association with the chromosomal masses in these cells is observed, the strongest labeling is in a subcompartment of the nucleolus (Chen et al. 2005; Metcalf and Wassarman 2007). The action of tMAC is important for localization of tTAFs to the nucleolus, as in aly mutant testes Sa protein localizes in a peri-nucleolar ring (Metcalf and Wassarman 2007). Only tTAFs and the testis-specific isoform TAF1-2 have this nucleolar localization, other TFIID subunits are found associated with euchromatin.
Polycomb and other PRC1 components localize exclusively to chromatin in spermatogonia, but significantly relocalize to the nucleolus in primary spermatocytes (Chen et al. 2005). This nucleolar localization of PRC1 depends on the action of the tTAFs, as Pc remains exclusively on euchromatin in sa mutant testes. Analysis by chromatin immuno precipitation (ChIP) revealed that tTAFs are normally associated with the promoters of target spermiogenesis genes, and that Pc preferentially accumulates at these regions in tTAF mutant testes. Together these observations suggest that tTAFs in primary spermatocytes act to remove the PRC1 repressor complex from target promoters and sequester it in the nucleolus (i.e., they repress the repressors), but they may also directly function as transcriptional activators, replacing the ubiquitous TFIID at target promoters.
SPECIALIZED TRANSCRIPTION MECHANISMS IN MURINE MALE GERM CELLS
This next section will describe several examples of specialized transcription mechanism in murine male germ cells, and highlight the observation that, although mouse germ cells also express paralogs of TAFs and other general transcription factors, their mechanism of action is quite distinct from that in Drosophila.
CREM, ACT, and KIF17b Cooperate in an Integrated Regulatory Mechanism Unique to Haploid Cells
Cyclic AMP response element (CRE) binding protein (CREB) and cyclic AMP response element modulator (CREM) are highly related proteins that regulate transcription in response to various stress, metabolic and developmental signals (Hummler et al. 1994; Sassone-Corsi 1995). CREB and CREM bind to the consensus palindromic CRE 5′-TGACGTCA-3′ or half-CRE 5′-TGACG-3′ and 5′-CGTCA-3′ elements present in the promoters of target genes. CREM exists in multiple isoforms that act either as repressors or activators (Foulkes and Sassone-Corsi 1992). In male germ cells of prepubertal animals, low expression of the repressor isoforms are observed. At puberty, follicle stimulating hormone (FSH), by an as yet undefined mechanism modulates the usage of alternative polyadenylation sites such that several destabilizer signals in the 3′ untranslated region of the CREM τ activator isoform mRNA are eliminated leading to increased stability and the accumulation of the CREM τ protein to high levels in postmeiotic round spermatids (Foulkes et al. 1992, 1993).
CREM knockout leads to early apoptosis of haploid cells, around step 4 of their development, and male sterility showing that CREM plays an essential role in spermiogenesis (Blendy et al. 1996; Nantel et al. 1996; Nantel and Sassone-Corsi 1996). Amongst the direct CREM targets are the genes encoding protamines, transition proteins, and outer dense fiber protein whose expression initiates in round spermatids, but whose gene products are required only later during the elongation stage. Diminished expression of these genes may account for the reduced sperm count and abnormalities seen in heterozygous CREM mutant mice, but do not readily explain the apoptosis. ChIP from adult testis coupled to high throughput sequencing shows that CREM occupies the promoters of more than 6000 target genes in haploid cells (Martianov et al. 2010). Comparison with transcriptome data shows that only a small subset of these genes are deregulated following CREM inactivation. The deregulated genes include the antiapoptotic factor Bcl6b and genes involved in signal transduction and metabolic processes (Beissbarth et al. 2003; Martianov et al. 2010). It is rather the deregulation of these genes that accounts for the apoptotic phenotype seen on CREM inactivation.
In somatic tissues, a variety of stimuli and several different kinases converge to phosphorylate a serine residue (S133 or S117) in the activation domains of CREB and CREM, respectively (Johannessen et al. 2004). Serine phosphorylation results in recruitment of the p300 and CREB binding protein (CBP) coactivators (Chrivia et al. 1993; Nakajima et al. 1997). Alternatively, CREB can also activate transcription of a subset of target genes through a phosphorylation-independent interaction with the TORC (transducers of regulated CREB activity) family of coactivators via the CREB bZIP domain that enhances interaction with the TFIID subunit TAF4 (Fig. 2) (Conkright et al. 2003; Xu et al. 2007). In haploid cells, CREM functions by yet another mechanism that bypasses the requirement for phosphorylation through interaction with the LIM-only domain protein ACT (activator of CREM in testis) encoded by the Fhl5 gene (Fimia et al. 1999) specifically expressed in haploid cells. ACT functions as a coactivator in yeast and in mammalian cells in which it stimulates CREM transcriptional activity. Inactivation of ACT in mice does not phenocopy loss of CREM as there is no block in spermatogenesis and males are fertile (Kotaja et al. 2004). ACT knockout does, however, lead to a severe reduction in the number of mature sperm and the appearance of frequent malformations of the sperm head and flagellum. These observations suggest only a subset of CREM target genes are regulated using ACT as a coactivator, or that ACT is required for a general fine-tuning of CREM target transcript levels.
