Abstract
The first lineage segregation in the pre-implantation mouse embryo gives rise to cells of the inner cell mass and the trophectoderm. Segregation into these two lineages during the 8-cell to 32-cell stages is accompanied by a significant amount of cell displacement, and as such it has been difficult to accurately track cellular behavior using conventional imaging techniques. Consequently, how cellular behaviors correlate with cell fate choices is still not fully understood. To achieve the high spatial and temporal resolution necessary for tracking individual cell lineages, we utilized two-photon light-scanning microscopy (TPLSM) to visualize and follow every cell in the embryo using fluorescent markers. We found that cells undergoing asymmetric cell fate divisions originate from a unique population of cells that have been previously classified as either outer or inner cells. This imaging technique coupled with a tracking algorithm we developed allows us to show that these cells, which we refer to as intermediate cells share features of inner cells but exhibit different dynamic behaviors and tendency to expose their cell surface in the mouse embryo between the fourth and fifth cleavages. We provide an accurate description of the correlation between cell division order and cell fate, and demonstrate that cell cleavage angle is a more accurate indicator of cellular polarity than cell fate. Our studies demonstrate the utility of two-photon imaging in answering questions in the pre-implantation field that have previously been difficult or impossible to address. Our studies provide a framework for the future use of specific markers to track cell fate molecularly and with high accuracy.
Keywords: Two-photon microscopy, pre-implantation, asymmetric division, cell-fate determination, lineage tracing, trophectoderm, inner cell mass
Introduction
The mouse pre-implantation embryo has been a useful model in which to study how cell fate determinations are made. After five rounds of division, the 32-cell embryo possesses the first committed cell lineages, the inner cell mass (ICM) and the trophectoderm (TE). The ICM contains apolar cells that further segregate into the epiblast (EPI) and primitive endoderm (PE), while the outer, polar cells form the TE. During and prior to the 8-cell stage all cells have a large amount of exposed surface as well as areas of contact with neighboring cells. It is only during the transition from an 8-cell to a 16-cell embryo that some cells become completely enclosed by other cells (Johnson and McConnell, 2004; Marikawa and Alarcon, 2009; Zernicka-Goetz, 2006). Although these initial inside cells are biased toward the ICM and outside cells toward the TE, their identities are not yet fixed.
Much effort has been devoted to study how cells in the pre-implantation embryo gradually sort into distinct and committed lineages. However, the origin of outer and inner cells found in the 32-cell embryo still remains a subject of debate. Depending on the methods and criteria used, previous studies have found anywhere from 6–8 (Fleming, 1987; Suwinska et al., 2008) to only 1–2 (Dietrich and Hiiragi, 2007) inner cells at the 16-cell stage. Additionally, to what extent the outer cells at the 16-cell stage can give rise to inner cells is unclear. Some studies have reported that asymmetric cell divisions generally occur perpendicular to the embryo surface and give rise to inner and outer cells, whereas symmetric divisions that occur tangential to the embryo surface only result in outer cells (Bischoff et al., 2008; Zernicka-Goetz, 2005). Yet other studies did not find such a correlation (Dard et al., 2009b). Resolving these controversies will rely on live-imaging techniques that allow for the tracking of individual cells in the entire embryo at much higher spatial and temporal resolutions than has previously been achieved.
Imaging the pre-implantation mouse embryo is challenging however, as it is very sensitive to prolonged periods of light exposure. To ensure proper development, 15–30 min time intervals have been used to acquire time-lapse movies in various studies (Dard et al., 2009a; Jedrusik et al., 2008; Morris et al., 2010). Furthermore, given the depth (~100 μm) of the mouse embryo and dense cytoplasmic content, conventional confocal microscopy techniques suffer considerably in their ability to obtain a consistent signal throughout the entire depth of the embryo. This poses a significant problem when trying to reconstruct and track individual cells along with their lineages and fates, and do so for every cell in the embryo. Additionally, the sensitivity and dense nature of the pre-implantation embryo make it difficult to track endogenous fluorescent markers that have insufficient brightness, while the use of micro-injected dyes or markers has the potential to compromise developmental integrity.
