Abstract
A decline in mitochondrial activity has been associated with aging and is a hallmark of many neurological diseases. Surveillance mechanisms acting at the molecular, organellar, and cellular level monitor mitochondrial integrity and ensure the maintenance of mitochondrial proteostasis. Here we will review the central role of mitochondrial chaperones and proteases, the cytosolic ubiquitin-proteasome system, and the mitochondrial unfolded response in this interconnected quality control network, highlighting the dual function of some proteases in protein quality control within the organelle and for the regulation of mitochondrial fusion and mitophagy.
Mitochondrial dysfunction, associated with aging and neurological diseases, is combated by chaperones and proteases, the cytosolic ubiquitin-proteasome system, and the mitochondrial unfolded protein response.
In all cellular compartments, correct protein folding is critical to maintain cellular homeostasis. In cases where proteins become misfolded or damaged, it is imperative that they are turned over and removed to prevent the formation of toxic folding intermediates or the accumulation of aggregates to levels that can be deleterious for the cell. Several neurodegenerative diseases share a common pathogenic mechanism, which involves the formation of fibrillar aggregates of a particular protein that can accumulate in the cytosol, the nucleus, or the mitochondria. Examples of this include accumulation of the amyloid-β peptide in Alzheimer’s disease (Kayed et al. 2003; Tanzi and Bertram 2005), accumulation of α-synuclein in Parkinson’s disease (Spillantini et al. 1997; Zarranz et al. 2004), and aggregation of a mutant form of the huntingtin protein caused by extended polyglutamine stretches in Huntington’s disease (DiFiglia et al. 1997). Although the exact mechanism of pathogenesis for these diseases remains unresolved, mitochondrial dysfunction is implicated in their progression, which may in turn be responsible for the loss of neurological cell populations because of their sensitivity and requirement for functional mitochondria (Rodolfo et al. 2010).
The evolution of mitochondria began approximately 1.5 billion years ago after an α-proteobacterium was engulfed by a preeukaryotic cell (Gray et al. 1999). Since that time, mitochondria have retained two phospholipid bilayers that segregate two aqueous compartments, the mitochondrial intermembrane space (IMS) and the mitochondrial matrix (Palade 1953). Mitochondria are found in essentially all eukaryotic cells and play integral roles in a number of the cell's metabolic pathways. For example, mitochondria are the key players in cellular ATP production through an elaborate respiratory chain network found in the organelles inner membrane (IM) (Mitchell 1961; Leonard and Schapira 2000). Mitochondria are also required for the β-oxidation of fatty acids, Fe-S biosynthesis, and Ca2+ homeostasis (Pinton et al. 1998; Rizzuto et al. 2000; Lill 2009; Modre-Osprian et al. 2009). Moreover, mitochondria are key regulators of programmed cell death and they participate in developmental processes as well as aging (Singh 2004; Green 2005).
In contrast to early depictions of mitochondria as singular kidney bean shaped entities, it is now well established that mitochondria form elaborate, reticular networks in many tissues (Bereiter-Hahn 1990). The ability of mitochondria to form such networks arises from two major factors: (1) Specialized machineries in the mitochondrial outer membrane (OM) and the IM allow mitochondria to fuse and divide and (2) mitochondria are able to be shuttled along cytoskeletal elements (Anesti and Scorrano 2006; Hoppins et al. 2007). This plasticity of mitochondria ensures that they are able to respond to different cellular cues, which is potentially important for their numerous functions. In different cell types, mitochondria adopt varying morphologies (Kuznetsov et al. 2009). For example, in cultured fibroblasts mitochondria form extensive reticular networks, whereas in neuronal cells, mitochondria can be found enriched at areas of high-energy demand, including presynaptic termini, axon initial segments, and growth cones. Furthermore, in muscle cells, mitochondria adopt a very uniform intermyofibrillar conformation (Vendelin et al. 2005). The dynamic nature of mitochondria provides an explanation as to how they adopt varying organizations in different cell populations. The importance of mitochondrial networks is highlighted by the fact that mutations in components involved in maintaining mitochondrial dynamics results in neurodegenerative diseases (Chan 2006; Olichon et al. 2006; Knott et al. 2008; Martinelli and Rugarli 2010; Winklhofer and Haass 2010).
MITOCHONDRIAL QUALITY CONTROL
A decrease in mitochondrial activity is closely linked with cellular dysfunction and aging, highlighting the importance of functional mitochondria for cell survival (Lin and Beal 2006; Rodolfo et al. 2010; Seo et al. 2010). Neuronal cell populations rely heavily on correct mitochondrial function because of their increased requirement for Ca2+ buffering and ATP at their synaptic termini. Therefore, it is not surprising that impaired mitochondrial function results in neurodegenerative diseases. Mitochondria have developed several mechanisms that act to survey and maintain organelle homeostasis (Fig. 1) (Tatsuta and Langer 2008). The first line of defense occurs on the molecular level and involves conserved intraorganellar protein quality control machinery. This includes chaperones that have been conserved from bacteria to higher eukaryotes as well as various proteolytic enzymes. In addition, recent studies have shown that mitochondrial protein quality control is also influenced by the ubiquitin-proteasome system (UPS) and that a mitochondria-specific unfolded protein response (UPR) is also operating in higher eukaryotes to attenuate build up of misfolded proteins in the organelle (Haynes and Ron 2010; Livnat-Levanon and Glickman 2010).
Figure 1.
Mitochondrial surveillance mechanisms. Low levels of mitochondrial damage are dealt with on the molecular level by conserved proteolytic machineries. Proteins that are unfolded may be refolded by chaperones or degraded by proteases. Mitochondrial dynamics contributes to the functional integrity of mitochondria. Mitochondrial fusion permits content mixing, and stress-induced mitochondrial hyperfusion (SIMH) may alleviate mild mitochondrial stress. Severe mitochondrial damage or depolarization of the mitochondrial membrane potential inhibits fusion, and ongoing fission will segregate the damaged mitochondria from the healthy network, allowing its turnover through mitophagy. Finally, if the level of damaged mitochondria becomes too high and mitochondrial outer membrane permeablization (MOMP) takes place, cytochrome c is released initiating cell death and thus turnover of the entire cell.
The second level of mitochondrial quality control arises because of the dynamic nature of the organelle itself. Mitochondria are constantly fusing and dividing, mediated by dynamin-like GTPases in the OM and IM (Hoppins et al. 2007; Otera and Mihara 2011). Mitochondrial fusion facilitates quality control by allowing a damaged mitochondrion, deficient in certain components to fuse with a healthy neighbor and replenish its stores. Furthermore, mild stresses that inhibit cytosolic translation have been shown to result in stress-induced mitochondrial hyperfusion, which may alleviate the deleterious effects of such insults (Tondera et al. 2009). The role of fusion in mitochondrial quality control has been highlighted by studies demonstrating that loss of the fusion mediators result in a decline in respiratory chain activity (Chen et al. 2005, 2007). In cases where the damage becomes too extreme and is accompanied by dissipation of the mitochondrial membrane potential, mitochondria are rendered fusion incompetent, preventing their fusion with the healthy network, limiting the damage. Similarly, it has been observed that when mitochondria divide, some contain a lower membrane potential across the IM (Twig et al. 2008). In these cases, the damaged mitochondria are turned over through mitophagy (Wang and Klionsky 2011; Youle and Narendra 2011). Interference with mitochondrial fission inhibits the autophagic degradation of damaged mitochondria (Tanaka et al. 2010), illustrating the intimate relationship of mitochondrial dynamics and quality control. The third level of quality control occurs on a cellular level, whereby extensive mitochondrial damage promotes release of proapoptotic factors, resulting in turnover of the entire cell through apoptosis (Wasilewski and Scorrano 2009; Martin 2010).
In this article, we will discuss mechanisms that govern mitochondrial quality control. We will focus on the components that ensure mitochondrial proteostasis at the molecular level and maintain the complement of mitochondrial proteins. Additional functions of some of these components, in particular mitochondrial proteases, are emerging for mitochondrial fusion, mitophagy, and apoptosis, pointing to an intricate network of cellular mechanisms maintaining the integrity and functionality of mitochondria.