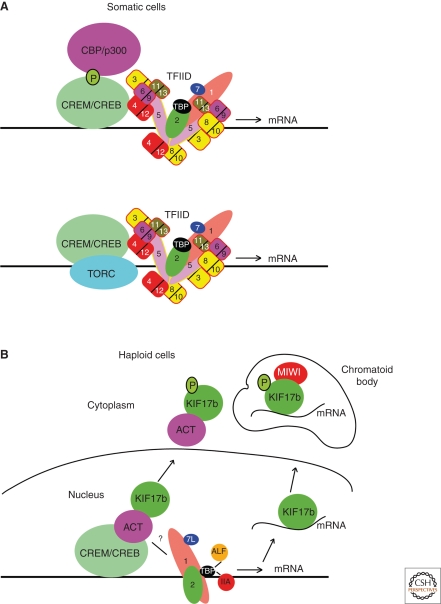
Figure 2.
Schematic model of the CREM-ACT-KIF17b regulatory pathway. (A) In somatic cells, CREB and CREM activate transcription following phosphorylation of a conserved serine residue and recruitment of the CBP and p300 coactivators. CREB and CREM can also interact with TAF4 that is required as a coactivator for their activity. Alternatively, CREB can also activate transcription independent of CBP/p300 via the TORC coactivators. (B) In germ cells, CREM interacts with ACT. At the elongation phase ACT is exported to the cytoplasm by KIF17B to repress CREM activation of target genes. KIF17B also transports mRNAs to the chromatoid body where it interacts with the MIWI protein. Phosphorylation of KIF17B by PKA increases its cytoplasmic localization.
The activity of the CREM-ACT pathway is controlled by KIF17b, a testis-specific form of the kinesin protein KIF17, that interacts with ACT (Macho et al. 2002). KIF17b colocalizes with ACT in the nucleus of haploid cells, but at the onset of the elongation phase, KIF17b and ACT cotranslocate to the cytoplasm. In ex vivo models, KIF17b-mediated export of ACT from the nucleus represses CREM activation of target genes. Intracellular translocation of KIF17b is dependent neither on its motor domain nor on microtubules, revealing a novel microtubule-independent function for kinesins (Kotaja et al. 2005). In cell culture assays, cyclic AMP-dependent protein kinase A (PKA) phosphorylates KIF17b to regulate its subcellular localization. Inhibition of the PKA pathway affects nuclear-cytoplasmic shuttling and promotes cytoplasmic accumulation of KIF17b. If such a mechanism were to operate in germ cells, it would provide a mechanism in which cyclic AMP signaling could modulate CREM-mediated transcription in male germ cells through control of KIF17b rather than direct phosphorylation of the CREM activation domain.
KIF17b is a multifunctional protein that is involved not only in the transport of CREM, but also of a set of CREM-regulated mRNAs (Chennathukuzhi et al. 2003). KIF17b physically interacts with a testis-specific member of the PIWI/Argonaute family, MIWI, a component of the chromatoid body implicated in RNA metabolism (Kotaja et al. 2006; Kotaja and Sassone-Corsi 2007). This interaction may play a role in the transportation of mRNAs to the chromatoid body. Thus, KIF17b links the processes of transcription and transport of specific mRNAs in male germ cells.
Together the above studies describe a unique and integrated mechanism by which CREM, ACT, and KIF17b act together to regulate transcription in developing male germ cells (summarized in Fig. 2).
TRF2 and TRF3: Two TBP-Related Proteins with Specific Roles in Male and Female Germ Cell Development
As described above, the tTAFs play an important role in Drosophila spermatogenesis. In mouse, several testis-expressed paralogs of TFIID components have been identified, such as TAF4b that plays an as yet undefined role in maintaining spermatogonia self-renewal (Falender et al. 2005), and TAF7L (see below). One of the most striking examples is, however, that of the TBP-related factors TRF2 and TRF3.
TBP has a bipartite structure with a poorly conserved amino-terminal region and a highly conserved carboxy-terminal domain that folds as a molecular saddle-shaped structure and binds DNA via the concave surface (Burley 1996). In vertebrates, two TBP-related proteins, TRF2 and TRF3 (also designated as TLF and TBP2, respectively), share this conserved carboxy-terminal domain (Dantonel et al. 1999; Rabenstein et al. 1999; Teichmann et al. 1999; Persengiev et al. 2003; Bartfai et al. 2004; Jallow et al. 2004). In mouse, TRF2 is highly expressed in male germ cells, whereas TRF3 expression is restricted to the ovary (Martianov et al. 2001; Xiao et al. 2006).
Genetic experiments in mice show a specific function of these factors in germ cells. TRF2-null mice are viable and females are fertile (Martianov et al. 2001; Zhang et al. 2001). In contrast, male animals are sterile with an almost complete arrest of spermiogenesis because of apoptosis of step 7 round spermatids. TRF2 mutant round spermatids are characterized by the fragmentation of the chromocenter, a nuclear structure comprising the centromeric heterochromatin from each chromosome (Martianov et al. 2002). In normal spermatids, a single dense nuclear structure is observed, whereas in TRF2 mutant cells, 3–4 large foci are found. The chromocenter is proposed to act as a chromatin organizer prior to the elongation phase (Zalensky et al. 1995). The TRF2 mutant spermatids undergo apoptosis at the beginning of the elongation phase suggesting that chromatin disorganization activates a checkpoint at this step.