Here we report the use of two-photon light scanning microscopy (TPLSM) to achieve superior time-resolution, complete depth of penetration, high viability, and non-invasive imaging of the pre-implantation mouse embryo. Coupled with tracking algorithms specifically tailored for the dividing mouse embryo we are able to extract detailed behaviors and create a complete lineage map for every cell in the embryo, providing new insights into how cell position is tied to cell fate, and definitively assign the origins of inner and outer cells.
Results
Two-photon imaging maintains viability and developmental competency
We chose to study the period of development between the 3rd and 6th cleavages as these are the stages in which the embryo segregates into inner and outer cell layers, and the first cell fate decisions are made. To maintain the developmental integrity and viability of embryos in culture while still being able to track individual cells we created a transgenic mouse model expressing Histone-2B-GFP (see Materials and Methods). Embryos possessing this transgene begin to express Histone-2B-GFP at a 4-cell stage, allowing us to trace cell lineages from their early origins.
Embryos were collected at 1.5 days post coitum (d.p.c.) at a 2-cell or 4-cell stage and allowed to develop overnight in a tissue-culture incubator before being placed on the stage the next morning after reaching the 8-cell stage. While GFP has a broad two-photon spectrum with several excitation peaks we chose to image with a wavelength of 820 nm, as this provided the highest signal intensity with the least amount of noise on our particular system. Time-lapse movies were taken every 6–7 min for a full z-stack with 2 μm sections (~53 sections total) until the embryos developed to the blastocyst stage. This allowed us to resolve individual nuclei in every cell with great clarity and accuracy (Fig. 1A, Supplementary Movie 1). We observed that time intervals longer than 10 minutes lead to difficulties in tracking and inaccuracies in calculating the cell division angle as cellular movements during mitosis are too dynamic to capture accurately with long time intervals (data not shown). Embryos averaged a delay of 6–10 hours to reach the blastocyst stage compared to those cultured without laser irradiation. Notably, embryos that did not express the H2B-GFP transgene but were imaged alongside their fluorescent litter-mates often reached the blastocyst stage faster and in some cases showed almost no delay compared to non-irradiated culture.
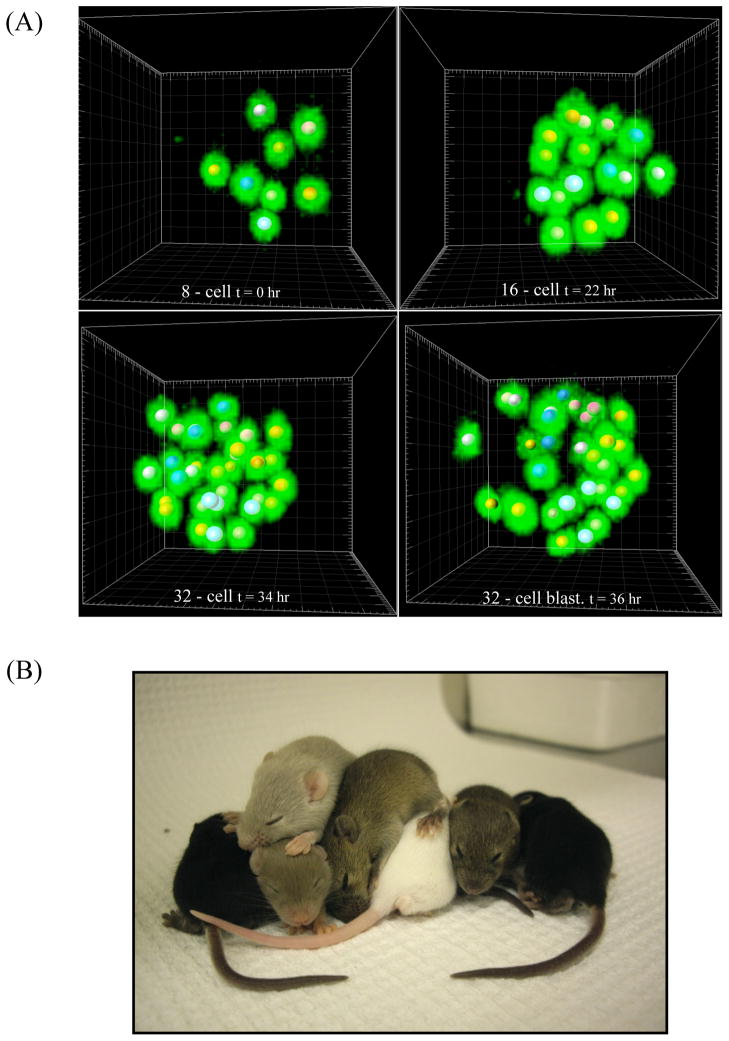
Figure 1.
