Abstract
In mammals, intracellular levels of cholesterol and fatty acids are controlled through a feedback regulatory system mediated by a family of transcription factors called sterol regulatory element-binding proteins (SREBPs). SREBPs are synthesized as inactive precursors bound to membranes of the endoplasmic reticulum. When cells are deprived of cholesterol and fatty acids, NH2-terminal fragments of SREBPs become proteolytically released from membranes and migrate to the nucleus to activate transcription of genes required for lipid synthesis and uptake. Conversely, lipid repletion inhibits proteolytic processing of SREBPs and thereby suppresses lipid accumulation. We review here studies in cultured cells that reveal the mechanism for regulation of SREBP proteolytic activation, and those in animal models in which SREBP proteolysis has been either activated or inhibited to show the essential role of SREBPs in regulating hepatic lipid homeostasis.
Sterol-regulatory-element-binding proteins (SREBPs) are cleaved and released from membranes when cells are deprived of cholesterol or fatty acids. They migrate to the nucleus and up-regulate genes required for lipid synthesis and uptake.
REGULATION OF CHOLESTEROL AND FATTY ACID SYNTHESIS
Cholesterol and fatty acids are important building blocks for animal cell membranes and their synthesis is essential for life. However, overproduction of cholesterol and fatty acids can be toxic to cells as well as to the whole animal, which evokes the need for regulatory mechanisms that control intracellular levels of these lipids. This homeostatic control is achieved by a feedback regulatory system that senses intracellular levels of cholesterol and fatty acids and modulates transcription of genes encoding lipogenic enzymes. The modulators are a family of membrane-bound transcription factors called sterol regulatory element-binding proteins (SREBPs) (Brown and Goldstein 1997).
Mammalian cells produce three isoforms of SREBPs. SREBP-1a and SREBP-1c are produced from a single gene using alternate promoters that produce transcripts with a different first exon, whereas SREBP-2 is encoded by a separate gene (Hua et al. 1995). Unlike typical transcription factors, SREBPs are synthesized as integral membrane proteins localized in the endoplasmic reticulum (ER). The NH2-terminal and the COOH-terminal domains of the proteins project into the cytosol (Fig. 1). They are anchored to membranes by a central domain containing two membrane-spanning sequences separated by a short loop that projects into the lumen of the ER (Fig. 1). The NH2-terminal domains of SREBPs are transcription factors of the basic-loop-helix-leucine zipper family that bind to enhancer sequences located in the promoters of lipogenic genes to activate transcription (Smith et al. 1990; Horton et al. 2003). These enhancer sequences are known as sterol response elements or SREs. Among the three isoforms of SREBPs, the NH2-terminal domains of SREBP-1a and SREBP-1c are more active in driving transcription of genes involved in fatty acid synthesis, whereas that of SREBP-2 is more active in stimulating transcription of genes involved in cholesterol biosynthesis (Pai et al. 1998; Horton et al. 2003). However, in order for the NH2-terminal domains of SREBPs to activate transcription in the nucleus, they have to be first released from the membrane. The proteolytic pathway that liberates the NH2-terminal fragment of SREBPs from membranes in response to intracellular levels of cholesterol and fatty acids is known as the SREBP pathway (Brown and Goldstein 1997).
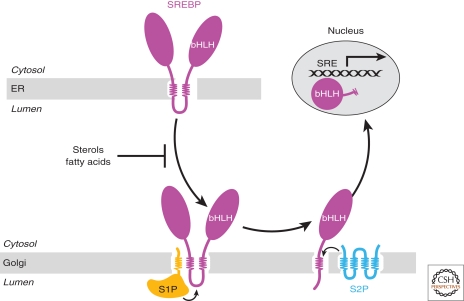
Figure 1.
The SREBP pathway. When cells are depleted of sterols and fatty acids, SREBPs are transported from the ER to the Golgi apparatus, in which they are first cleaved by the Golgi-localized Site-1 Protease (S1P). S1P cleaves SREBPs in the luminal loop between the two membrane-spanning sequences. Once the two halves of the SREBP are separated, a second Golgi protease, Site-2 Protease (S2P), cleaves the NH2-terminal bHLH-Zip domain of SREBPs at a site located three residues within the membrane-spanning region. After the second cleavage, the NH2-terminal domain is released from the membrane and enters the nucleus, in which it activates genes controlling lipid synthesis.
REGULATION OF THE SREBP PATHWAY IN CULTURED CELLS
Regulated Intramembrane Proteolysis of SREBPs
In cells that are depleted of cholesterol, the NH2-terminal domains of the SREBPs are released from membranes by two sequential proteolytic cleavages. The first proteolytic reaction cleaves the SREBPs at a site within the lumen of the ER (Fig. 1). In SREBP-2, this cleavage occurs between the leucine and serine residue of the sequence RSVLS (Duncan et al. 1997). This cleavage is mediated by Site-1 protease (S1P), a membrane-bound serine protease whose active site projects into the lumen (Sakai et al. 1998). S1P absolutely requires a basic residue at the P4 position, and it strongly prefers a leucine at the P1 position (Duncan et al. 1997). Cleavage by S1P separates SREBPs into two membrane-bound halves. The NH2-terminal half is cleaved by Site-2 protease (S2P), a membrane-bound metalloprotease whose active site is buried within the membrane (Fig. 1) (Rawson et al. 1997; Feng et al. 2007). Site-2 cleavage releases the NH2-terminal domains of the SREBPs from membranes, allowing them to enter the nucleus and activate transcription of their target genes (Fig. 1). S2P cleaves SREBP-2 at a site that is three residues within the putative transmembrane segment (Duncan et al. 1998). This cleavage does not occur until the bulk of the luminal portion of SREBPs is first removed by S1P-mediated proteolysis (Sakai et al. 1996). Cleavage by S2P also requires helix-breaking residues distal to the cleavage site in the transmembrane domain (Ye et al. 2000). These residues may partially unwind the α-helical structure surrounding the cleavage site so that the scissile peptide bond is accessible to the protease.