MITOCHONDRIAL CHAPERONES—MEDIATING PROTEIN IMPORT AND FOLDING
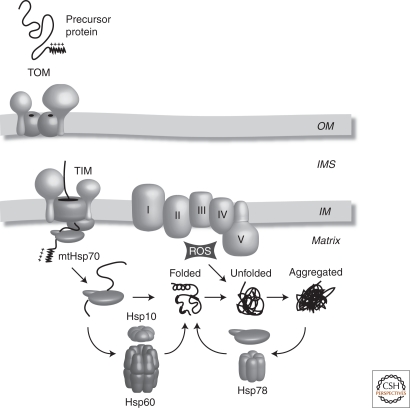
Although mitochondria contain their own genome, it only encodes for 13 polypeptides in humans. The vast majority of the mitochondrial protein complement is actually synthesized in the cytosol and subsequently imported into the organelle (Neupert and Herrmann 2007; Chacinska et al. 2009). For efficient translocation of proteins across the mitochondrial membranes, mitochondrial precursor proteins are kept in a relatively unfolded state to squeeze through the narrow pores formed by tightly gated translocons (Kovermann et al. 2002; Paschen et al. 2003; Rehling et al. 2003). In this unfolded state, precursor proteins have an increased chance of exposing hydrophobic regions promoting unwanted protein–protein interactions or cause them to aggregate, however these unwanted effects are typically overcome through the action of chaperones (Fig. 2). Members of the heat shock family, including Hsp70 and Hsp90, have been shown to guide newly synthesized proteins to the mitochondrial surface and additional factors are likely to exist (Young et al. 2003; Fan et al. 2006). Once proteins enter mitochondria, an additional set of chaperones must adopt this role. A member of the heat shock family, mtHsp70 (Ssc1 in Saccharomyces cerevisiae), has been shown to mediate the ATP-dependent import of mitochondrial proteins that contain a cleavable amino-terminal mitochondrial targeting signal (Kang et al. 1990). Once the proteins have been imported into mitochondria, mtHsp70 adopts a secondary role, in which it maintains proteins in an unfolded conformation allowing the protein to find its correct tertiary structure. A subset of mitochondrial matrix proteins require additional assistance to fold following their import and interaction with mtHsp70. These proteins interact with the chaperonin Hsp60 that, in conjunction with Hsp10, encapsulates the protein removing it from the crowded milieu of the mitochondrial matrix allowing the protein to fold (Horwich et al. 1999). The role of this chaperone family is to provide kinetic assistance to proteins in their quest to fold into their functional form utilizing energy derived from ATP hydrolysis. The importance of the Hsp chaperone system in mitochondria has been highlighted in yeast, in which loss of Ssc1, Hsp60, or Hsp10 results in a lethal phenotype (Craig et al. 1989; Reading et al. 1989; Höhfeld and Hartl 1994). Furthermore, mutations in Hsp60 in humans have been implicated in the pathogenesis of hereditary spastic paraplegia (HSP) 13 (Hansen et al. 2002).
Figure 2.
Chaperone systems in the mitochondrial matrix. mtHsp70 drives the import of nuclear encoded preproteins through the TOM and TIM (translocases in the OM and IM, respectively). In addition, mtHsp70 facilitates protein folding, although some proteins require an additional chaperonin complex, Hsp60/10, to fold. Stress-induced unfolding renders proteins prone to aggregation. The Hsp78 chaperone acts as a disaggregase and in conjunction with mtHsp70 can restore a proteins tertiary structure. OM, mitochondrial outer membrane; IMS, mitochondrial intermembrane space; IM, mitochondrial inner membrane; I–V, respiratory chain complexes, ROS, reactive oxygen species.
Hsp78, a member of the ClpB/Hsp104 family, is an additional chaperone found in the mitochondrial matrix of yeast but not higher eukaryotes. It is required at elevated temperatures, conferring compartment-specific thermotolerance to mitochondria and is necessary for the resolubilization of aggregated proteins in vivo (Schmitt et al. 1996; Doyle and Wickner 2009). Hsp78 also functions in collaboration with proteolytic enzymes to facilitate protein turnover (Schmitt et al. 1995; Voos 2009).
MITOCHONDRIAL PROTEASES—MAINTAINING ORGANELLAR PROTEOSTASIS
Under nonstress conditions, 5%–10% of the yeast mitochondrial proteome is degraded per generation time showing that mitochondrial proteins are continually recycled (Augustin et al. 2005). However, additional reasons for a particular protein becoming a substrate for proteolysis exist. Unfolded proteins no longer capable of being refolded by chaperones are turned over. In addition, proteins that function as part of the mitochondrial respiratory chain are encoded by two genomes. An imbalance in the expression of nuclear or mitochondrial encoded subunits leads to an increase in unassembled subunits, which must be turned over to prevent mitochondrial stress. Furthermore, an inevitable by-product of the respiratory chain and ATP generation in mitochondria is the unwanted production of reactive oxygen species (ROS) from complexes I and III. Proteins can be modified by ROS, which may result in loss of protein function (Stadtman and Berlett 1998). Mitochondria have evolved machineries to alleviate the production of ROS such as manganese superoxide dismutase (MnSOD), which act to detoxify these free radicals (Miao and St Clair 2009). However, when ROS causes protein modifications and incorrect folding, such damaged proteins must be turned over.
ATP-Dependent Proteolysis in the Mitochondrial Matrix
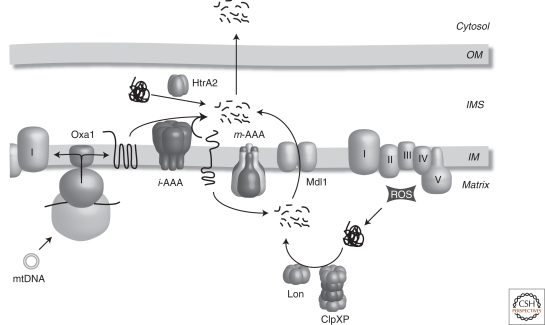
The turnover of misfolded or damaged proteins in mitochondria is accomplished by a specific mitochondrial complement of proteases (Fig. 3). The mitochondrial matrix harbors members of the Lon protease family, which is conserved among prokaryotes and eukaryotes (Desautels and Goldberg 1982; Van Dyck et al. 1994; Gottesman 1996). Lon (Pim1 in yeast) is required for the elimination of denatured or oxidatively damaged proteins (Suzuki et al. 1994; Bota and Davies 2002). Although Lon can degrade mildly oxidized hydrophobic substrates, severely oxidized substrates aggregate, rendering them resistant to degradation (Bota and Davies 2002). Lon functions in collaboration with the chaperone system, which keeps client proteins in an unfolded state until their proteolytic removal can be initiated (Wagner et al. 1994; Bender et al. 2011). The reduced level of unfoldase activity in Lon restricts its ability to degrade proteins that are folded and functional in the mitochondrial matrix (Koodathingal et al. 2009). In general, most substrates of Lon adopt a complex structural organization with bulky prosthetic groups such as the Fe/S cluster. Perhaps such substrates are more prone to misfolding or to damage in conditions of mitochondrial stress. In mammalian mitochondria, LON has been shown to degrade aconitase, an Fe/S protein, the steroidogenic acute regulatory protein (StAR), and the mitochondrial transcription factor A (TFAM) (Bota and Davies 2002; Granot et al. 2007; Matsushima et al. 2010). LON appears to play a role in cellular aging as in aged mice, the levels of functional LON decline, concomitant with an increase in oxidatively damaged mitochondrial proteins and increased mitochondrial dysfunction (Bota et al. 2002, 2005). Furthermore, overexpression of Lon in the fungal aging model, Podospora anserina, results in an increased lifespan without affecting respiration, growth, or fertility (Luce and Osiewacz 2009). It remains to be shown whether the quality control function of Lon or its regulatory role in mtDNA stability is responsible for these effects (Matsushima et al. 2010).
Figure 3.
Proteolytic systems in mitochondria. Newly synthesized subunits of the mitochondrial respiratory chain are inserted into the IM by Oxa1 and assemble with newly imported nuclear encoded subunits. Unassembled subunits as well as unfolded or damaged IM proteins are turned over by membrane embedded AAA protease complexes. Proteins that become damaged or unfolded in the mitochondrial matrix are degraded by Lon or ClpXP (in higher eukaryotes). Peptides that are generated in the mitochondrial matrix are exported from matrix through the peptide transporter Mdl1 and eventually out of the organelle. HtrA2 may play a role in turnover of misfolded or unassembled protein in the IMS. OM, mitochondrial outer membrane; IMS, mitochondrial intermembrane space; IM, mitochondrial inner membrane; I–V, respiratory chain complexes; ROS, reactive oxygen species.