At present, it is not clear whether TRF2 acts as a transcription factor or has a more direct structural role on the chromatin and evidence exists for both possibilities. TRF2, like TBP, interacts with TFIIA (Teichmann et al. 1999) and with a paralog of the TFIIA α/β subunit, ALF (TFIIA-like factor) (Upadhyaya et al. 1999; Ozer et al. 2000), that is specifically expressed in late pachytene spermatocytes and in haploid round spermatids (Catena et al. 2005). Like TFIIA, ALF is cleaved into α and β subunits that associate with TFIIAγ. Immunoprecipitation experiments show that male germ cells contain not only TFIIAα/β/γ and ALFα/β-TFIIAγ complexes, but also the full combination of hybrid complexes between the TFIIA/ALF α and/or β subunits (Catena et al. 2005). The TRF2-TFIIA/ALF interaction suggests that TRF2 may function as a transcription factor. However, TRF2 does not bind to TATA sequences and no alternative TRF2 binding motif or response element has been identified on the promoters of germ cell expressed genes. A demonstration that murine TRF2 acts as a bona fide transcription factor will require the identification of such elements and the demonstration that TRF2 is bound in vivo to the target promoters.
TRF2 may also have a more direct role on chromatin as two hybrid experiments have shown an interaction with TIPT (TRF2 interacting protein in testis), a novel uncharacterized testis-specific protein that is specifically expressed in late pachytene and haploid round spermatids (Brancorsini et al. 2008). TIPT also interacts with the heterochromatin proteins HP1α and HP1γ and colocalizes with HP1γ in the euchromatin compartment. Although the function of TIPT in testis remains to be determined, these observations provide a potential direct link between TRF2 and chromatin organization.
It is noteworthy that in Drosophila, TRF2 is also required for spermatogenesis. Mutation of TRF2 leads to defects in meiotic chromatin condensation and chromosome segregation (Kopytova et al. 2006). TRF2 forms a high molecular weight protein complex with the DNA binding factor DREF and proteins such as ISWI involved in chromatin remodeling providing further evidence that TRF2 may directly modulate chromatin organization (Hochheimer et al. 2002).
The specific role of TRF2 in male germ cells is mirrored by that of TRF3 in female germ cells. Oogenesis in mammals, comprises a complex series of events within the ovarian follicles. Oocytes are generated from oogonia and then undergo the early stages of meiotic prophase I. During reproductive life, oocytes are continuously selected from a pool of primordial follicles to develop via a growth phase during which they are arrested at diplotene of prophase I. Studies of TBP and TRF3 expression show a remarkable developmental switch. TBP is expressed in the primordial follicles, but is rapidly down-regulated prior to arrest in metaphase I (Gazdag et al. 2007). In contrast, TRF3 is strongly expressed throughout folliculogenesis, declines at the oocyte stage and disappears following fertilization. These results suggest that TRF3 replaces TBP to direct the specific gene expression program required for oocyte development. This is indeed the case as inactivation of the TRF3 gene results in female infertility, deregulation of oocyte-specific genes and altered chromatin organization (Gazdag et al. 2009). In contrast, inactivation of TBP in the female germ line does not affect oocyte formation demonstrating that it is TRF3 and not TBP that is essential for oocyte development.
There is thus, a striking parallel between the roles of TRF2 and TRF3 in the male and female germ lines. Both proteins show a specific and dynamically regulated expression in male or female germ cells, they are required to direct specific gene expression programs and they play a role in chromatin organization. Interestingly, however, these factors play more general roles in embryonic development in other vertebrates such as the zebrafish Danio rerio or Xenopus (Veenstra et al. 2000; Muller et al. 2001; Bartfai et al. 2004; Hart et al. 2007; Jacobi et al. 2007). Evolution has therefore selected a more specialized role for these proteins in germ cell function in mammals, where embryonic development takes place within the mother’s body.
Evidence for “Promiscuous” Transcription in Male Haploid Germ Cells
Although there is a remarkable parallel between the roles of TRF2 and TRF3 in male and female germ cells, there is also a major difference. In female germ cells, TRF3 replaces TBP, but in male germ cells, TBP is strongly up-regulated and coexpressed with TRF2 (Schmidt and Schibler 1995; Martianov et al. 2002). TBP mRNA levels are almost 100-fold up-regulated in haploid cells compared to somatic tissues, principally because of use of alternate promoters (Schmidt et al. 1997; Schmidt and Schibler 1997). TBP protein levels are 8- to 10-fold higher in testis than in somatic cells. Similarly, TFIIB, RNA polymerase II, TFIIA, and ALF are also strongly expressed in haploid cells (Schmidt and Schibler 1995). It has been proposed that overexpression of the general transcription machinery at this stage is required to produce the large amounts of some mRNAs that have to be synthesized and stored for translation during the elongation and remodeling phase. There is also evidence that overall mRNA levels are generally higher in haploid cells than in somatic cells because of “promiscuous” transcription where in addition to transcription of highly expressed testis-specific genes and of normal “housekeeping” genes, many other genes are also transcribed albeit at lower levels (Schmidt and Schibler 1995). It has been suggested that this may facilitate DNA repair of the transcribed genes before transmission to the next generation, because transcription and DNA repair are coupled (Citterio et al. 2000; Laine and Egly 2006).