Two-photon live imaging of pre-implantation development. (A) 3D reconstruction of mouse embryos expressing H2B-GFP (green) imaged by TPLSM in IMARIS software at different time-points during the 8, 16, 32, and 32-cell blastocyst stages. Z resolution 2 μm, 820 nm wavelength. Each 8-cell nucleus is marked by a ball of a distinct color, which is also used to mark all of that 8-cell’s progeny. (B) P12 mouse pups born from embryo transfer after being imaged by TPLSM for 48 hours from an 8 to 32/64-cell stage under the same conditions as in (A).
To test the developmental competency of embryos exposed to these imaging conditions, we transplanted imaged embryos into a pseudo-pregnant CD1 wild-type female. Pups carrying the H2B-GFP transgene were born naturally 18 days later and reared to P12 with normal appearance and behavior (Fig. 1B). Additionally, we repeated our results on a second TPLSM system with an identical Ti:Sapphire laser using the same growth conditions. This second system allowed for acquisition of a z-series at more than twice the speed of the first, and under these conditions embryos experienced negligible delay compared to non-irradiated in vitro culture (data not shown). This demonstrates that TPLSM provides superior temporal and spatial resolution as well as high viability for studying pre-implantation development.
Cells giving rise to both outer and ICM cell fates occupy unique positions
We reconstructed time-lapse movies into 3D using IMARIS software (Bitplane, AG), which allowed us to clearly visualize and follow embryo development over the entire course of the time-series. Additionally, by using the Surfaces function coupled with image overlays from the bright-field channel we were able to model an estimated projection of the embryo surface. While this is not a completely accurate or quantitative prediction of the embryo surface it allowed us to visually estimate the position of a cell’s nucleus relative to the embryo surface and to construct lineages trees from the 8-cell to 32-cell stage. We defined the outer cells as those whose nuclei are closest to the outer surface of the embryos. Using this criterion, we found that at the 16-cell stage 72.3% of cells clearly localized to the outer layer and would contribute to the extra-embryonic lineages. Of these outer cells 81.9% underwent symmetric cell division to only give rise to TE cells. Interestingly, while the progeny from the remaining 18.1% of 16-cell outer parents initially localized to the outer surface of the embryo one daughter from these parents would suddenly fall inward and re-localize to the inside of the embryo just prior to or during cavitation of the 32-cell embryo (Fig. 2A and Supplemental Movie 2). Typically only 1–2 outer cells in the 32-cell stage embryo experienced this internalization. Since these relocated cells appear at the surface of the ICM facing the blastocoel cavity, they are likely giving rise to the primitive endoderm (PE) lineages. We will refer to these cells as transient-outer cells to distinguish them from the TE cells. However, longer-term imaging beyond the 32-cell stage using genetic markers is needed to define their cell fate.