A second protease exists in the mitochondrial matrix, ClpP, which is conserved from bacteria to higher eukaryotes (de Sagarra et al. 1999; Santagata et al. 1999). Interestingly, no homolog of ClpP has been detected in yeast. ClpP functions in a complex with ClpX, a AAA+ chaperone protein in the mitochondrial matrix and it is postulated that ClpX is required for substrate recognition (Kang et al. 2002, 2005). The function of ClpXP within mitochondria is not yet clearly defined, however, ClpP is able to degrade a folding impaired mutant of the mitochondrial enzyme, medium chain acyl-CoA dehydrogenase (Hansen et al. 2005). Moreover, a role for ClpP in the mitochondrial UPR has been proposed recently and will be discussed later in the article (Haynes et al. 2007).
Protein Quality Control across the Mitochondrial IM
The mitochondrial IM is a very proteinaceous environment and considered the most protein rich biological membrane. It houses respiratory chain complexes required for oxidative phosphorylation, numerous carrier proteins that facilitate the transfer of small metabolites across the IM and the machinery that is required for the import of many mitochondrial proteins. The assembly of the respiratory chain complexes requires the coordinated expression of proteins from two genomes, therefore these proteins can accumulate without their partner subunits. In addition, mitochondrial IM proteins are prime targets for oxidative damage because of their proximal location of ROS production. This means that quality control across the IM is critical, as there are many opportunities for protein misfolding or damage in this compartment. Unlike all other membranes in the cell, the mitochondrial IM is the only membrane that is not exposed to the cytosol, therefore proteins of the mitochondrial IM are not easily accessible to and most likely not directly turned over through the UPS.
The mitochondrial IM harbors two AAA+ metalloprotease complexes to ensure protein quality control (Koppen and Langer 2007). The two-protease complexes expose their catalytic domains to opposing faces of the mitochondrial IM with the m-AAA protease exerting its activity in the mitochondrial matrix and the i-AAA protease being active in the IMS (Leonhard et al. 2000). First structural information for a eukaryotic AAA+ protease was recently obtained for the yeast m-AAA protease using single particle electron cryomicroscopy (Lee et al. 2010). The m-AAA protease forms a hexamer in solution, resulting in a large, ring-like molecular machine, with a central pore for substrates to enter into a proteolytic chamber. In humans, there are two subunits that comprise the m-AAA protease, Afg3l2 and paraplegin. Although both assemble into a heterooligomer, it has also been shown that Afg3l2 is capable of assembling into an additional homooligomeric isoform (Koppen et al. 2007). The situation is more complicated in mouse as there is an additional subunit, Afg3l1, which can also form a homooligomeric complex as well as heterooligomeric complexes with Afg3l2 and/or paraplegin (Koppen et al. 2007). The i-AAA protease on the other hand, is composed of only a single subunit, YME1L (or Yme1 in yeast). Mgr1 and Mgr3 have been proposed to act as substrate adaptors of the yeast i-AAA protease, required for the turnover of misfolded substrates (Dunn et al. 2008). The importance of quality control across the IM is highlighted by the finding that mutations in both of the genes encoding for paraplegin and Afg3l2 lead to the neurological diseases hereditary spastic paraplegia (HSP) and spinocerebellar ataxia type 28 (SCA28), respectively (Rugarli and Langer 2006; Martinelli et al. 2009; Di Bella et al. 2010; Martinelli and Rugarli 2010).
Substrates of the i-AAA and m-AAA protease include various nonassembled IM proteins that are peripherally associated or integrated into the mitochondrial IM (Arlt et al. 1996; Leonhard et al. 1996; Korbel et al. 2004). In yeast, the m-AAA protease has additional roles, including processing of the mitochondrial ribosomal subunit Mrpl32 to the mature form. Maturation of Mrpl32 is required for its assembly into the mitochondrial ribosome and the subsequent translation of mitochondrial encoded proteins (Nolden et al. 2005). Notably, impaired Mrpl32 maturation was identified as the primary cause of the respiratory deficiency of yeast cells lacking the m-AAA protease (Nolden et al. 2005). The m-AAA protease also mediates the dislocation of Ccp1 from the IM to allow its maturation by the rhomboid protease Pcp1, a function that does not require proteolytic activity of the m-AAA protease (Esser et al. 2002; Tatsuta et al. 2007). Specific substrates of the i-AAA protease have remained more elusive. Recently however, yeast Yme1 was shown to degrade two IMS proteins that are involved in maintenance of mitochondrial phospholipid levels, Ups1 and Ups2 (Potting et al. 2010). Both proteins are intrinsically unstable and degraded by Yme1 under physiological conditions, controlling their accumulation within mitochondria. Furthermore, in higher eukaryotes, YME1L affects cleavage of the fusion mediator OPA1 and controls mitochondrial morphology (Griparic et al. 2007; Song et al. 2007).
A number of additional proteases are localized to the mitochondrial IM. Although some proteases appear to exert regulatory functions during mitochondrial biogenesis, a role for others in quality control is just emerging. These include the metallopeptidase Oma1, which was originally discovered in yeast to cleave a mutant IM protein in the absence of the m-AAA protease and was thus termed Oma1 because of its overlapping activity with the m-AAA protease (Käser et al. 2003). Oma1 is evolutionarily conserved and is also found in higher eukaryotes, although a role in turnover/processing of misfolded proteins substrates has not yet been assigned. Instead, mammalian OMA1 has been shown to be the protease responsible for induced cleavage of OPA1 and will be discussed in more detail later (Ehses et al. 2009; Head et al. 2009). Atp23 represents yet another conserved peptidase in the IM that may contribute to the maintenance of protein quality control. Yeast Atp23 acts not only as a peptidase for the maturation of Atp6 but also as a chaperone for its assembly into the F1FO-ATP synthase (Osman et al. 2007; Zeng et al. 2007). More recently, Atp23 was also shown to play a role in concert with Yme1 in the turnover of Ups1 (Potting et al. 2010). Although a mammalian Atp23 homolog does exist, its function remains uncharacterized.
Protein Quality Control in the Mitochondrial IMS
Many proteins that perform essential functions are localized to the mitochondrial IMS, however the mechanisms of protein quality control in this subcompartment remains poorly understood. IM peptidases like the i-AAA protease, which expose their proteolytic domains to the IMS, can degrade nonnative IMS proteins in proximity to the IM, yet additional peptidases exist in this compartment. HtrA2/Omi is a highly conserved serine protease originally identified as the homolog of the Escherichia coli stress-responsive protease HtrA/DegP (Pallen and Wren 1997). Bacterial HtrA proteins play key roles in protein quality control in the periplasm at elevated temperatures (Kim et al. 1999). The HtrA2 homolog, DegP, has a role of channeling unfolded proteins into repair, assembly, or degradation pathways independently of ATP (Spiess et al. 1999; Krojer et al. 2008). Interestingly, loss of HtrA2 produces a large number of damaged mitochondria and increased levels of unfolded respiratory chain subunits, indicating that HtrA2 may represent an important component that maintains proteostasis in the IMS, however, its precise role remains unclear (Martins et al. 2004; Moisoi et al. 2009). HtrA2 levels increase in response to various stresses and it has been proposed to play a role in autophagy (Li et al. 2010). Moreover, HtrA2 was proposed to be involved in the progression of apoptosis, in which the active form of the protease is released into the cytosol to cleave antiapoptotic proteins (Martins et al. 2002; Suzuki et al. 2004). However, this role was challenged when a mouse lacking HtrA2 did not display decreased levels of apoptosis but instead displayed neurological defects with Parkinsonian features (Jones et al. 2003; Martins et al. 2004; Strauss et al. 2005).