Proteolytic activation of SREBPs is the first example of a signal transduction pathway that involves the generation of regulatory molecules from membrane proteins through proteolytic cleavage. In addition to lipid metabolism, this signaling mechanism, which is termed Regulated Intramembrane Proteolysis (RIP), influences processes as diverse as cellular differentiation, immune defense and the response to unfolded proteins (Brown et al. 2000).
Sterols and fatty acids inhibit RIP of SREBPs by separating SREBPs from S1P and S2P. S1P and S2P are active in the Golgi complex, whereas SREBPs are localized in the ER of lipid-loaded cells (Fig. 1) (DeBose-Boyd et al. 1999). Following lipid-depletion, SREBPs are transported from the ER to the Golgi complex in which they encounter active S1P and S2P (DeBose-Boyd et al. 1999; Nohturfft et al. 2000). This lipid-regulated, ER to Golgi transport of SREBPs is controlled by a pair of polytopic ER membrane proteins: Scap and Insig.
Sterols Control RIP of SREBPs by Two Intracellular Sterol Sensors: Scap and Insig
Scap is a polytopic membrane protein that contains an NH2-terminal domain with eight transmembrane helices and a cytosolic COOH-terminal domain that mediates complex formation with the COOH-terminal domain of SREBPs (Fig. 2) (Sakai et al. 1997). In sterol-depleted cells, the Scap/SREBP complex exits the ER in COPII-coated vesicles that bud from ER membranes (Fig. 2) (Nohturfft et al. 2000; Espenshade et al. 2002; Sun et al. 2005). As a result, SREBPs are cleaved by S1P and S2P in the Golgi complex and the transcription factor domain is released from membranes. When cholesterol builds up in ER membranes, the sterol binds to Scap (Radhakrishnan et al. 2004), and this triggers a conformational change in its cytosolic loop located between TM helices six and seven (Fig. 2) (Brown et al. 2002; Sun et al. 2005). This conformational change causes Scap to bind Insig proteins, which disrupts the interaction of Scap with components of the COP-II coat and results in the retention of the Scap/SREBP complex in the ER (Sun et al. 2005). Scap responds cooperatively to ER cholesterol levels to control SREBP activation in a switch-like manner so that SREBP-2 transport is abruptly blocked when ER cholesterol exceeds 5% of total ER lipids (molar basis) (Radhakrishnan et al. 2008).
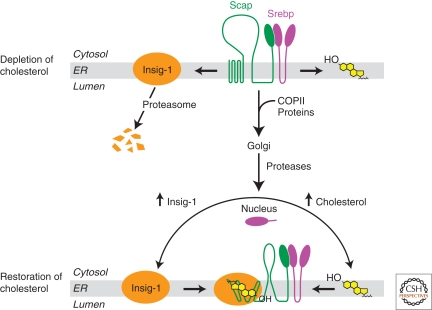
Figure 2.
Cholesterol controls transport of SREBPs from the ER to Golgi complex by regulating the binding between Insig-1 and Scap. In cells depleted of cholesterol, Insig-1 is dissociated from Scap and degraded by proteasome. This allows the Scap/SREBP complex to be incorporated into COP-II-coated vesicles and transported to the Golgi complex, in which SREBPs are proteolytically activated. The NH2-terminal domain of SREBPs enters the nucleus to activate genes required for cholesterol synthesis as well as the gene encoding Insig-1. The proteolytic processing of SREBPs will not be terminated until two SREBP-induced products converge on Scap simultaneously: (1) Newly synthesized cholesterol accumulated in the ER that induces a conformational change in Scap, resulting in its increased affinity with Insig-1; and (2) Newly synthesized Insig-1 that interacts with Scap. In these cells with cholesterol restoration, binding between Scap and Insig-1 stabilizes Insig-1 and prevents incorporation of the Scap/SREBP complex into COP-II-coated vesicles.
Mammalian cells contain two isoforms of Insig proteins, namely Insig-1 and Insig-2 (Yabe et al. 2002; Yang et al. 2002). Both proteins are polytopic membrane proteins with six transmembrane helices (Feramisco et al. 2004) that bind to cholesterol-loaded Scap to retain the Scap/SREBP complex in the ER (Yabe et al. 2002; Yang et al. 2002). Insig proteins also play an important role in oxysterol-regulated cleavage of SREBPs. In addition to cholesterol, oxysterols such as 25-hydroxycholesterol potently inhibit cleavage of SREBPs (Hua et al. 1996a). However, this oxysterol does not bind Scap, nor does it induce the conformational change in Scap (Adams et al. 2004; Radhakrishnan et al. 2004). Instead, 25-hydroxycholesterol binds to purified recombinant Insig proteins (Radhakrishnan et al. 2007). Remarkably, Insigs do not bind to cholesterol. These observations suggest that 25-hydroxycholesterol and cholesterol trigger Scap-Insig through distinct mechanisms: 25-hydroxycholesterol binds to the Insig proteins, whereas cholesterol binds to Scap. Both of these binding reactions produce the same result: ER retention of the Scap/SREBP complex.