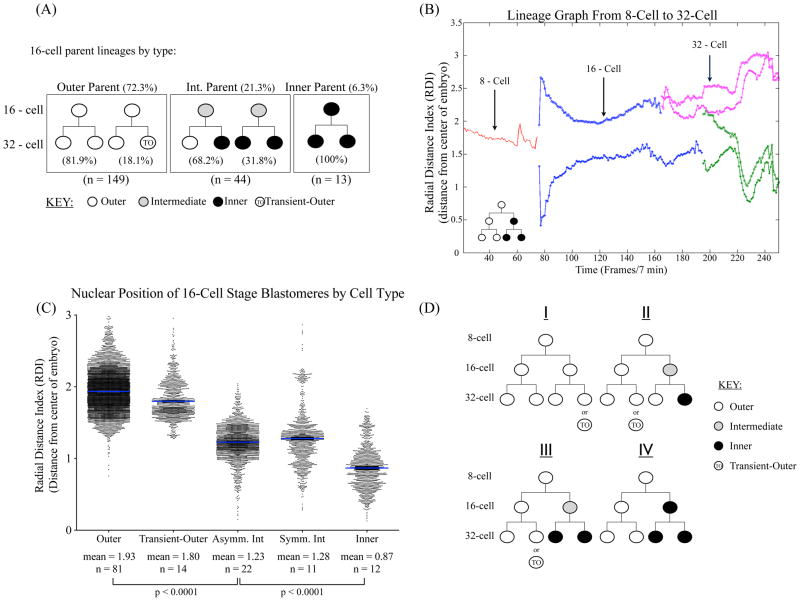
Figure 2.
Three cell populations in 16-cell embryos identified by lineage tracing (A) Numbers and percentages for each of the three 16-cell stage cell types. Outer 16-cell parents account for 72.3% of all 16-cell stage cells, while intermediate parents constitute 21.3% and inner parents 6.3%. 16-cell stage outer cells divide symmetrically to produce two TE 32-cell daughters 81.9% of the time, and the remaining 18.1% give rise to one TE and one transient-outer, putative PE daughter cell. Intermediate cells found at the 16-cell stage divide to give rise to asymmetric outer and inner 32-cell daughters 68.18% or entirely inner progeny 31.82% of the time. By contrast, inner 16-cell stage cells only give rise to inner daughters at the 32-cell stage (100% of the time).. (B) Example of a lineage graph depicting the position of a cell’s nuclei from the center of the embryo. Radial distance index (RDI) plotted for each time-point over the course of the entire time-lapse (see Materials and Methods). RDI 0 represents the center of the embryo. The lineage in this example follows a single 8-cell parent (red line) into two 16-cell daughters (blue lines) and 4 32-cell granddaughters (pink and green lines). This particular lineage gave rise to two outer TE cells (pink) and two inner ICM cells (green). (C) The radial distance index (RDI) was calculated for each cell at each time point during the 16-cell stage from the first frame in which the 16-cell appeared after the 4th cleavage to the first frame when the cell undergoes the 5th cleavage. The RDI is calculated as the distance of each nucleus from the center of the embryo, 0 (see Materials and Methods). The RDI for each time point for each cell during the 16-cell stage was then grouped according to that cell’s lineage: outer, transient-outer, asymmetric and symmetric intermediate cells, and inner cells as determined by lineage tracing and visual analysis, resulting in a dot-plot of all cell positions for each time point during the 16-cell stage for each category. There is a significant difference in RDI between outer cells and the intermediate cells, as well as inner and intermediate cells (p-value <0.0001). (D) Lineage trees for the 8–32 cell stage. Four different lineages starting from the 8-cell stage are described in the text, giving rise to both inner and outer progeny by the 32-cell blastocyst stage. n refers to the number of cells analyzed in each category.
We defined inner cells as those that have their nuclei clearly surrounded by the nuclei of its neighbors. By this criterion, we found that only 6.3% of cells occupied this position in 16-cell embryos, and that these cells only gave rise to ICM progenies (Fig. 2A). This is consistent with previous reports that found only 1–2 inner cells in the 16-cell embryo (Dietrich and Hiiragi, 2007). We found that the remaining 21.4% of cell nuclei occupied a position between the inner cells and the outer cells as defined above, and that while they were predicted to expose at least some of their cell surface during the 16-cell stage they were located more inward compared to nuclei of the solely outer lineages. Of these cells, 68.2% underwent asymmetric divisions to produce both TE and ICM daughters, whereas the remainder underwent symmetric divisions to give rise to two ICM daughters (Fig. 2A). Importantly, we found that none of these cells produced two symmetric outer daughters. These analyses suggest that in the 16-cell stage embryo cells that give rise to both inner and outer progeny occupy a unique intermediate position between the inner and outer cells.