TURNOVER OF MITOCHONDRIAL OM PROTEINS—EXPLOITING THE UPS
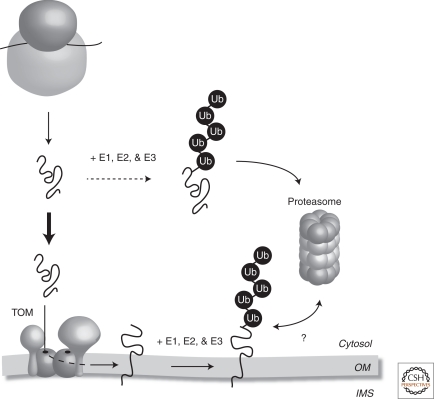
Proteins of the mitochondrial OM play important roles in the regulation of metabolism, apoptosis, and other signaling events. Therefore, quality control of mitochondrial OM proteins is of very high importance. However, information regarding how the quality control of proteins in the mitochondrial OM is maintained is just beginning to emerge. The UPS is a primarily cytosolic multicomponent system responsible for the removal of proteins from multiple cellular compartments (Finley 2009). Several proteomic screens have identified a number of ubiquitinated mitochondrial proteins (Sickmann et al. 2003; Jeon et al. 2007). Mistargeting of mitochondrial precursor proteins may partially explain the ubiquitination of mitochondrial proteins, however, it is conceivable that continual proteasomal degradation of mitochondrial precursor proteins takes place (Fig. 4). This was recently observed for the dynamin-like GTPase OPA1 in the IM, in which addition of the protease inhibitor MG132 stabilized the precursor forms of newly synthesized OPA1 resulting in the accumulation of mature OPA1 within mitochondria (Ehses et al. 2009). Proteins that reside in the OM are accessible to UPS machinery and increasing evidence suggests that certain mitochondrial OM proteins are ubiquitinated and subjected to turnover via the UPS pathway (Fig. 4) (Livnat-Levanon and Glickman 2010). This suggests that a pathway for mitochondrial OM turnover exists that is analogous to that observed for endoplasmic reticulum proteins. In yeast, at least one pathway of Fzo1 degradation involves ubiquitination and degradation by the 26S proteasome, although proteasome-independent pathways of Fzo1 degradation have also been proposed (Neutzner and Youle 2005; Escobar-Henriques et al. 2006). Mitofusin 1 (Mfn1) and Mfn2 have been shown to be degraded in a proteasome-dependent fashion (Tanaka et al. 2010). In line with the concept of the UPS operating at the mitochondrial OM, several ubiquitin ligases have been localized to this compartment (Livnat-Levanon and Glickman 2010).
Figure 4.
Mitochondrial proteins and the ubiquitin-proteasome system. Following their synthesis in the cytosol, mitochondrial precursor proteins follow two pathways. The first (bold line) ensures that the precursor protein is efficiently targeted to the translocase of the outer membrane (TOM). Some proteins that reside in the mitochondrial OM can be ubiquitinated (Ub) by a cascade of E1, E2, and E3 enzymes and subsequently turned over in an ubiquitin-proteasome-dependent pathway. It remains unclear if mitochondrial OM proteins are first extracted from the OM and delivered to the proteasome (indicated by the ?). It is possible that components are recruited to the mitochondrial surface to extract the protein or that the proteasome itself is recruited to the mitochondrial surface to facilitate protein turnover. The second pathway (dashed line) dictates that a fraction of newly synthesized mitochondrial precursor proteins will be ubiquitinated and degraded by the 26S proteasome in the cytosol before reaching the mitochondria. Precursor proteins that contain mutations or have been mistargeted may also be degraded in the cytosol. OM, mitochondrial outer membrane; IMS, mitochondrial intermembrane space.
Although mounting evidence favors the turnover of mitochondrial OM proteins via the UPS, the molecular events that govern this process are poorly understood. In yeast, Vms1 has been shown to recruit Cdc48/VCP/p97 and Npl4 to the mitochondrial surface in response to mitochondrial stress (Heo et al. 2010). It is conceivable that Vms1 is required to link the components of the UPS to mitochondria to mediate the turnover of mitochondrial OM proteins. Clearly, further studies are required to define the role and the molecular mechanism of the UPS in the turnover of mitochondrial OM proteins. The identification of mitochondria-specific components of the UPS, like Vms1, will be instrumental to distinguish the degradation of mitochondrial precursor proteins prior to their targeting to mitochondria in the cytosol from the turnover of proteins already inserted into the OM.
PROTEOLYTIC CONTROL OF MITOCHONDRIAL FUSION AND MITOPHAGY
The dynamic nature of the mitochondrial network provides an additional level of quality control. Mutations in components of the mitochondrial fusion and fission machineries are associated with various neurodegenerative disorders, highlighting the importance of mitochondrial dynamics and quality control for neuronal survival (Chan 2006; Chen and Chan 2010). In addition to maintaining mitochondrial proteostasis, some mitochondrial proteases play central roles in mitochondrial fusion. It is therefore conceivable that protein damage within mitochondria impacts directly on mitochondrial dynamics, pointing to an interconnected network of different quality control mechanisms.
Processing of the Dynamin-like GTPase OPA1
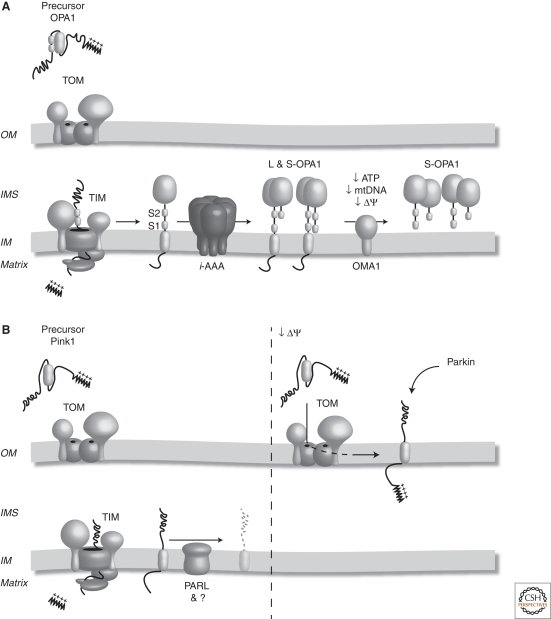
Fusion of the mitochondrial IM requires the dynamin-like GTPase Optic atrophy 1 (OPA1) (Olichon et al. 2003) or its homolog Mgm1 in yeast (Wong et al. 2000). Both proteins are subjected to proteolytic processing, illustrating that mitochondrial proteases regulate IM fusion. Long and short forms of Mgm1 are required to maintain the mitochondrial network and cristae morphology (Herlan et al. 2003). In mammalian mitochondria, long and short OPA1 isoforms are also observed, which arise because of alternative splicing as well as posttranslational protein processing of long isoforms at two sites, S1 and S2 (Fig. 5A) (Delettre et al. 2001; Satoh et al. 2003; Ishihara et al. 2006; Griparic et al. 2007). Similar to Mgm1, long and short OPA1 isoforms are required for mitochondrial fusion and normal cristae structures (Song et al. 2007). Cleavage of OPA1 and Mgm1 as well as the assembly of long and short isoforms is stimulated by cardiolipin, resulting in the stimulation of the GTPase activity, which promotes mitochondrial fusion (DeVay et al. 2009; Ban et al. 2010).
Figure 5.
Mitochondrial proteases influence mitochondrial morphology and turnover. (A) OPA1 is synthesized in the cytosol and targeted to mitochondria via its amino-terminal mitochondrial targeting sequence. Following import into the organelle, the targeting signal is cleaved and the hydrophobic transmembrane domain anchors OPA1 in the IM. Following lateral release into the lipid bilayer, OPA1 is subjected to constitutive processing at site 1 (S1) and by Yme1 at site 2 (S2), generating long and short forms of the protein (L and S OPA1) that ensure mitochondrial fusion. Under conditions of mitochondrial dysfunction, including decreased ATP, mtDNA, or membrane potential (ΔΨ), the long OPA1 isoforms are degraded by OMA1, resulting in mitochondrial fragmentation. (B) Pink1 is synthesized in the cytosol with a cleavable mitochondrial targeting sequence. Following its import into the IM, the mitochondrial targeting sequence is cleaved. Pink1 is then processed by PARL and turned over. Under conditions of mitochondrial dysfunction, such as a decrease in membrane potential (ΔΨ), Pink1 is alternately sorted to the mitochondrial OM where it can recruit Parkin to the mitochondrial surface and trigger mitophagy.