In agreement with this idea, ChIP-seq experiments show widespread occupancy by CREM of target genes that have no specific role in spermatogenesis, but whose promoters are active in haploid cells (Martianov et al. 2010). The euchromatin of round spermatids therefore seems rather permissive for transcription factor binding which, together with high levels of the basal transcription machinery, may lead to the expression of a large set of genes and generally higher mRNA levels. This suggests a model in which haploid cells express germ cell–specific paralogs of the general factors for two distinct purposes, to orchestrate the expression of a specific gene expression program and at the same time to ensure sufficient levels of basal factors to promote elevated levels of transcription.
Additional evidence for the above hypothesis comes from the study of TAF7L, a germ cell–specific paralog of the TFIID subunit TAF7 that plays an important role in transcription initiation (Gegonne et al. 2006). TAF7L was identified in a screen for spermatogonial expressed genes located on the X chromosome (Wang et al. 2001). The TAF7L protein is localized in the cytoplasm in spermatogonia and early primary spermatocytes, but from mid-pachytene stage onward, TAF7L is imported into the nucleus and accumulates strongly in postmeiotic cells (Pointud et al. 2003). The import of TAF7L into the nucleus is coordinated with both a loss of TAF7 expression, and a potent up-regulation of TBP. In contrast, the TAF4 subunit of TFIID that is essential for TFIID stability is strongly down-regulated at the same stage. Haploid cells therefore appear to contain an abundance of the TBP-TAF1-TAF7 module, but low amounts of the core domain complex and hence low levels of TFIID. Furthermore, the genomes of old world monkeys, apes, and humans include a retrotransposed copy of TAF1 encoding TAF1L that displays the same functional properties as TAF1 (Wang and Page 2002). This may represent a strategy for human germ cells to evade meiotic sex chromosome inactivation of TAF1 on the X chromosome, and maintain high levels of TAF1 activity in haploid cells. These observations suggest that high levels of specific and/or promiscuous gene expression are mediated via the TBP-TAF1-TAF7L module of TFIID.
This model has been tested by TAF7L inactivation in mouse (Cheng et al. 2007) in which its loss does not lead to arrested spermatogenesis, but to a strongly reduced sperm count and reduced fertility. Sperm from mutant animals are characterized by abnormal morphology, reduced motility, and folded tails. Transcriptome analysis, however, identified only six transcripts with mildly reduced expression. These relatively minor changes in gene expression are unlikely to fully account for the observed phenotype. Although further transcriptome studies on purified germ cell populations may reveal more profound effects on gene expression indicative of a specific function of TAF7L for a subset of target genes, these observations are open to an alternative interpretation. Rather than regulating expression of specific genes in haploid cells, TAF7L is required to ensure generally elevated transcription. An overall reduction in transcript levels is more difficult to observe in transcriptome array experiments compared to the selective loss or induction of a set of genes. This interpretation would explain the paucity of changes on the Affymetrix array, and account for a phentotype with a generalized reduction in spermatogenesis rather than arrest at a specific stage.
The above results lend weight to the idea that paralogs of the general transcription machinery are required in male germ cells for two different, but related functions. Overexpressed components of the basal transcription machinery, including ALF, act together with the TBP-TAF1-TAF7l sub-module of TFIID to direct high levels of mRNA transcription in haploid cells required for normal spermatogenesis. TRF2 on the other hand has evolved to direct a specific aspect of the gene expression program and/or modulate chromatin structure in haploid cells (summarized in Fig. 1). These ideas can be tested by, for example, knockout of ALF, which one would predict to have a general effect on transcription and a phenotype similar to that of TAF7L rather than leading to arrest at a specific stage. Further experiments of this type will confirm or infirm the above model and lead to a better understanding of the mechansims required for correct spermatogenesis and male fertility.
CONCLUDING REMARKS
The studies described above provide insights into transcriptional regulatory mechanisms that are unique to male germ cells. TAF paralogs play important, but mechanistically distinct roles in both Drosophila and mouse spermatogenesis. In Drosophila, these proteins act to sequester the PRC1 repressor complex to facilitate expression of genes required for spermatogenesis in spermatocytes. In contrast, in mice, they may be required to ensure general high levels of mRNA synthesis in haploid cells. A recurrent theme, that has now also found an echo in female germ cells, is the importance of germ cell–specific paralogs of somatically expressed factors and of components of the general transcription machinery illustrated by the tTAFs, TRF2, TRF3, and ALF. Consider also TCEA2, a murine paralog of TFIIS that is selectively expressed at the late pachytene and round spermatid stages (Ito et al. 1996; Umehara et al. 1997). TFIIS was long considered as a transcription elongation factor, however, it has now been shown to be also generally required at the preinitiation stage (Guglielmi et al. 2007; Kim et al. 2007; Sikorski and Buratowski 2009). Here is yet another example of a germ cell–specific paralog of the general transcription machinery, whose function remains to be determined. An unresolved question is why male and female germ cells have adopted this strategy to direct their specific gene expression programs. The diversity of mechanisms discussed above suggests that there is probably no single answer. Future studies should provide further insights into this question.