In addition to retaining the Scap/SREBP complex in the ER, Scap-Insig binding modulates degradation of Insig-1 protein. In sterol-replete cells, Insig-1 binds to Scap, and the interaction stabilizes the Insig-1 protein (Gong et al. 2006). Sterol deprivation decreases the affinity of the Scap/Insig-1 complex; Insig-1 dissociates from Scap and becomes rapidly ubiquitinated and degraded by 26S proteasomes (Fig. 2). This degradation further enhances proteolytic activation of SREBPs by stimulating ER to Golgi transport of Scap/SREBP. When SREBPs reach the nucleus, they activate transcription, causing increased cholesterol synthesis. Inasmuch as Insig-1 is also a target gene of SREBPs (Yabe et al. 2002; Horton et al. 2003), transcription of its mRNA is also stimulated (Fig. 2). SREBPs will continue to be sent to the nucleus until two criteria have been met: (1) the synthesis of Insig-1 mRNA has been increased sufficiently to allow new Insig-1 molecules to be made; and (2) sufficient cholesterol has been produced in order for newly synthesized Insig-1 to bind Scap thereby stabilizing Insig-1 and retaining the Scap/SREBP complex in the ER (Fig. 2). The requirement for this convergence may serve to even out oscillations that might occur as cholesterol synthesis is turned on and off (Brown and Goldstein 2009).
Unlike Insig-1, Insig-2 is not a transcriptional target of SREBPs (Yabe et al. 2002). The protein also has a long half-life that is not regulated by sterols (Lee et al. 2006a). In cultured cells like CHO cells, the amount of Insig-2 is so low as not to interfere with the convergent regulatory scheme mediated by Insig-1 (Sever et al. 2004). However, in liver Insig-2 may play an important role in regulating the SREBP pathway. The role of Insig-2 in controlling hepatic lipid homeostasis will be discussed in detail below.
Regulation of SREBP-1 by Fatty Acids
SREBP-1 is more active to drive genes involved in fatty acid synthesis than those participated in cholesterol synthesis (Pai et al. 1998; Horton et al. 2003). Thus, SREBP-1 is also subjected to feedback inhibition by fatty acids. Polyunsaturated fatty acids (PUFA) inhibit transcription of SREBP-1 but not SREBP-2 (Ou et al. 2001). This is because transcription of SREBP-1 is activated by liver X receptors (LXRs) (Repa et al. 2000a; DeBose-Boyd et al. 2001), nuclear receptors that are antagonized by PUFA (Ou et al. 2001). In mice, PUFA inhibits transcription of the SREBP-1c, but not of SREBP-1a, owing to the presence of the LXR response element in the promoter of the SREBP-1c gene (Repa et al. 2000a). This is not the case in human cells in which the LXR response element is found in the promoter regions of both SREBP-1a and SREBP-1c. As a result, transcription of SREBP-1a is also inhibited by PUFA in human cells (Hannah et al. 2001).
Unsaturated fatty acids also inhibit proteolytic activation of SREBP-1 (Hannah et al. 2001). This activity is not limited to PUFA; mono-unsaturated fatty acids such as oleate inhibit SREBP processing as well. In contrast, saturated fatty acids are unable to suppress proteolytic activation of SREBP-1. Unsaturated fatty acids inhibit SREBP-1 cleavage by blocking the proteasomal degradation of Insig-1 (Lee et al. 2008). The excess Insig-1 has a more profound effect in blocking cleavage of SREBP-1 than that of SREBP-2 because more Insig-1 is required to inhibit proteolytic activation of SREBP-1 (Engelking et al. 2005; Lee et al. 2008). Because unsaturated fatty acids inhibit SREBP-1 cleavage by increasing the amount of Insig-1 protein, this inhibition is not independent of sterols, which are required for Insig-1 to bind Scap. Rather, by increasing the amount of Insig-1, unsaturated fatty acids make SREBP-1 more sensitive to the inhibitory effects of sterols (Lee et al. 2008).
Remarkably, unsaturated fatty acids and sterols inhibit Insig-1 degradation by different mechanisms. In sterol- and fatty acid-depleted cells, Insig-1 binds to gp78, a membrane-bound E3 ubiquitin ligase that attaches polyubiquitin chains to Insig-1 (Fig. 3) (Lee et al. 2006b). Insig-1 also binds to Ubxd8, a protein that recruits p97 complex to Insig-1 through its bridging interaction with both proteins (Fig. 3) (Lee et al. 2008). Ubiquitination of Insig-1 and recruitment of p97 to the protein lead to recognition and subsequent degradation of Insig-1 by proteasomes (Fig. 3) (Ikeda et al. 2009). Sterols cause Insig-1 to bind to Scap, and this precludes the binding of gp78, thereby inhibiting ubiquitination of Insig-1 (Fig. 3) (Lee et al. 2008). In contrast, sterols do not inhibit binding of the Ubxd8/p97 complex to Insig-1 (Lee et al. 2006b), as this interaction does not require ubiquitination of Insig-1 (Fig. 3) (Ikeda et al. 2009). Unsaturated fatty acids do not block this ubiquitination, but they prevent the recruitment of p97 to Insig-1 by blocking the interaction between Ubxd8 and Insig-1 (Fig. 3) (Lee et al. 2008). Inasmuch as ubiquitination and recruitment of p97 to Insig-1 is required for the protein to be recognized by proteasomes, either sterols or unsaturated fatty acids can block the degradation pathway. Thus, the regulated degradation of Insig-1 outlined in Figure 3 plays an important role in feedback inhibition in synthesis of cholesterol and fatty acids.