It is presently not understood how a balanced formation of long and short isoforms of OPA1 is ensured. The IM rhomboid protease, Pcp1, is responsible for cleavage of Mgm1 in yeast (McQuibban et al. 2003; Sesaki et al. 2003; Herlan et al. 2004). The mammalian homolog of Pcp1, PARL, has been linked to the release of short OPA1 isoforms from mitochondria during apoptosis and suggested to play a role in OPA1 processing (Cipolat et al. 2006). However, studies in cell lines lacking PARL, revealed that it is dispensable for OPA1 cleavage (Cipolat et al. 2006; Duvezin-Caubet et al. 2007; Ehses et al. 2009). Rather, the i-AAA protease Yme1 has been implicated in generation of at least one short form of OPA1 by constitutive cleavage at the processing site S2 (Song et al. 2007). Loss of Yme1 results in loss of a particular short isoform of OPA1 and is accompanied by a highly fragmented mitochondrial morphology (Griparic et al. 2007; Song et al. 2007). Components of the m-AAA protease have also been shown to influence OPA1 processing. In a heterologous system, subunits of the mammalian m-AAA protease were able to cleave OPA1 when coexpressed in yeast (Duvezin-Caubet et al. 2007). Unexpectedly, loss of the m-AAA protease in mammalian cells results in enhanced processing of the long OPA1 isoforms, accompanied by fragmentation of the mitochondrial network and loss of cristae structures (Ehses et al. 2009). How the m-AAA protease affects OPA1 processing therefore remains enigmatic.
Under conditions of mitochondrial dysfunction, including lowered ATP levels, loss of mtDNA and dissipation of the mitochondrial membrane potential across the IM, the short forms of OPA1 accumulate at the expense of the long OPA1 isoforms (Ishihara et al. 2004; Duvezin-Caubet et al. 2006; Baricault et al. 2007; Olichon et al. 2007; Guillery et al. 2008). Stress-induced degradation of long OPA1 isoforms was found to be mediated by OMA1 (Ehses et al. 2009; Head et al. 2009). This raises the exciting possibility that OMA1 is able to sense mitochondrial dysfunction and in doing so, acts to process OPA1 to promote mitochondrial fragmentation and possibly turnover through mitophagy. The increased cleavage of OPA1 by OMA1 occurs rapidly in response to different mitochondrial insults, ensuring that a damaged mitochondrion can be efficiently segregated from the network and removed through mitophagy in a rapid fashion.
PARL Processes Pink1 within Mitochondria
Early steps of mitophagy are under proteolytic control. Two proteins that have been associated with familial Parkinson's disease, PTEN-induced mitochondrial kinase Pink1 and the E3 ubiquitin ligase Parkin, are required for the selective removal of dysfunctional mitochondria through mitophagy (Kitada et al. 1998; Valente et al. 2004; Narendra et al. 2008). Although the precise role of Pink1 is unclear, recent work has shown that under physiological conditions Pink1 is imported into mitochondria and cleaved by the mitochondrial IM rhomboid protease PARL (Fig. 5B) (Deas et al. 2010; Jin et al. 2010). Following its cleavage, Pink1 is constitutively turned over by an as yet unknown protease that is sensitive to MG132. Under conditions of mitochondrial dysfunction, Pink1 is not directed to the IM and instead is alternatively sorted to the OM, where it can recruit Parkin to the mitochondrial surface (Deas et al. 2010; Jin et al. 2010; Narendra et al. 2010). Mitochondrial Parkin activates the UPS leading to a widespread degradation of OM proteins including Mfn1 and Mfn2 and finally triggers mitophagy (Narendra et al. 2008; Tanaka et al. 2010; Chan et al. 2011). Thus, stabilization of Pink1 at the OM and recruitment of Parkin to the mitochondrial OM is crucial to induce mitophagy (Narendra et al. 2010; Chan et al. 2011). It will be interesting to define the contribution of blocked OM protein degradation and mitophagy to the progression of Parkinson’s disease, especially with the recent observation that Parkin does not appear to be recruited to mitochondria in neuronal cells (Van Laar et al. 2011).
A MITOCHONDRIAL-SPECIFIC UNFOLDED PROTEIN RESPONSE
Although it is clear that a number of conserved chaperones and proteases are required to maintain mitochondrial proteostasis, a mitochondrial-specific UPR has evolved to cope with the increased unfolded protein load in mitochondria under stress conditions. In mammalian cells, deletion of mitochondrial DNA results in increased Hsp60 and Hsp70 levels, probably because of the accumulation of nuclear encoded subunits of the respiratory chain that are no longer able to assemble with their partner proteins encoded by the mitochondrial genome (Martinus et al. 1996). Furthermore, overexpression of a folding impaired protein in the mitochondrial matrix results in an increased expression of mitochondrial Hsp60, Hsp70, and also ClpP (Zhao et al. 2002). Conditions that cause mitochondrial dysfunction do not result in increased expression of ER chaperones, demonstrating a mitochondrial-specific UPR. CHOP was the first component identified to be required for the up-regulation of Hsp60 (Zhao et al. 2002; Horibe and Hoogenraad 2007). Proteins up-regulated by the mtUPR are activated through a CHOP element and transcriptional up-regulation involves the hetero-dimerization of the C/EBP homology protein CHOP and C/EBPβ (Zhao et al. 2002). CHOP itself is up-regulated in response to mitochondrial stress, as well as in response to ER stress, which suggests that additional transcription factors and promoter elements must be present to delineate between the two types of stress. Further analysis of the promoter region of proteins up-regulated in response to mitochondrial stress reveals two additional conserved mitochondrial unfolded response elements (MURE’s) (Aldridge et al. 2007). The transcription factors that bind to the additional MURE’s remain to be identified.
Studies in Caenorhabditis elegans have been useful in confirming the presence and defining the mechanism of a mitochondrial-specific UPR (Yoneda et al. 2004). A genome-wide RNAi screen revealed three genes involved in the mtUPR: a transcription factor, DVE-1, a ubiquitin-like protein, UBL-5, and a homolog of E. coli ClpP, CLPP-1 (Haynes et al. 2007). An additional transcription factor, ZC376.7 as well as the mitochondrial IM peptide exporter, HAF-1, are also involved in the mtUPR in C. elegans (Haynes et al. 2010). Identification of these components has allowed a model to be proposed for how the mtUPR is sensed, and the signal transduced to the nucleus for the up-regulation of mitochondrial chaperones in C. elegans (Haynes and Ron 2010). Unfolded proteins in the mitochondrial matrix may accumulate, where they can be degraded by the ATP-dependent protease ClpXP. In yeast, Mdl1 was shown to be required for the release of peptides from mitochondria generated by proteolysis (Young et al. 2001). Similar to Mdl1, the C. elegans homolog, HAF-1, is required for the efflux of peptides generated by ClpXP from the mitochondria. These peptides may act as a signal, which causes the transcription factor ZC376.7 to accumulate in the nucleus (Arnold et al. 2006; Haynes and Ron 2010). ZC376.7 and CHOP are both leucine zipper proteins and it is therefore conceivable that they play similar roles in the mtUPR in mammals and C. elegans. Upon mitochondrial stress, UBL-5 expression is enhanced and it associates with DVE-1 and the complex then interacts with the Hsp60 promoter. How the interaction of DVE-1 and UBL-5 affects ZC376.7 to facilitate mitochondrial chaperone expression is unclear and will certainly be the focus of future studies. Strikingly, recent studies in C. elegans suggest that the mtUPR is cell-nonautonomous (Durieux et al. 2011). When mitochondrial perturbation occurs in one tissue, it can be sensed and acted on in another distal tissue. It will be interesting to see if this is conserved in higher eukaryotes and to unravel how signaling is mediated.
CONCLUDING REMARKS—MITOCHONDRIAL QUALITY CONTROL IN AGING AND DISEASE
Mitochondria are essential organelles because of the numerous anabolic and catabolic roles that they play within the cell. The importance of functional mitochondria has been highlighted by the fact that situations that lead to mitochondrial dysfunction are often associated with aging and disease. Many of the diseases associated with mitochondrial dysfunction are progressive, manifesting later in life. Examples of this include Parkinson’s disease and Alzheimer’s disease, suggesting that for a period of time, mitochondria are able to cope with the challenges imposed on them. What happens over time for the rapid and in some cases insidious onset of the disease is not clear, however it is often related to mitochondrial dysfunction. By looking at the levels of mitochondrial quality control, we can begin to understand how disease progression may occur in such a fashion. Over time, ROS may begin to accumulate in cells, which results in a vicious cycle as the increased levels of damage promote increased levels of mitochondrial dysfunction, further increasing the production of ROS. At the early stages of disease, minor amounts of damage to mitochondrial proteins can be dealt with rapidly and efficiently because of the presence of the conserved molecular chaperones and proteolytic machineries. As the damage increases, the molecular quality control machinery may become overwhelmed and the second level of defense is put into action, using mitochondrial fusion and fission as a means to either complement mitochondrial function or target them for elimination through mitophagy. If the level of mitochondrial damage becomes too high, the pathway of mitophagy is blocked and mitochondria begin to release proapoptotic factors and the entire cell is removed. This may help to explain the gradual onset observed in neurological disease, in which the mechanisms that govern mitochondrial quality control become overwhelmed, eventually resulting in cell death. Neurons are postmitotic and depend on functional mitochondria for energy production. This renders neurons very susceptible to mitochondrial dysfunction, which may account for the large number of neurological diseases associated with mitochondrial dysfunction. We are gaining valuable insight into the mechanisms that govern mitochondrial quality control, however there many aspects remain poorly understood. Although there are distinct levels of mitochondrial quality control, it is clear that there is overlap and cross talk between the machineries. It is critical that we continue to study and unravel the mechanisms of mitochondrial quality control to further our understanding mitochondrial dysfunction in disease.