ACKNOWLEDGMENTS
H.W.-C. is a Royal Society University Research Fellow. Funding for the analysis of tMAC has been from the Wellcome Trust. Funding for in situ hydridizations and FlyTED has been from the BBSRC. The laboratory of I.D. is supported by grants from the CNRS, the INSERM, the ANR, the Association pour la Recherche contre le Cancer, the Ligue Nationale et Départementale Région Alsace contre le Cancer. I.D. is an équipe labéllisée of the Ligue Nationale contre le Cancer.
Footnotes
Editors: Paolo Sassone-Corsi, Margaret T. Fuller, and Robert Braun
Additional Perspectives on Germ Cells available at www.cshperspectives.org
REFERENCES
- Albert M, Peters AH 2009. Genetic and epigenetic control of early mouse development. Curr Opin Genet Dev 19: 113–121 [DOI] [PubMed] [Google Scholar]
- Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM 1992. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell 69: 977–988 [DOI] [PubMed] [Google Scholar]
- Ayyar S, Jiang J, Collu A, White-Cooper H, White RA 2003. Drosophila TGIF is essential for developmentally regulated transcription in spermatogenesis. Development 130: 2841–2852 [DOI] [PubMed] [Google Scholar]
- Barreau C, Benson E, Gudmannsdottir E, Newton F, White-Cooper H 2008. Post-meiotic transcription in Drosophila testes. Development 135: 1897–1902 [DOI] [PubMed] [Google Scholar]
- Bartfai R, Balduf C, Hilton T, Rathmann Y, Hadzhiev Y, Tora L, Orban L, Muller F 2004. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr Biol 14: 593–598 [DOI] [PubMed] [Google Scholar]
- Beall EL, Bell M, Georlette D, Botchan MR 2004. Dm-myb mutant lethality in Drosophila is dependent upon mip130: Positive and negative regulation of DNA replication. Genes Dev 18: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EL, Lewis PW, Bell M, Rocha M, Jones DL, Botchan MR 2007. Discovery of tMAC: A Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev 21: 904–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T, Borisevich I, Horlein A, Kenzelmann M, Hergenhahn M, Klewe-Nebenius A, Klaren R, Korn B, Schmid W, Vingron M, et al. 2003. Analysis of CREM-dependent gene expression during mouse spermatogenesis. Mol Cell Endocrinol 212: 29–39 [DOI] [PubMed] [Google Scholar]
- Blendy JA, Kaestner KH, Weinbauer GF, Nieschlag E, Schutz G 1996. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 380: 162–165 [DOI] [PubMed] [Google Scholar]
- Brancorsini S, Davidson I, Sassone-Corsi P 2008. TIPT, a male germ cell–specific partner of TRF2, is chromatin-associated and interacts with HP1. Cell Cycle 7: 1415–1422 [DOI] [PubMed] [Google Scholar]
- Burley SK 1996. The TATA box binding protein. Curr Opin Struct Biol 6: 69–75 [DOI] [PubMed] [Google Scholar]
- Catena R, Argentini M, Martianov I, Parello C, Brancorsini S, Parvinen M, Sassone-Corsi P, Davidson I 2005. Proteolytic cleavage of ALF into α- and β-subunits that form homologous and heterologous complexes with somatic TFIIA and TRF2 in male germ cells. FEBS Lett 579: 3401–3410 [DOI] [PubMed] [Google Scholar]
- Chen X, Hiller M, Sancak Y, Fuller MT 2005. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science 310: 869–872 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Buffone MG, Kouadio M, Goodheart M, Page DC, Gerton GL, Davidson I, Wang PJ 2007. Abnormal sperm in mice lacking the taf7l gene. Mol Cell Biol 27: 2582–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennathukuzhi V, Morales CR, El-Alfy M, Hecht NB 2003. The kinesin KIF17b and RNA-binding protein TB-RBP transport specific cAMP-responsive element modulator-regulated mRNAs in male germ cells. Proc Natl Acad Sci 100: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39: 715–720 [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365: 855–859 [DOI] [PubMed] [Google Scholar]
- Citterio E, Vermeulen W, Hoeijmakers JH 2000. Transcriptional healing. Cell 101: 447–450 [DOI] [PubMed] [Google Scholar]
- Cler E, Papai G, Schultz P, Davidson I 2009. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell Mol Life Sci 66: 2123–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M 2003. TORCs: Transducers of regulated CREB activity. Mol Cell 12: 413–423 [DOI] [PubMed] [Google Scholar]
- Cui M, Chen J, Myers TR, Hwang BJ, Sternberg PW, Greenwald I, Han M 2006. SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev Cell 10: 667–672 [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Wurtz JM, Poch O, Moras D, Tora L 1999. The TBP-like factor: An alternative transcription factor in metazoa? Trends Biochem Sci 24: 335–339 [DOI] [PubMed] [Google Scholar]
- de Rooij DG 2001. Proliferation and differentiation of spermatogonial stem cells. Reproduction 121: 347–354 [DOI] [PubMed] [Google Scholar]
- Dorus S, Busby SA, Gerike U, Shabanowitz J, Hunt DF, Karr TL 2006. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat Genet 38: 1440–1445 [DOI] [PubMed] [Google Scholar]
- Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, Morris PL, Tjian R, Richards JS 2005. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev 19: 794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Yochem J 2007. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev Biol 306: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, De Cesare D, Sassone-Corsi P 1999. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature 398: 165–169 [DOI] [PubMed] [Google Scholar]