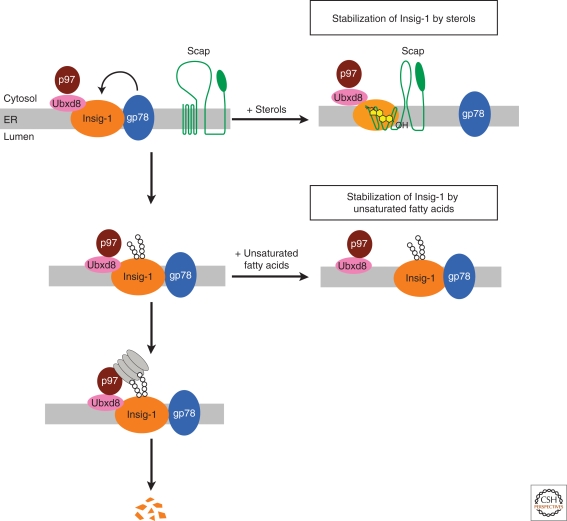
Figure 3.
Unsaturated fatty acids and sterols inhibit Insig-1 degradation independently. In sterol- and fatty acid-depleted cells, Insig-1 is ubiquitinated by gp78. Ubxd8 recruits the ATPase p97 to Insig-1. These two signals on Insig-1 lead to recognition and subsequent degradation of Insig-1 by proteasomes. Sterols induce binding of Scap to Insig-1, a reaction that displaces gp78 from Insig-1. As a result, ubiquitination of Insig-1 is inhibited. In contrast, sterols do not inhibit binding of the Ubxd8/p97 complex to Insig-1. Unsaturated fatty acids do not affect ubiquitination of Insig-1; instead, they block the interaction between Insig-1 and Ubxd8, thereby preventing the recruitment of p97 to Insig-1. Inasmuch as proteasome binding requires both ubiquitination and p97, either sterols or unsaturated fatty acids can block the degradation of Insig-1.
REGULATION OF THE SREBP PATHWAY IN THE LIVER
In mammals, the liver plays a major role in metabolism; the organ is one of the most active sites of cholesterol and fatty acid synthesis in the body. Consistent with studies in cultured cells, in vivo studies reveal that the SREBP pathway drives expression of a genetic program in the liver that leads to enhanced synthesis and uptake of cholesterol, fatty acids, triglycerides, and phospholipids. In this section, we review the role of individual SREBP isoforms (SREBP-1a, -1c, and -2) and their membrane protein regulators (Scap and Insigs) in control of lipid synthesis in the liver. These roles were elucidated through the analysis of genetically manipulated mice in which the SREBP pathway was either activated or inactivated.
Animal Models in which the SREBP Pathway is Activated
The three SREBP isoforms are produced in culture cells as well as in the liver of animals, but at different ratios. In nonhepatic cells, the SREBP-1a transcript is much more abundant than the SREBP-1c transcript (Shimomura et al. 1997). The SREBP-1c transcript predominates in the liver; its level can be up to 10-fold more abundant than that of the SREBP-1a transcript in the organ. The SREBP-2 transcript is present in all cells and tissues at a relatively constant level. To determine the function of the individual SREBP isoforms in the liver, transgenic mice overexpressing SREBPs that are truncated prior to the first transmembrane domain were generated. These proteins, designated nuclear SREBPs (nSREBPs), are synthesized in the cytosol rather than on ER membranes. As a result, they constitutively migrate to the nucleus and activate gene transcription without a requirement for proteolysis. Notably, nSREBPs are immune to feedback regulation by sterols.
Overexpression of nSREBP-1a had dramatic consequences on lipid metabolism in the liver (Shimano et al. 1996). Transgenic nSREBP-1a mice presented markedly elevated levels of mRNAs encoding genes required for cholesterol and fatty acid synthesis. As a result, the cholesterol and fatty acid synthetic rates were increased five-fold and 20-fold, respectively, which led to the progressive development of an enlarged liver filled with cholesterol and triglycerides. The preference of fatty acid synthesis in favor of cholesterol synthesis was preserved in nSREBP-1c transgenic mice; however, the effect was more pronounced (Shimano et al. 1997a). The livers of these animals overaccumulated triglycerides, but showed no increase in cholesterol. Consistent with this observation, mRNAs for hepatic fatty acid biosynthetic enzymes and fatty acid synthetic rates are elevated about fourfold, whereas rates of cholesterol synthesis and expression of cholesterol biosynthetic genes were unchanged (Shimano et al. 1997a). In contrast to results in nSREBP-1 transgenic livers, hepatic overexpression of nSREBP-2 resulted in preferential activation of cholesterol synthesis (20-fold) over fatty acid synthesis (4-fold) (Horton et al. 1998).