ACKNOWLEDGMENTS
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 635) and the European Research Council (REC-AdG-233078) to T.L. and a fellowship of the Alexander-von-Humboldt foundation to M.J.B.
Footnotes
Editors: Richard Morimoto, Dennis Selkoe, and Jeffrey Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Aldridge JE, Horibe T, Hoogenraad NJ 2007. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS ONE 2: e874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anesti V, Scorrano L 2006. The relationship between mitochondrial shape and function and the cytoskeleton. Biochim Biophys Acta 1757: 692–699 [DOI] [PubMed] [Google Scholar]
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T 1996. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85: 875–885 [DOI] [PubMed] [Google Scholar]
- Arnold I, Wagner-Ecker M, Ansorge W, Langer T 2006. Evidence for a novel mitochondria-to-nucleus signalling pathway in respiring cells lacking i-AAA protease and the ABC-transporter Mdl1. Gene 367: 74–88 [DOI] [PubMed] [Google Scholar]
- Augustin S, Nolden M, Muller S, Hardt O, Arnold I, Langer T 2005. Characterization of peptides released from mitochondria: Evidence for constant proteolysis and peptide efflux. J Biol Chem 280: 2691–2699 [DOI] [PubMed] [Google Scholar]
- Ban T, Heymann JA, Song Z, Hinshaw JE, Chan DC 2010. OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum Mol Genet 19: 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baricault L, Segui B, Guegand L, Olichon A, Valette A, Larminat F, Lenaers G 2007. OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp Cell Res 313: 3800–3808 [DOI] [PubMed] [Google Scholar]
- Bender T, Lewrenz I, Franken S, Baitzel C, Voos W 2011. Mitochondrial enzymes are protected from stress-induced aggregation by mitochondrial chaperones and the Pim1/LON protease. Mol Biol Cell 22: 541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn J 1990. Behavior of mitochondria in the living cell. Int Rev Cytol 122: 1–63 [DOI] [PubMed] [Google Scholar]
- Bota DA, Davies KJ 2002. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol 4: 674–680 [DOI] [PubMed] [Google Scholar]
- Bota DA, Van Remmen H, Davies KJ 2002. Modulation of Lon protease activity and aconitase turnover during aging and oxidative stress. FEBS Lett 532: 103–106 [DOI] [PubMed] [Google Scholar]
- Bota DA, Ngo JK, Davies KJ 2005. Downregulation of the human Lon protease impairs mitochondrial structure and function and causes cell death. Free Radic Biol Med 38: 665–677 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N 2009. Importing mitochondrial proteins: Machineries and mechanisms. Cell 138: 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC 2006. Mitochondria: Dynamic organelles in disease, aging, and development. Cell 125: 1241–1252 [DOI] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC 2011. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet 20: 1726–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC 2010. Physiological functions of mitochondrial fusion. Ann NY Acad Sci 1201: 21–25 [DOI] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC 2005. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem 280: 26185–26192 [DOI] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC 2007. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell 130: 548–562 [DOI] [PubMed] [Google Scholar]
- Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D’Adamio L, et al. 2006. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126: 163–175 [DOI] [PubMed] [Google Scholar]
- Craig EA, Kramer J, Shilling J, Werner-Washburne M, Holmes S, Kosic-Smithers J, Nicolet CM 1989. SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol Cell Biol 9: 3000–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sagarra MR, Mayo I, Marco S, Rodriguez-Vilarino S, Oliva J, Carrascosa JL, Castaño JG 1999. Mitochondrial localization and oligomeric structure of HClpP, the human homologue of E. coli ClpP. J Mol Biol 292: 819–825 [DOI] [PubMed] [Google Scholar]
- Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, Renton AE, Harvey RJ, Whitworth AJ, Martins LM, et al. 2010. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet 20: 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C, Griffoin JM, Kaplan J, Dollfus H, Lorenz B, Faivre L, Lenaers G, Belenguer P, Hamel CP 2001. Mutation spectrum and splicing variants in the OPA1 gene. Hum Genet 109: 584–591 [DOI] [PubMed] [Google Scholar]
- Desautels M, Goldberg AL 1982. Demonstration of an ATP-dependent, vanadate-sensitive endoprotease in the matrix of rat liver mitochondria. J Biol Chem 257: 11673–11679 [PubMed] [Google Scholar]
- DeVay RM, Dominguez-Ramirez L, Lackner LL, Hoppins S, Stahlberg H, Nunnari J 2009. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J Cell Biol 186: 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bella D, Lazzaro F, Brusco A, Plumari M, Battaglia G, Pastore A, Finardi A, Cagnoli C, Tempia F, Frontali M, et al. 2010. Mutations in the mitochondrial protease gene AFG3L2 cause dominant hereditary ataxia SCA28. Nat Genet 42: 313–321 [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277: 1990–1993 [DOI] [PubMed] [Google Scholar]
- Doyle SM, Wickner S 2009. Hsp104 and ClpB: Protein disaggregating machines. Trends Biochem Sci 34: 40–48 [DOI] [PubMed] [Google Scholar]
- Dunn CD, Tamura Y, Sesaki H, Jensen RE 2008. Mgr3p and Mgr1p are adaptors for the mitochondrial i-AAA protease complex. Mol Biol Cell 19: 5387–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A 2011. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144: 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Jagasia R, Wagener J, Hofmann S, Trifunovic A, Hansson A, Chomyn A, Bauer MF, Attardi G, Larsson NG, et al. 2006. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J Biol Chem 281: 37972–37979 [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Koppen M, Wagener J, Zick M, Israel L, Bernacchia A, Jagasia R, Rugarli EI, Imhof A, Neupert W, et al. 2007. OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA protease in mitochondria. Mol Biol Cell 18: 3582–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, Langer T 2009. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol 187: 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M, Westermann B, Langer T 2006. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J Cell Biol 173: 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser K, Tursun B, Ingenhoven M, Michaelis G, Pratje E 2002. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the m-AAA complex and the putative rhomboid protease Pcp1. J Mol Biol 323: 835–843 [DOI] [PubMed] [Google Scholar]
- Fan AC, Bhangoo MK, Young JC 2006. Hsp90 functions in the targeting and outer membrane translocation steps of Tom70-mediated mitochondrial import. J Biol Chem 281: 33313–33324 [DOI] [PubMed] [Google Scholar]
- Finley D 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S 1996. Proteases and their targets in Escherichia coli. Annu Rev Genet 30: 465–506 [DOI] [PubMed] [Google Scholar]
- Granot Z, Kobiler O, Melamed-Book N, Eimerl S, Bahat A, Lu B, Braun S, Maurizi MR, Suzuki CK, Oppenheim AB, et al. 2007. Turnover of mitochondrial steroidogenic acute regulatory (StAR) protein by Lon protease: The unexpected effect of proteasome inhibitors. Mol Endocrinol 21: 2164–2177 [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF 1999. Mitochondrial evolution. Science 283: 1476–1481 [DOI] [PubMed] [Google Scholar]
- Green DR 2005. Apoptotic pathways: Ten minutes to dead. Cell 121: 671–674 [DOI] [PubMed] [Google Scholar]
- Griparic L, Kanazawa T, van der Bliek AM 2007. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol 178: 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillery O, Malka F, Landes T, Guillou E, Blackstone C, Lombes A, Belenguer P, Arnoult D, Rojo M 2008. Metalloprotease-mediated OPA1 processing is modulated by the mitochondrial membrane potential. Biol Cell 100: 315–325 [DOI] [PubMed] [Google Scholar]
- Hansen JJ, Durr A, Cournu-Rebeix I, Georgopoulos C, Ang D, Nielsen MN, Davoine CS, Brice A, Fontaine B, Gregersen N, et al. 2002. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet 70: 1328–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Gregersen N, Bross P 2005. Differential degradation of variant medium-chain acyl-CoA dehydrogenase by the protein quality control proteases Lon and ClpXP. Biochem Biophys Res Commun 333: 1160–1170 [DOI] [PubMed] [Google Scholar]