- Foulkes NS, Sassone-Corsi P 1992. More is better: Activators and repressors from the same gene. Cell 68: 411–414 [DOI] [PubMed] [Google Scholar]
- Foulkes NS, Mellstrom B, Benusiglio E, Sassone-Corsi P 1992. Developmental switch of CREM function during spermatogenesis: From antagonist to activator. Nature 355: 80–84 [DOI] [PubMed] [Google Scholar]
- Foulkes NS, Schlotter F, Pevet P, Sassone-Corsi P 1993. Pituitary hormone FSH directs the CREM functional switch during spermatogenesis. Nature 362: 264–267 [DOI] [PubMed] [Google Scholar]
- Fuller MT 1998. Genetic control of cell proliferation and differentiation in Drosophila spermatogenesis. Semin Cell Dev Biol 9: 433–444 [DOI] [PubMed] [Google Scholar]
- Gangloff Y, Romier C, Thuault S, Werten S, Davidson I 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci 26: 250–257 [DOI] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, Schmid CW, Bradbury EM 1990. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J Biol Chem 265: 20662–20666 [PubMed] [Google Scholar]
- Gazdag E, Rajkovic A, Torres-Padilla ME, Tora L 2007. Analysis of TATA-binding protein 2 (TBP2) and TBP expression suggests different roles for the two proteins in regulation of gene expression during oogenesis and early mouse development. Reproduction 134: 51–62 [DOI] [PubMed] [Google Scholar]
- Gazdag E, Santenard A, Ziegler-Birling C, Altobelli G, Poch O, Tora L, Torres-Padilla ME 2009. TBP2 is essential for germ cell development by regulating transcription and chromatin condensation in the oocyte. Genes Dev 23: 2210–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegonne A, Weissman JD, Zhou M, Brady JN, Singer DS 2006. TAF7: A possible transcription initiation check-point regulator. Proc Natl Acad Sci 103: 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georlette D, Ahn S, MacAlpine DM, Cheung E, Lewis PW, Beall EL, Bell SP, Speed T, Manak JR, Botchan MR 2007. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev 21: 2880–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C, Casal J, Ripoll P 1988. Functional monopolar spindles caused by mutation in mgr, a cell division gene of Drosophila melanogaster. J Cell Sci 89: 39–47 [DOI] [PubMed] [Google Scholar]
- Guglielmi B, Soutourina J, Esnault C, Werner M 2007. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci 104: 16062–16067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Ceol CJ, Lu X, Horvitz HR 2006. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc Natl Acad Sci 103: 16782–16787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart DO, Raha T, Lawson ND, Green MR 2007. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature 450: 1082–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT 2004. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131: 5297–5308 [DOI] [PubMed] [Google Scholar]
- Hiller M, Lin T-Y, Wood C, Fuller MT 2001. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev 15: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R 2002. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420: 439–445 [DOI] [PubMed] [Google Scholar]
- Hummler E, Cole TJ, Blendy JA, Ganss R, Aguzzi A, Schmid W, Beermann F, Schutz G 1994. Targeted mutation of the CREB gene: Compensation within the CREB/ATF family of transcription factors. Proc Natl Acad Sci 91: 5647–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Xu Q, Takeuchi H, Kubo T, Natori S 1996. Spermatocyte-specific expression of the gene for mouse testis-specific transcription elongation factor S-II. FEBS Lett 385: 21–24 [DOI] [PubMed] [Google Scholar]
- Jacobi UG, Akkers RC, Pierson ES, Weeks DL, Dagle JM, Veenstra GJ 2007. TBP paralogs accommodate metazoan- and vertebrate-specific developmental gene regulation. EMBO J 26: 3900–3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ 2004. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci 101: 13525–13530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaramaiah Raja S, Renkawitz-Pohl R 2005. Replacement by Drosophila melanogaster protamines and Mst77F of histones during chromatin condensation in late spermatids and role of sesame in the removal of these proteins from the male pronucleus. Mol Cell Biol 25: 6165–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, White-Cooper H 2003. Transcriptional activation in Drosophila spermatogenesis involves the mutually dependent function of aly and a novel meiotic arrest gene cookie monster. Development 130: 563–573 [DOI] [PubMed] [Google Scholar]
- Jiang J, Benson E, Bausek N, Doggett K, White-Cooper H 2007. Tombola, a tesmin/TSO1-family protein, regulates transcriptional activation in the Drosophila male germline and physically interacts with always early. Development 134: 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M, Delghandi MP, Moens U 2004. What turns CREB on? Cell Signal 16: 1211–1227 [DOI] [PubMed] [Google Scholar]
- Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA 2007. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc Natl Acad Sci 104: 16068–16073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolthur-Seetharam U, Martianov I, Davidson I 2008. Specialization of the general transcriptional machinery in male germ cells. Cell Cycle 7: 3493–3498 [DOI] [PubMed] [Google Scholar]