Development of a cell-tracking algorithm to analyze dynamic nuclear positions in the embryo
The high temporal and spatial resolutions provided by TPLSM allowed us to more quantitatively analyze dynamic cell behaviors from the 8-cell to 32-cell stages using nuclei positions. We reconstructed the time-lapse images into 3D movies using IMARIS software and marked nuclei positions using the Spot Identification Function. Each embryo was then manually checked to ensure proper identification of nuclei for every frame in the movie. Coordinates for each nuclear position at each frame were then exported for tracking. To trace and calculate the position of each individual nucleus within an embryo for each frame, we developed an automated tracking algorithm and convex hull model, which is used to predict the embryo surface and calculate the center of the embryo at each frame (see Materials and Methods for details). This allowed us to not only trace cell lineages but to also analyze the dynamics of individual cell positioning based on their nuclear positions. Tracking and reconstruction results were then verified manually for every cell in each embryo.
In order to quantitatively resolve whether or not a cell’s nuclear position correlates with its lineage we calculated the radial distance index (RDI) for each cell nucleus for each time point from the beginning of the time-lapse to the end. Briefly, the RDI is defined as the distance from the center of the embryo to the center of a cell’s nucleus (see Materials and Methods). This allowed us to construct lineage graphs tracking nuclear position over time from the 8-cell to 32-cell stages for each 8-cell and all of its progeny, an example of which is shown in Fig. 2A. We then grouped and plotted the RDI during the 16-cell stage for each 16-cell stage blastomere as follows: those 16-cell parents that gave rise to two outer TE progenies are denoted as “Outer”, and those outer 16-cell parents that give rise to putative PE progenies “Transient-Outer”. Cells whose nuclei were determined visually during the 16-cell stage to occupy an intermediate position between the innermost and outermost nuclei were grouped according to whether or not they gave rise to asymmetric TE and ICM or symmetric ICM progenies. The inner lineages are denoted simply as “Inner” (Fig. 2C).
The 16-cell parents that gave rise to outer or transient-outer cells share similar nuclear positions on the outer surface of the embryo with a mean RDI of 1.93 ± 0.25 and 1.80 ± 0.25, respectively. Inner nuclei reside closest to the center of the embryo averaging an RDI of 0.87 ± 0.29, and intermediate cell nuclei occupy a middle position in the embryo regardless of whether they give rise to asymmetric or symmetric inner 32-cell progeny (mean RDI of 1.23 ± 0.23 and 1.28 ± 0.37, respectively) (Fig. 2C). These quantitative analyses demonstrate that the intermediate cells that we identified visually indeed occupy a position between the inner cells and outer cells during the 16-cell stage of embryo development.
We used this information to construct hierarchical lineage trees and describe four different lineages originating from the 8-cell stage (Fig. 2D). The first lineage (I) gives rise to an entirely outer lineage or outer with one transient-outer daughter. Lineage II has one outer and one bi-potent intermediate cell at the 16-cell stage in which the outer 16-cell will give rise to outer or transient-outer, and the bi-potent intermediate cell divides to give outer and inner 32-cell daughters. Lineage III has a similar 16-cell stage combination as Lineage II, only in this lineage the intermediate cell gives rise to two inner 32-cell daughters. Lineage IV possesses the “true” inner cells, which only give rise to ICM progeny.
Consistent with previous reports (Yamanaka et al., 2010) we found that approximately 56% of 8-cells underwent asymmetric cell division to contribute to both inner and outer cell lineages. These consist of 42% that divided into one outer and one intermediate 16-cell and 14% that gave rise to one outer and one inner 16-cell daughter. The remaining 44% of 8-cell parents divided to give rise to two outer 16-cell daughters, and of these 18% give rise to transient-outer progenies that would later become internalized to form what appear to be initial PE cells.