- Haynes CM, Ron D 2010. The mitochondrial UPR–protecting organelle protein homeostasis. J Cell Sci 123: 3849–3855 [DOI] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D 2007. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell 13: 467–480 [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D 2010. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell 37: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM 2009. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol 187: 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J-M, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, et al. 2010. A stress-responsive system for mitochondrial protein degradation. Molecular Cell 40: 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlan M, Vogel F, Bornhövd C, Neupert W, Reichert AS 2003. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem 278: 27781–27788 [DOI] [PubMed] [Google Scholar]
- Herlan M, Bornhovd C, Hell K, Neupert W, Reichert AS 2004. Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J Cell Biol 165: 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhfeld J, Hartl FU 1994. Role of the chaperonin cofactor Hsp10 in protein folding and sorting in yeast mitochondria. J Cell Biol 126: 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Lackner L, Nunnari J 2007. The machines that divide and fuse mitochondria. Annu Rev Biochem 76: 751–780 [DOI] [PubMed] [Google Scholar]
- Horibe T, Hoogenraad NJ 2007. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS ONE 2: e835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL, Weber-Ban EU, Finley D 1999. Chaperone rings in protein folding and degradation. Proc Natl Acad Sci 96: 11033–11040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Eura Y, Mihara K 2004. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci 117: 6535–6546 [DOI] [PubMed] [Google Scholar]
- Ishihara N, Fujita Y, Oka T, Mihara K 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J 25: 2966–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon HB, Choi ES, Yoon JH, Hwang JH, Chang JW, Lee EK, Choi HW, Park ZY, Yoo YJ 2007. A proteomics approach to identify the ubiquitinated proteins in mouse heart. Biochem Biophys Res Commun 357: 731–736 [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ 2010. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 191: 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, Davies E, Hajnoczky G, Saunders TL, Van Keuren ML, et al. 2003. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature 425: 721–727 [DOI] [PubMed] [Google Scholar]
- Kang PJ, Ostermann J, Shilling J, Neupert W, Craig EA, Pfanner N 1990. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348: 137–143 [DOI] [PubMed] [Google Scholar]
- Kang SG, Ortega J, Singh SK, Wang N, Huang NN, Steven AC, Maurizi MR 2002. Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J Biol Chem 277: 21095–21102 [DOI] [PubMed] [Google Scholar]
- Kang SG, Dimitrova MN, Ortega J, Ginsburg A, Maurizi MR 2005. Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX. J Biol Chem 280: 35424–35432 [DOI] [PubMed] [Google Scholar]
- Käser M, Kambacheld M, Kisters-Woike B, Langer T 2003. Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J Biol Chem 278: 46414–46423 [DOI] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300: 486–489 [DOI] [PubMed] [Google Scholar]
- Kim KI, Park SC, Kang SH, Cheong GW, Chung CH 1999. Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein in Escherichia coli. J Mol Biol 294: 1363–1374 [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392: 605–608 [DOI] [PubMed] [Google Scholar]
- Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E 2008. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci 9: 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koodathingal P, Jaffe NE, Kraut DA, Prakash S, Fishbain S, Herman C, Matouschek A 2009. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J Biol Chem 284: 18674–18684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M, Langer T 2007. Protein degradation within mitochondria: Versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol 42: 221–242 [DOI] [PubMed] [Google Scholar]
- Koppen M, Metodiev MD, Casari G, Rugarli EI, Langer T 2007. Variable and tissue-specific subunit composition of mitochondrial m-AAA protease complexes linked to hereditary spastic paraplegia. Mol Cell Biol 27: 758–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel D, Wurth S, Käser M, Langer T 2004. Membrane protein turnover by the m-AAA protease in mitochondria depends on the transmembrane domains of its subunits. EMBO Rep 5: 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovermann P, Truscott KN, Guiard B, Rehling P, Sepuri NB, Muller H, Jensen RE, Wagner R, Pfanner N 2002. Tim22, the essential core of the mitochondrial protein insertion complex, forms a voltage-activated and signal-gated channel. Mol Cell 9: 363–373 [DOI] [PubMed] [Google Scholar]
- Krojer T, Sawa J, Schafer E, Saibil HR, Ehrmann M, Clausen T 2008. Structural basis for the regulated protease and chaperone function of DegP. Nature 453: 885–890 [DOI] [PubMed] [Google Scholar]
- Kuznetsov AV, Hermann M, Saks V, Hengster P, Margreiter R 2009. The cell-type specificity of mitochondrial dynamics. Int J Biochem Cell Biol 41: 1928–1939 [DOI] [PubMed] [Google Scholar]
- Lee S, Augustin S, Tatsuta T, Gerdes F, Langer T, Tsai FT 2010. Electron cryomicroscopy structure of a membrane-anchored mitochondrial AAA protease. J Biol Chem 286: 4404–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JV, Schapira AH 2000. Mitochondrial respiratory chain disorders II: Neurodegenerative disorders and nuclear gene defects. Lancet 355: 389–394 [DOI] [PubMed] [Google Scholar]
- Leonhard K, Herrmann JM, Stuart RA, Mannhaupt G, Neupert W, Langer T 1996. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J 15: 4218–4229 [PMC free article] [PubMed] [Google Scholar]
- Leonhard K, Guiard B, Pellecchia G, Tzagoloff A, Neupert W, Langer T 2000. Membrane protein degradation by AAA proteases in mitochondria: Extraction of substrates from either membrane surface. Mol Cell 5: 629–638 [DOI] [PubMed] [Google Scholar]
- Li B, Hu Q, Wang H, Man N, Ren H, Wen L, Nukina N, Fei E, Wang G 2010. Omi/HtrA2 is a positive regulator of autophagy that facilitates the degradation of mutant proteins involved in neurodegenerative diseases. Cell Death Differ 17: 1773–1784 [DOI] [PubMed] [Google Scholar]
- Lill R 2009. Function and biogenesis of iron-sulphur proteins. Nature 460: 831–838 [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF 2006. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443: 787–795 [DOI] [PubMed] [Google Scholar]
- Livnat-Levanon N, Glickman MH 2010. Ubiquitin-proteasome system and mitochondria–reciprocity. Biochim Biophys Acta 1809: 80–87 [DOI] [PubMed] [Google Scholar]
- Luce K, Osiewacz HD 2009. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat Cell Biol 11: 852–858 [DOI] [PubMed] [Google Scholar]
- Martin SJ 2010. Cell biology. Opening the cellular poison cabinet. Science 330: 1330–1331 [DOI] [PubMed] [Google Scholar]
- Martinelli P, Rugarli EI 2010. Emerging roles of mitochondrial proteases in neurodegeneration. Biochim Biophys Acta 1797: 1–10 [DOI] [PubMed] [Google Scholar]
- Martinelli P, La Mattina V, Bernacchia A, Magnoni R, Cerri F, Cox G, Quattrini A, Casari G, Rugarli EI 2009. Genetic interaction between the m-AAA protease isoenzymes reveals novel roles in cerebellar degeneration. Hum Mol Genet 18: 2001–2013 [DOI] [PubMed] [Google Scholar]
- Martins LM, Iaccarino I, Tenev T, Gschmeissner S, Totty NF, Lemoine NR, Savopoulos J, Gray CW, Creasy CL, Dingwall C, et al. 2002. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J Biol Chem 277: 439–444 [DOI] [PubMed] [Google Scholar]
- Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P, Abuin A, Grau E, Geppert M, Livi GP, et al. 2004. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol 24: 9848–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Hoj PB, Hoogenraad NJ 1996. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur J Biochem 240: 98–103 [DOI] [PubMed] [Google Scholar]
- Matsushima Y, Goto Y, Kaguni LS 2010. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM). Proc Natl Acad Sci 107: 18410–18415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuibban GA, Saurya S, Freeman M 2003. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423: 537–541 [DOI] [PubMed] [Google Scholar]
- Miao L, St Clair DK 2009. Regulation of superoxide dismutase genes: Implications in disease. Free Radic Biol Med 47: 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P 1961. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191: 144–148 [DOI] [PubMed] [Google Scholar]
- Modre-Osprian R, Osprian I, Tilg B, Schreier G, Weinberger KM, Graber A 2009. Dynamic simulations on the mitochondrial fatty acid β-oxidation network. BMC Syst Biol 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisoi N, Klupsch K, Fedele V, East P, Sharma S, Renton A, Plun-Favreau H, Edwards RE, Teismann P, Esposti MD, et al. 2009. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ 16: 449–464 [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM 2007. Translocation of proteins into mitochondria. Annu Rev Biochem 76: 723–749 [DOI] [PubMed] [Google Scholar]