Transgenic mice that express a mutant version of Scap in the liver containing an asparagine substitution for aspartic acid-443 (D443N) in the sterol-sensing domain represents another animal model in which the SREBP pathway is activated (Korn et al. 1998). In cultured cells, the D443N mutation in Scap abolishes its binding to Insigs, which renders the protein refractory to sterol-mediated ER retention (Hua et al. 1996b; Yang et al. 2002). Thus, D443N Scap continues to facilitate transport-dependent activation of SREBPs in the presence of sterols and cells expressing the mutant protein overproduce cholesterol. Similarly, transgenic mice expressing Scap D443N in the liver produce elevated levels of nSREBPs as compared to wild type controls (Korn et al. 1998). Correspondingly, expression of genes required for synthesis and uptake of cholesterol and fatty acids were increased and the livers accumulated large amounts of cholesterol and triglycerides. When subjected to a diet enriched in cholesterol, processing of SREBPs was markedly resistant to suppression, demonstrating an essential role for Scap as a sterol sensor in the liver (Korn et al. 1998).
The final animal model for SREBP activation was created through germline disruption of the Insig-2 gene and liver-specific disruption of the Insig-1 gene through Cre-mediated recombination (Engelking et al. 2005). On a normal chow diet, these mice (designated L-Insig-1−/−; Insig-2−/−) overaccumulate cholesterol and triglycerides in the liver, but levels of nSREBPs and their target genes were not reduced. Cholesterol feeding failed to inhibit SREBP processing, reduce expression of lipogenic genes, and block lipid synthesis in Insig-1/-2 double knockout mice. Moreover, the level of HMG CoA reductase protein, the rate-limiting enzyme in cholesterol synthesis, was disproportionately elevated compared to its mRNA in the livers. This observation is consistent with studies in cultured cells showing that Insigs not only mediate sterol-mediated ER retention of Scap, but they also mediate sterol-accelerated degradation of HMG CoA reductase (Sever et al. 2003; Goldstein et al. 2006; DeBose-Boyd 2008). Like Scap, HMG CoA reductase contains a sterol-sensing domain within its NH2-terminal membrane attachment region that precedes a large cytosolic domain; the cytosolic domain of HMG CoA reductase exerts all enzymatic activity (Liscum et al. 1985; Roitelman et al. 1992). Accumulation of sterols causes Insigs to bind to the membrane domain of HMG CoA reductase, initiating a series of reactions that lead to the polyubiquitination of the enzyme and its subsequent degradation by proteasomes. Together, these observations highlight the importance of Insigs in the regulation of lipid synthesis in the liver.
Animal Models in which the SREBP Pathway is Inactivated
In addition to the five mouse models in which the SREBP pathway is activated in the liver, six models have been generated to achieve the opposite effect: Inactivation of the SREBP pathway in the liver through the elimination of genes encoding either SREBP, S1P, or Scap and transgenic overexpression of Insig-1.
Complete lethality was observed following germline deletion of the SREBP-2 gene in mice with mortality occurring at embryonic days 7–8 (Shimano et al. 1997b). Homozygous disruption of the SREBP-1 gene, which eliminated both the SREBP-1a and SREBP-1c transcripts, led to partial lethality; 15%–45% of the mice survive past embryonic day 11. The surviving SREBP-1 knockout mice appeared normal at birth and throughout adulthood. Their livers showed reduced synthesis of fatty acids, owing to reduced expression of fatty acid biosynthetic genes. To compensate for the loss of SREBP-1, livers of the knockout mice manifest elevated levels of SREBP-2 mRNA and protein leading to increased transcription of cholesterol biosynthetic genes and increased cholesterol synthesis. In contrast to SREBP-1 and SREBP-2 knockout mice, selective disruption of SREBP-1c did not cause embryonic lethality; the animals showed reduced expression of fatty acid biosynthetic genes and reduced fatty acid synthesis (Liang et al. 2002). In addition, the compensatory increase in SREBP-2 activity and cholesterol synthesis was also observed in SREBP-1c knockouts.
Studies of the SREBP-1 and SREBP-2 knockout mice indicate that germline elimination of all nSREBPs is an embryonic lethal event. To overcome this lethality, animals were produced in which the SREBP pathway is inactivated in the livers of adults through Cre-mediated recombination. The liver-specific disruption of the S1P and Scap genes led to the dramatic reduction in levels of nSREBPs (Matsuda et al. 2001; Yang et al. 2001). Consequently, expression of SREBP target genes and rates of cholesterol and fatty acid synthesis were markedly diminished (20%–30% of wild type controls). The S1P and Scap knockout mice appear relatively normal in contrast to the embryonic lethality associated with SREBP-1 and SREBP-2 deficient animals.
In cultured cells, overexpression of Insigs renders Scap-mediated processing of SREBPs much more sensitive to sterols (Yang et al. 2002). When overexpressed at high levels, Insig-1 blocks SREBP processing, even in the absence of sterols. Similarly, overexpression of Insig-1 in the mouse liver caused a modest decrease (40%–50%) in the amount of nSREBPs and increased sensitivity of SREBP processing to inhibition by dietary cholesterol (Engelking et al. 2004). Consistent with this, the expression of SREBP target genes and rates of lipid synthesis are decreased in Insig-1 transgenic animals.