Intermediate cells tend to have exposed outer surfaces but express low levels of Cdx2
We next sought to determine whether the intermediate cells identified above had a higher probability of exposing their cell-surface membranes during the 16-cell stage than those of inner cells. Since cell membranes cannot be visualized live by available methods without introducing potential artifacts by manipulations involving microinjection or dye staining, we chose to isolate embryos throughout the 16-cell stage and stain them with Alexa Fluor Phalloidin, which reveals the whole outline of individual cells in the embryo. This allowed us to clearly visualize outer cell surfaces and cell boundaries and correlate surface exposure with nuclear RDI through 3D reconstruction. We grouped cells in each embryo based on their nuclear RDI into three categories: RDIs between 2.3 and 1.56 (correlating with > 90% of all nuclear positions of outer-cell lineages, including transient-outer cells), RDIs from 1.55 to 1.0 (correlating with 90% of nuclear positions of the asymmetric intermediate cell lineages), and RDIs less than 1.0. While visual classification of nuclear position is somewhat arbitrary, by using nuclear RDI we are able to set discrete cut-offs according to cell lineages. Whereas it is possible that some cells from the inner cell lineage may over-lap with those of intermediate nuclear position, it is important to note that those 16-cells which undergo asymmetric cell fate division are found within this 1.55 – 1.0 RDI range at least 90% of the time.
We found that of nuclei occupying an RDI between 2.3 and 1.56, corresponding with the outer lineages, 100% had exposed outer cell surface. Those nuclei found between 1.55 and 1.0 displayed some membrane exposure 72.19% of the time. Cells with nuclei below an RDI of 1.0 exposed their surfaces only 22.46% of the time (Fig. 3A). Notably, in nearly half (11/24) of the embryos analyzed 15 out of 16 cells in the embryo had membrane exposure. Examples of intermediate cell positions and exposures are presented in Fig. 3B. Additionally, we observed that cells with nuclear positions between 1.55 and 1.0 had cell shapes similar to those with RDIs of less than 1.0, regardless of membrane exposure. Cells within these ranges had relatively spherical, polyhedral-like shapes. Furthermore, their nuclei tended to be positioned near the center of the cell mass. These results clearly show that intermediate cells have a higher probability to have some outer membrane exposure than do inner cells, and that this is not due to irregular cell-shapes. However, using immunostaining analyses we cannot explore the possibility that individual intermediate cells experience periods of membrane exposure followed by complete enclosure. Future analysis with a viable membrane-marker and real-time imaging will be critical in determining this dynamicity.
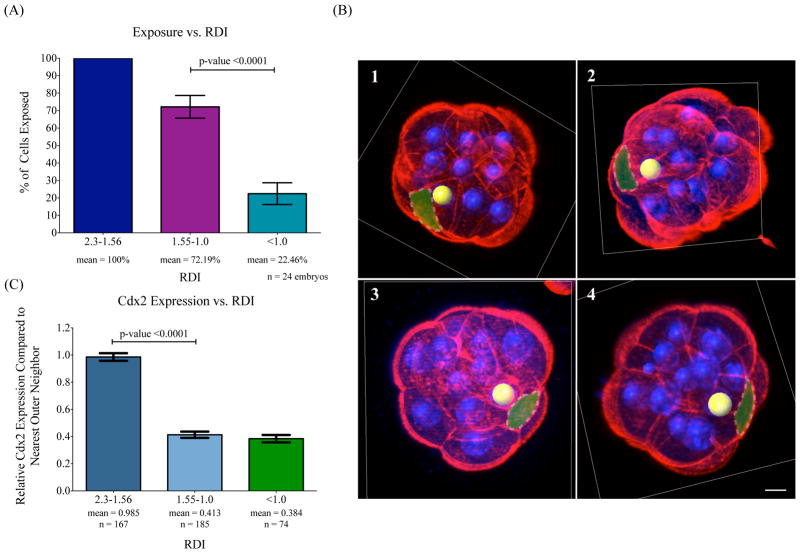
Figure 3.