- Neutzner A, Youle RJ 2005. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. J Biol Chem 280: 18598–18603 [DOI] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T 2005. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123: 277–289 [DOI] [PubMed] [Google Scholar]
- Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G 2003. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem 278: 7743–7746 [DOI] [PubMed] [Google Scholar]
- Olichon A, Guillou E, Delettre C, Landes T, Arnaune-Pelloquin L, Emorine LJ, Mils V, Daloyau M, Hamel C, Amati-Bonneau P, et al. 2006. Mitochondrial dynamics and disease, OPA1. Biochim Biophys Acta 1763: 500–509 [DOI] [PubMed] [Google Scholar]
- Olichon A, Elachouri G, Baricault L, Delettre C, Belenguer P, Lenaers G 2007. OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ 14: 682–692 [DOI] [PubMed] [Google Scholar]
- Osman C, Wilmes C, Tatsuta T, Langer T 2007. Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1FO-ATP synthase. Mol Biol Cell 18: 627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Mihara K 2011. Molecular mechanisms and physiologic functions of mitochondrial dynamics. J Biochem 149: 241–251 [DOI] [PubMed] [Google Scholar]
- Palade GE 1953. An electron microscope study of the mitochondrial structure. J Histochem Cytochem 1: 188–211 [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Wren BW 1997. The HtrA family of serine proteases. Mol Microbiol 26: 209–221 [DOI] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W 2003. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426: 862–866 [DOI] [PubMed] [Google Scholar]
- Pinton P, Brini M, Bastianutto C, Tuft RA, Pozzan T, Rizzuto R 1998. New light on mitochondrial calcium. Biofactors 8: 243–253 [DOI] [PubMed] [Google Scholar]
- Potting C, Wilmes C, Engmann T, Osman C, Langer T 2010. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J 29: 2888–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading DS, Hallberg RL, Myers AM 1989. Characterization of the yeast HSP60 gene coding for a mitochondrial assembly factor. Nature 337: 655–659 [DOI] [PubMed] [Google Scholar]
- Rehling P, Pfanner N, Meisinger C 2003. Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane—A guided tour. J Mol Biol 326: 639–657 [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Bernardi P, Pozzan T 2000. Mitochondria as all-round players of the calcium game. J Physiol 529: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodolfo C, Ciccosanti F, Giacomo GD, Piacentini M, Fimia GM 2010. Proteomic analysis of mitochondrial dysfunction in neurodegenerative diseases. Expert Rev Proteomics 7: 519–542 [DOI] [PubMed] [Google Scholar]
- Rugarli EI, Langer T 2006. Translating m-AAA protease function in mitochondria to hereditary spastic paraplegia. Trends Mol Med 12: 262–269 [DOI] [PubMed] [Google Scholar]
- Santagata S, Bhattacharyya D, Wang FH, Singha N, Hodtsev A, Spanopoulou E 1999. Molecular cloning and characterization of a mouse homolog of bacterial ClpX, a novel mammalian class II member of the Hsp100/Clp chaperone family. J Biol Chem 274: 16311–16319 [DOI] [PubMed] [Google Scholar]
- Satoh M, Hamamoto T, Seo N, Kagawa Y, Endo H 2003. Differential sublocalization of the dynamin-related protein OPA1 isoforms in mitochondria. Biochem Biophys Res Commun 300: 482–493 [DOI] [PubMed] [Google Scholar]
- Schmitt M, Neupert W, Langer T 1995. Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-hsp70. EMBO J 14: 3434–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M, Neupert W, Langer T 1996. The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J Cell Biol 134: 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo AY, Joseph AM, Dutta D, Hwang JC, Aris JP, Leeuwenburgh C 2010. New insights into the role of mitochondria in aging: Mitochondrial dynamics and more. J Cell Sci 123: 2533–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Southard SM, Hobbs AE, Jensen RE 2003. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem Biophys Res Commun 308: 276–283 [DOI] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, et al. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK 2004. Mitochondrial dysfunction is a common phenotype in aging and cancer. Ann NY Acad Sci 1019: 260–264 [DOI] [PubMed] [Google Scholar]
- Song Z, Chen H, Fiket M, Alexander C, Chan DC 2007. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol 178: 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Beil A, Ehrmann M 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97: 339–347 [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M 1997. α-Synuclein in Lewy bodies. Nature 388: 839–840 [DOI] [PubMed] [Google Scholar]
- Stadtman ER, Berlett BS 1998. Reactive oxygen-mediated protein oxidation in aging and disease. Drug Metab Rev 30: 225–243 [DOI] [PubMed] [Google Scholar]
- Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D, Gasser T, Wszolek Z, Muller T, Bornemann A, et al. 2005. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson’s disease. Hum Mol Genet 14: 2099–2111 [DOI] [PubMed] [Google Scholar]
- Suzuki CK, Suda K, Wang N, Schatz G 1994. Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science 264: 273–276 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Takahashi-Niki K, Akagi T, Hashikawa T, Takahashi R 2004. Mitochondrial protease Omi/HtrA2 enhances caspase activation through multiple pathways. Cell Death Differ 11: 208–216 [DOI] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol 191: 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L 2005. Twenty years of the Alzheimer’s disease amyloid hypothesis: A genetic perspective. Cell 120: 545–555 [DOI] [PubMed] [Google Scholar]
- Tatsuta T, Augustin S, Nolden M, Friedrichs B, Langer T 2007. m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J 26: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T, Langer T 2008. Quality control of mitochondria: Protection against neurodegeneration and ageing. EMBO J 27: 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, et al. 2009. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J 28: 1589–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Hyde B, Shirihai OS 2008. Mitochondrial fusion, fission and autophagy as a quality control axis: The bioenergetic view. Biochim Biophys Acta 1777: 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, et al. 2004. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304: 1158–1160 [DOI] [PubMed] [Google Scholar]
- Van Dyck L, Pearce DA, Sherman F 1994. PIM1 encodes a mitochondrial ATP-dependent protease that is required for mitochondrial function in the yeast Saccharomyces cerevisiae. J Biol Chem 269: 238–242 [PubMed] [Google Scholar]
- Van Laar VS, Arnold B, Cassady SJ, Chu CT, Burton EA, Berman SB 2011. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum Mol Genet 20: 927–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendelin M, Beraud N, Guerrero K, Andrienko T, Kuznetsov AV, Olivares J, Kay L, Saks VA 2005. Mitochondrial regular arrangement in muscle cells: A “crystal-like” pattern. Am J Physiol Cell Physiol 288: C757–C767 [DOI] [PubMed] [Google Scholar]
- Voos W 2009. Mitochondrial protein homeostasis: The cooperative roles of chaperones and proteases. Res Microbiol 160: 718–725 [DOI] [PubMed] [Google Scholar]
- Wagner I, Arlt H, van Dyck L, Langer T, Neupert W 1994. Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO J 13: 5135–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Klionsky DJ 2011. Mitochondria removal by autophagy. Autophagy 7: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewski M, Scorrano L 2009. The changing shape of mitochondrial apoptosis. Trends Endocrinol Metab 20: 287–294 [DOI] [PubMed] [Google Scholar]
- Winklhofer KF, Haass C 2010. Mitochondrial dysfunction in Parkinson’s disease. Biochim Biophys Acta 1802: 29–44 [DOI] [PubMed] [Google Scholar]
- Wong ED, Wagner JA, Gorsich SW, McCaffery JM, Shaw JM, Nunnari J 2000. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J Cell Biol 151: 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D 2004. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci 117: 4055–4066 [DOI] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP 2011. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T 2001. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science 291: 2135–2138 [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112: 41–50 [DOI] [PubMed] [Google Scholar]
- Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, et al. 2004. The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann Neurol 55: 164–173 [DOI] [PubMed] [Google Scholar]
- Zeng X, Neupert W, Tzagoloff A 2007. The metalloprotease encoded by ATP23 has a dual function in processing and assembly of subunit 6 of mitochondrial ATPase. Mol Biol Cell 18: 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ 2002. A mitochondrial specific stress response in mammalian cells. EMBO J 21: 4411–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]