Studies of the 11 lines of genetically manipulated mice described in this section highlight the individual roles of SREBP isoforms in hepatic lipid metabolism. The major SREBP-1 isoform in the mouse liver, SREBP-1c, preferentially activates transcription of genes required for fatty acid synthesis, whereas SREBP-2 preferentially activates genes for cholesterol synthesis. Notably, SREBP-1c is relatively weak in stimulating transcription of target genes as compared to SREBP-1a and SREBP-2. This is likely because of differences in the length of an acidic sequence at the NH2-terminus of the proteins that mediates transcriptional activation. The transcriptional activation domain is longer in SREBP-1a and SREBP-2 than in SREBP-1c. The feedback regulation of SREBP processing in the liver, much like the situation in cultured cells, is achieved through the sterol-induced binding of the sterol-sensing domain of Scap to Insigs, which traps the Scap/SREBP complex in the ER. These observations indicate that general mechanisms that regulate SREBP processing operate similarly in the livers of mice as they do in cultured cells.
Transcriptional Regulation of the SREBP Pathway
In addition to elaborate control at the posttranslational level (ER to Golgi transport and proteolysis of SREBPs and proteasomal degradation of Insig-1 and HMG CoA reductase); the SREBP pathway is also subject to transcriptional regulation. Although posttranslational regulation of the SREBP pathway occurs in both cultured cells and livers of whole animals, transcriptional regulation is more evident in the liver.
At least three mechanisms operate in the liver to control transcription of SREBP genes. The first mechanism involves feed-forward transcriptional regulation of the SREBP-1c and SREBP-2 genes, which is mediated by sterol response elements (SREs) present in the promoter/enhancer region of each gene (Sato et al. 1996; Amemiya-Kudo et al. 2000). The SRE maintains the basal expression of the SREBP-1c gene in the liver (Fig. 4) (Chen et al. 2004). The feed-forward transcriptional control explains increased levels of SREBP mRNAs observed in nSREBP transgenic animals and decreased levels of the mRNAs in S1P- and Scap-deficient animals (Matsuda et al. 2001; Yang et al. 2001). In the second mechanism, LXRα and LXRβ, nuclear receptors that heterodimerize with retinoic X receptors and become activated by a variety of sterols such as oxysterols (Repa et al. 2000), selectively regulate the expression of the SREBP-1c gene (Repa et al. 2000; DeBose-Boyd et al. 2001; Liang et al. 2002). In LXRα- and LXRβ-deficient mice, expression of SREBP-1c and fatty acid biosynthetic genes are reduced and do not become up-regulated by synthetic LXR agonists (Repa et al. 2000). Similarly, fatty acid biosynthetic genes are not up-regulated by LXR agonists in SREBP-1c knockout mice (Liang et al. 2002). LXR-mediated regulation of SREBP-1c allows induction of fatty acid synthesis under conditions of sterol overload, which may have important implications for intracellular storage of esterified cholesterol.
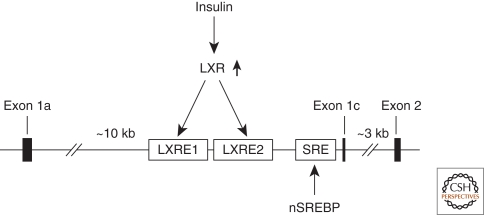
Figure 4.
Regulation of SREBP-1c expression in the liver. The promoter of SREBP-1c contains two LXR-response elements (LXRE) and a SRE. Nuclear SREBPs bind SRE to maintain basal transcription of SREBP-1c. Insulin stimulates the activity of LXR, which binds LXRE to activate transcription of SREBP-1c.
The third mechanism for transcriptional regulation is also selective for the SREBP-1c gene. A key action of the hormone insulin is to stimulate synthesis of fatty acids in the liver. Transcription of the SREBP-1c gene in the livers of mice is reduced following fasting, which suppresses insulin levels, and elevated following refeeding, which restores insulin (Horton et al. 1998). A similar response pattern is observed for mRNAs encoding SREBP-1c target genes. Importantly, elimination of the SREBP-1c gene abolishes insulin-induced activation of fatty acid biosynthetic genes (Liang et al. 2002). Although the exact mechanism through which insulin augments transcription of the SREBP-1c gene is not known, the reaction appears to require the action of LXR and SREBP itself, thereby creating a feed forward stimulation (Fig. 4) (Chen et al. 2004). The livers of Scap knockout mice or Insig-1 transgenic mice show a marked decrease in insulin-mediated stimulation of fatty acid biosynthetic genes that normally occurs in wild type animals following the fasting/refeeding regimen (Matsuda et al. 2001; Engelking et al. 2004). Thus, proteolytic activation of SREBP-1c appears to mediate the action of insulin on fatty acid synthesis in the liver.
Insulin also has a major effect on the expression on Insigs in the liver. Two transcripts are derived from the Insig-2 gene: the liver-specific transcript designated Insig-2a and the ubiquitous transcript designated Insig-2b (Yabe et al. 2003). Insig-2a differs from Insig-2b because of the utilization of an upstream promoter that produces a different 5′ noncoding exon that splices into the same coding exon that initiates Insig-2b. The Insig-2a and Insig-2b transcripts encode the same protein. Insig-2b is expressed at very low levels in the liver, whereas Insig-2a is expressed at high levels in the liver, but only when insulin levels are low.