- Kopytova DV, Krasnov AN, Kopantceva MR, Nabirochkina EN, Nikolenko JV, Maksimenko O, Kurshakova MM, Lebedeva LA, Yerokhin MM, Simonova OB, et al. 2006. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol Cell Biol 26: 7492–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A 2004. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119: 181–193 [DOI] [PubMed] [Google Scholar]
- Kotaja N, Sassone-Corsi P 2007. The chromatoid body: A germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol 8: 85–90 [DOI] [PubMed] [Google Scholar]
- Kotaja N, Macho B, Sassone-Corsi P 2005. Microtubule-independent and protein kinase A–mediated function of kinesin KIF17b controls the intracellular transport of activator of CREM in testis (ACT). J Biol Chem 280: 31739–31745 [DOI] [PubMed] [Google Scholar]
- Kotaja N, De Cesare D, Macho B, Monaco L, Brancorsini S, Goossens E, Tournaye H, Gansmuller A, Sassone-Corsi P 2004. Abnormal sperm in mice with targeted deletion of the act (activator of cAMP-responsive element modulator in testis) gene. Proc Natl Acad Sci 101: 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Lin H, Parvinen M, Sassone-Corsi P 2006. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J Cell Sci 119: 2819–2825 [DOI] [PubMed] [Google Scholar]
- Laine JP, Egly JM 2006. When transcription and repair meet: A complex system. Trends Genet 22: 430–436 [DOI] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR 2004. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev 18: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT 1996. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development 122: 1331–1341 [DOI] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell 26: 539–551 [DOI] [PubMed] [Google Scholar]
- Macho B, Brancorsini S, Fimia GM, Setou M, Hirokawa N, Sassone-Corsi P 2002. CREM-dependent transcription in male germ cells controlled by a kinesin. Science 298: 2388–2390 [DOI] [PubMed] [Google Scholar]
- Martianov I, Brancorsini S, Gansmuller A, Parvinen M, Davidson I, Sassone-Corsi P 2002. Distinct functions of TBP and TLF/TRF2 during spermatogenesis: Requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development 129: 945–955 [DOI] [PubMed] [Google Scholar]
- Martianov I, Choukrallah M-A, Legras S, Rijkers E, Van Ijcken W, Jost B, Sassone-Corsi P, Davidson I 2010. Developmentally regulated occupancy of an extended repertoire of CREB and CREM binding loci in male germ cells. BMC Genomics 11: 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Fimia GM, Dierich A, Parvinen M, Sassone-Corsi P, Davidson I 2001. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell 7: 509–515 [DOI] [PubMed] [Google Scholar]
- Matangkasombut O, Auty R, Buratowski S 2004. Structure and function of the TFIID complex. Adv Protein Chem 67: 67–92 [DOI] [PubMed] [Google Scholar]
- Metcalf CE, Wassarman DA 2007. Nucleolar colocalization of TAF1 and testis-specific TAFs during Drosophila spermatogenesis. Dev Dyn 236: 2836–2843 [DOI] [PubMed] [Google Scholar]
- Muller F, Lakatos L, Dantonel J, Strahle U, Tora L 2001. TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr Biol 11: 282–287 [DOI] [PubMed] [Google Scholar]
- Nakajima T, Uchida C, Anderson SF, Parvin JD, Montminy M 1997. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev 11: 738–747 [DOI] [PubMed] [Google Scholar]
- Nantel F, Sassone-Corsi P 1996. CREM: A transcriptional master switch during the spermatogenesis differentiation program. Front Biosci 1: pd266–269 [DOI] [PubMed] [Google Scholar]
- Nantel F, Monaco L, Foulkes NS, Masquilier D, LeMeur M, Henriksen K, Dierich A, Parvinen M, Sassone-Corsi P 1996. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 380: 159–162 [DOI] [PubMed] [Google Scholar]
- Osterloh L, von Eyss B, Schmit F, Rein L, Hubner D, Samans B, Hauser S, Gaubatz S 2007. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J 26: 144–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer J, Moore PA, Lieberman PM 2000. A testis-specific transcription factor IIA (TFIIA τ) stimulates TATA- binding protein-DNA binding and transcription activation. J Biol Chem 275: 122–128 [DOI] [PubMed] [Google Scholar]
- Papai G, Tripathi MK, Ruhlmann C, Werten S, Crucifix C, Weil PA, Schultz P 2009. Mapping the initiator binding Taf2 subunit in the structure of hydrated yeast TFIID. Structure 17: 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Edwards P, Minor J, Naiman D, Lu J, Doctolero M, Vainer M, Chan C, Malley J, et al. 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol 5: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perezgasga L, Jiang J, Bolival B Jr, Hiller M, Benson E, Fuller MT, White-Cooper H 2004. Regulation of transcription of meiotic cell cycle and terminal differentiation genes by the testis-specific Zn-finger protein matotopetli. Development 131: 1691–1702 [DOI] [PubMed] [Google Scholar]
- Persengiev SP, Zhu X, Dixit BL, Maston GA, Kittler EL, Green MR 2003. TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc Natl Acad Sci 100: 14887–14891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkinton M, Sandoval R, Song J, Ness SA, Colamonici OR 2007. Mip/LIN-9 regulates the expression of B-Myb and the induction of cyclin A, cyclin B, and CDK1. J Biol Chem 282: 168–175 [DOI] [PubMed] [Google Scholar]
- Pointud JC, Mengus G, Brancorsini S, Monaco L, Parvinen M, Sassone-Corsi P, Davidson I 2003. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci 116: 1847–1858 [DOI] [PubMed] [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jegou B, Kaessmann H 2008. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol 6: e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenstein MD, Zhou S, Lis JT, Tjian R 1999. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc Natl Acad Sci 96: 4791–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P 1995. Transcription factors responsive to cAMP. Annu Rev Cell Dev Biol 11: 355–377 [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P 2002. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296: 2176–2178 [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412: 655–660 [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Schibler U 1995. High accumulation of components of the RNA polymerase II transcription machinery in rodent spermatids. Development 121: 2373–2383 [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Schibler U 1997. Developmental testis-specific regulation of mRNA levels and mRNA translational efficiencies for TATA-binding protein mRNA isoforms. DevBiol 184: 138–149 [DOI] [PubMed] [Google Scholar]
- Schmit F, Korenjak M, Mannefeld M, Schmitt K, Franke C, von Eyss B, Gagrica S, Hanel F, Brehm A, Gaubatz S 2007. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle 6: 1903–1913 [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Ohbayashi T, Makino Y, Tamura T, Schibler U 1997. Spermatid-specific overexpression of the TATA-binding protein gene involves recruitment of two potent testis-specific promoters. J Biol Chem 272: 5326–5334 [DOI] [PubMed] [Google Scholar]
- Schultz N, Hamra FK, Garbers DL 2003. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci 100: 12201–12206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski TW, Buratowski S 2009. The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol 21: 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann M, Wang Z, Martinez E, Tjernberg A, Zhang D, Vollmer F, Chait BT, Roeder RG 1999. Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci 96: 13720–13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers HT, Tora L 2005. SAGA unveiled. Trends Biochem Sci 30: 7–10 [DOI] [PubMed] [Google Scholar]
- Tora L 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev 16: 673–675 [DOI] [PubMed] [Google Scholar]
- Umehara T, Kida S, Hasegawa S, Fujimoto H, Horikoshi M 1997. Restricted expression of a member of the transcription elongation factor S-II family in testicular germ cells during and after meiosis. J Biochem 121: 598–603 [DOI] [PubMed] [Google Scholar]
- Upadhyaya AB, Lee SH, DeJong J 1999. Identification of a general transcription factor TFIIAα/β homolog selectively expressed in testis. J Biol Chem 274: 18040–18048 [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Ramos L, Baart EB, van den Berg IM, Derijck AA, van der Vlag J, Martini E, de Boer P 2008. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev Biol 8: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Weeks DL, Wolffe AP 2000. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in xenopus. Science 290: 2312–2315 [DOI] [PubMed] [Google Scholar]
- Wang Z, Mann RS 2003. Requirement for two nearly identical TGIF-related homeobox genes in Drosophila spermatogenesis. Development 130: 2853–2865 [DOI] [PubMed] [Google Scholar]
- Wang PJ, Page DC 2002. Functional substitution for TAF(II)250 by a retroposed homolog that is expressed in human spermatogenesis. Hum Mol Genet 11: 2341–2346 [DOI] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC 2001. An abundance of X-linked genes expressed in spermatogonia. Nat Genet 27: 422–426 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang L, Wang Z 2008. Transgenic analyses of TGIF family proteins in Drosophila imply their role in cell growth. J Genet Genomics 35: 457–465 [DOI] [PubMed] [Google Scholar]
- White-Cooper H 2009. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction 139: 11–21 [DOI] [PubMed] [Google Scholar]
- White-Cooper H, Leroy D, MacQueen A, Fuller MT 2000. Transcription of meiotic cell cycle and terminal differentiation genes depends on a conserved chromatin associated protein, whose nuclear localisation is regulated. Development 127: 5463–5473 [DOI] [PubMed] [Google Scholar]
- White-Cooper H, Schafer MA, Alphey LS, Fuller MT 1998. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development 125: 125–134 [DOI] [PubMed] [Google Scholar]
- Xiao L, Kim M, DeJong J 2006. Developmental and cell type-specific regulation of core promoter transcription factors in germ cells of frogs and mice. Gene Expr Patterns 6: 409–419 [DOI] [PubMed] [Google Scholar]
- Xu W, Kasper LH, Lerach S, Jeevan T, Brindle PK 2007. Individual CREB-target genes dictate usage of distinct cAMP-responsive coactivation mechanisms. EMBO J 26: 2890–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalensky AO, Allen MJ, Kobayashi A, Zalenskaya IA, Balhorn R, Bradbury EM 1995. Well-defined genome architecture in the human sperm nucleus. Chromosoma 103: 577–590 [DOI] [PubMed] [Google Scholar]
- Zhang D, Penttila TL, Morris PL, Teichmann M, Roeder RG 2001. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292: 1153–1155 [DOI] [PubMed] [Google Scholar]