Reciprocal regulation of Insigs is observed in the livers of mice subjected to the fasting/refeeding protocol. In fasting mice when insulin levels are low, the SREBP-1c gene is not transcribed and nSREBP-1c is not produced. In addition, Insig-1 mRNA and protein levels are reduced and Insig-2 protein levels rise, owing to increased production of the Insig-2a transcript. When insulin levels rise after refeeding, Insig-2 mRNA, and protein rapidly disappears because of transcriptional repression. At the same time, insulin induces transcription of the SREBP-1c gene, nSREBP-1c activates the Insig-1 gene, and Insig-1 mRNA and protein levels are restored, effectively replacing Insig-2 with Insig-1. The significance of the reciprocal regulation of Insigs to the regulation of SREBP processing in the liver is still under investigation.
CONCLUSIONS AND PERSPECTIVES
As we have outlined in this article, the discovery of the SREBP pathway nearly 20 years ago has led to major advances in the understanding of molecular mechanisms that govern the synthesis and uptake of cholesterol and fatty acids. Despite these advances at both the cellular and whole-animal level, key questions remain. For example, what is the molecular basis for the interactions of cholesterol and oxysterols with the membrane domain of Scap and Insigs, respectively? What amino acid residues in these proteins mediate sterol binding? How do interactions with sterols trigger formation of the Scap-Insig complex? Answers to these questions can only be provided by detailed structural analysis of Scap and Insigs by X-ray crystallography. This is an especially challenging task considering the insolubility of sterols in aqueous solution and the requirement of detergents to solubilize the hydrophobic Scap and Insig proteins.
The other unresolved questions in the SREBP pathway pertain to the action of insulin on lipid metabolism through stimulation of SREBP-1c transcription and proteolytic activation. Recent studies have implicated a kinase complex designated the mammalian target of rapamycin complex 1 (mTORC1) in the insulin-mediated induction of SREBP-1c transcription (Li et al. 2010). Although the mechanism through which mTORC1 modulates SREBP-1c expression remains to be determined, the possibility exists that the kinase may activate SREBP-1c, LXR, or a coactivator that interacts with these transcription factors. Considering that SREBP-1c must be proteolytically activated to modulate target gene expression, an obvious question is whether insulin also stimulates cleavage of the protein. However, this requires the examination of transgenic animals expressing full-length SREBP-1c in the liver under transcriptional control of an insulin-independent, constitutive promoter.
Footnotes
Editor: Kai Simons
Additional Perspectives on The Biology of Lipids available at www.cshperspectives.org
REFERENCE
- Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL 2004. Cholesterol and 25-Hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and insigs 2. J Biol Chem 279: 52772–52780 [DOI] [PubMed] [Google Scholar]
- Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty AH, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, et al. 2000. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem 275: 31078–31085 [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL 1997. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89: 331–340 [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL 2009. Cholesterol feedback: From Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res 50: S15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL 2000. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell 100: 391–398 [DOI] [PubMed] [Google Scholar]
- Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL 2002. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell 10: 237–245 [DOI] [PubMed] [Google Scholar]
- Chen G, Liang G, Ou J, Goldstein JL, Brown MS 2004. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci 101: 11245–11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBose-Boyd RA 2008. Feedback regulation of cholesterol synthesis: Sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res 18: 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ 1999. Transport-dependent proteolysis of SREBP: Relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99: 703–712 [DOI] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Ou J, Goldstein JL, Brown MS 2001. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc Natl Acad Sci 98: 1477–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan EA, Brown MS, Goldstein JL, Sakai J 1997. Cleavage site for sterol-regulated protease localized to a leu-Ser bond in the lumenal loop of sterol regulatory element-binding protein-2. J Biol Chem 272: 12778–12785 [DOI] [PubMed] [Google Scholar]
- Duncan EA, Dave UP, Sakai J, Goldstein JL, Brown MS 1998. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J Biol Chem 273: 17801–17809 [DOI] [PubMed] [Google Scholar]
- Engelking LJ, Kuriyama H, Hammer RE, Horton JD, Brown MS, Goldstein JL, Liang G 2004. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J Clin Invest 113: 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelking LJ, Liang G, Hammer RE, Takaishi K, Kuriyama H, Evers BM, Li WP, Horton JD, Goldstein JL, Brown MS 2005. Schoenheimer effect explained - feedback regulation of cholesterol synthesis in mice mediated by Insig proteins 1. J Clin Invest 115: 2489–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade PJ, Li WP, Yabe D 2002. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc Natl Acad Sci 99: 11694–11699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, Shi Y 2007. Structure of a site-2 protease family intramembrane metalloprotease. Science 318: 1608–1612 [DOI] [PubMed] [Google Scholar]
- Feramisco JD, Goldstein JL, Brown MS 2004. Membrane topology of human insig-1, a protein regulator of lipid synthesis. J Biol Chem 279: 8487–8496 [DOI] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS 2006. Protein sensors for membrane sterols 1. Cell 124: 35–46 [DOI] [PubMed] [Google Scholar]
- Gong Y, Lee JN, Lee PC, Goldstein JL, Brown MS, Ye J 2006. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake 2. Cell Metab 3: 5–24 [DOI] [PubMed] [Google Scholar]
- Hannah VC, Ou J, Luong A, Goldstein JL, Brown MS 2001. Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J Biol Chem 276: 4365–4372 [DOI] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci 100: 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shimomura I, Brown MS, Hammer RE, Goldstein JL, Shimano H 1998. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest 101: 2331–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Nohturfft A, Goldstein JL, Brown MS 1996b. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell 87: 415–426 [DOI] [PubMed] [Google Scholar]
- Hua X, Sakai J, Brown MS, Goldstein JL 1996a. Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J Biol Chem 271: 10379–10384 [DOI] [PubMed] [Google Scholar]
- Hua X, Wu J, Goldstein JL, Brown MS, Hobbs HH 1995. Structure of the human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics 25: 667–673 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Demartino GN, Brown MS, Lee JN, Goldstein JL, Ye J 2009. Regulated endoplasmic reticulum-associated degradation of a polytopic protein: p97 recruits proteasomes to Insig-1 before extraction from membranes. J Biol Chem 284: 34889–34900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn BS, Shimomura I, Bashmakov Y, Hammer RE, Horton JD, Goldstein JL, Brown MS 1998. Blunted feedback suppression of SREBP processing by dietary cholesterol in transgenic mice expressing sterol-resistant SCAP(D443N). J Clin Invest 102: 2050–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JN, Gong Y, Zhang X, Ye J 2006a. Proteasomal degradation of ubiquitinated Insig proteins is determined by serine residues flanking ubiquitinated lysines 3. Proc Natl Acad Sci 103: 4958–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JN, Song B, DeBose-Boyd RA, Ye J 2006b. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase, gp78 2. J Biol Chem 281: 39308–39315 [DOI] [PubMed] [Google Scholar]
- Li S, Brown MS, Goldstein JL 2010. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci 107: 3441–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Yang J, Horton JD, Hammer RE, Goldstein JL, Brown MS 2002. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem 277: 9520–9528 [DOI] [PubMed] [Google Scholar]
- Liscum L, Finer-Moore J, Stroud RM, Luskey KL, Brown MS, Goldstein JL 1985. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J Biol Chem 260: 522–530 [PubMed] [Google Scholar]
- Matsuda M, Korn BS, Hammer RE, Moon YA, Komuro R, Horton JD, Goldstein JL, Brown MS, Shimomura I 2001. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev 15: 1206–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ 2000. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell 102: 315–323 [DOI] [PubMed] [Google Scholar]
- Ou J, Tu H, Shan B, Luk A, DeBose-Boyd RA, Bashmakov Y, Goldstein JL, Brown MS 2001. Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing ligand-dependent activation of the LXR. Proc Natl Acad Sci 98: 6027–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai JT, Guryev O, Brown MS, Goldstein JL 1998. Differential stimulation of cholesterol and unsaturated fatty acid biosynthesis in cells expressing individual nuclear sterol regulatory element-binding proteins. J Biol Chem 273: 26138–26148 [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS 2008. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: A delicate balance. Cell Metab 8: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL 2007. From the cover: Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Oxysterols block transport by binding to Insig. Proc Natl Acad Sci 104: 6511–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL 2004. Direct binding of cholesterol to the purified membrane tegion of SCAP; mechanism for a sterol-sensing domain 1. Mol Cell 15: 259–268 [DOI] [PubMed] [Google Scholar]
- Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL 1997. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell 1: 47–57 [DOI] [PubMed] [Google Scholar]
- Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev 14: 2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitelman J, Olender EH, Bar-Nun S, Dunn WA Jr, Simoni RD 1992. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: Implications for enzyme degradation in the endoplasmic reticulum. J Cell Biol 117: 959–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL 1996. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 85: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Sakai J, Nohturfft A, Cheng D, Ho YK, Brown MS, Goldstein JL 1997. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J Biol Chem 272: 20213–20221 [DOI] [PubMed] [Google Scholar]
- Sakai J, Rawson RB, Espenshade PJ, Cheng D, Seegmiller AC, Goldstein JL, Brown MS 1998. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol Cell 2: 505–514 [DOI] [PubMed] [Google Scholar]
- Sato R, Inoue J, Kawabe Y, Kodama T, Takano T, Maeda M 1996. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J Biol Chem 271: 26461–26464 [DOI] [PubMed] [Google Scholar]
- Sever N, Lee PCW, Song BL, Rawson RB, DeBose-Boyd RA 2004. Isolation of mtant cells lacking Insig-1 through selection with SR-12813, an agent that stimulates degradation of 3-Hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem 279: 43136–43147 [DOI] [PubMed] [Google Scholar]
- Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA 2003. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol Cell 11: 25–33 [DOI] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Hammer RE, Shimomura I, Brown MS, Goldstein JL 1996. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest 98: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL 1997a. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest 99: 846–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD 1997b. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest 100: 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS 1997. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest 99: 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Osborne TF, Goldstein JL, Brown MS 1990. Identification of nucleotides responsible for enhancer activity of sterol regulatory element in low density lipoprotein receptor gene. J Biol Chem 265: 2306–2310 [PubMed] [Google Scholar]
- Sun LP, Li L, Goldstein JL, Brown MS 2005. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J Biol Chem 280: 26483–26490 [DOI] [PubMed] [Google Scholar]
- Yabe D, Brown MS, Goldstein JL 2002. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci 99: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS 2003. Liver-specific mRNA for Insig-2 down-regulated by insulin: Implications for fatty acid synthesis. Proc Natl Acad Sci 100: 3155–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS 2002. Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110: 489–500 [DOI] [PubMed] [Google Scholar]
- Yang J, Goldstein JL, Hammer RE, Moon YA, Brown MS, Horton JD 2001. Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. Proc Natl Acad Sci 98: 13607–13612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Dave UP, Grishin NV, Goldstein JL, Brown MS 2000. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc Natl Acad Sci 97: 5123–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]