Intermediate cells in 16-cell stage embryos have outer surface exposure and low levels of Cdx2 expression. (A) Cell surface exposure versus RDI. Cells with outer membrane exposure based on phalloidin staining of actin are grouped according to their RDIs. Cells with an RDI between 2.3 and 1.56 have 100% exposure, RDIs between 1.55 and 1.0 are exposed 72.19% of the time, and RDIs of less than 1.0 are exposed 22.46% of the time. n refers to the number of embryos analyzed. Error bars represent standard error of the mean (SEM). Significantly higher percentages of cells with RDI between 1.55-1.0 are exposed than those with RDI of less than 1.0, p-value <0.0001. (B) Examples of intermediate cells with surface exposure. Four examples of individual 16-cell stage embryos are shown with Alexa Fluor 568 Phalloidin in red and DAPI staining in blue. Spots represent nuclear positions and embryos are cut with orthogonal slices in the IMARIS program for easier visualization. Yellow spots denote nuclei in intermediate positions whose cell-surface is highlighted using a transparent green layover. The RDI of each yellow-marked cell nucleus is as follows 1) 1.37, 2) 1.25, 3) 1.19, and 4) 1.38. Scale bar = 10 μm. (C) Cdx2 expression versus nuclear position. The relative Cdx2 expression of a cell’s closest, outer-most neighbor is compared between cells with an RDI of 2.3-1.56; mean of 0.985, 1.55-1.0; mean 0.413, and RDI values less than 1.0; mean 0.384. n refers to the number of cells analyzed in each category. Error bars represent SEM. Expression levels between RDIs of 2.3-1.56 and 1.55-1.0 are significantly different, p-value <0.0001. n refers to the number of cells analyzed.
To further analyze the intermediate cells, we characterized Cdx2 and Oct4 expression by immunohistochemistry in 16-cell embryos. Oct4 expression, essential for the inner cell fates, is uniform throughout the 16-cell stage embryo, and we found no correlation between RDI and Oct4 expression (data not shown). Similar to previous studies, we found that the expression of Cdx2, which is critical for TE cell fates, is already restricted to cells on the outside of 16-cell stage embryos (Strumpf et al., 2005). We found that cells with a nuclear RDI greater than 1.56, corresponding to the outer lineages, showed a greater than 2-fold average expression of Cdx2 than those in intermediate or inner positions (RDI 1.55-1.0, and <1.0) (Fig. 3C). Additionally, there was no significant difference in Cdx2 expression between cells with nuclear RDIs from 1.55 to 1.0 and those with RDIs of less than 1.0 (Fig. 3C). Together, the above analyses show that the intermediate cells represent a unique group of cells that, while appearing as outer cells based on their tendency to expose their cell-surface, appear as inner cells based on Cdx2 expression.
Cell-cycle timing and division order correlate with cell fate
With the identification of the intermediate cell population in 16-cell embryos, we next evaluated their behavior as compared to outer or inner cells. To this end, we compared the cell-cycle timing and division order of these cells to outer and inner cells. Previous studies have shown that inner cells take longer to divide from the 4th to 5th cleavages than outer cells, indicating a connection between cell position, identity and behavior (MacQueen and Johnson, 1983). To determine the cell-cycle timing of each of the three cell populations we identified during the 16-cell stage we calculated the time between the 4th and 5th cleavage as follows.
During the transition from 8 to 16-cells (4th cleavage), the first frame in which the dividing nuclei had separated into two distinct nuclei (usually 1–2 frames after the metaphase plate appeared) was marked as time-start for both daughter cells. Each daughter cell was then tracked for the entirety of the 16-cell stage and its lineage and position determined as described previously. During the next round of division from 16 to 32 cells (5th cleavage) the first frame in which the 16-cell daughter separated into two distinct nuclei was marked as time-final. Each daughter from this division was tracked and its final position determined to give a complete lineage from the 16-cell to 32-cell stages. The cell-cycle time for each cell during the 16-cell stage was then determined as the interval between time-start and time-final, and grouped according to lineage.
We found that, as previously described, inner cells had a significantly longer cell-cycle time during the 16-cell stage than outer cells (Fig. 4A). Interestingly, the outer cells in the 16-cell embryos that gave rise to the transient-outer cells in the 32-cell embryo (Fig. 2B) showed no difference in their cell-cycle time from the outer 16-cell parents that gave rise to two TE 32-cell daughters. The intermediate cells behaved like inner cells, having comparable cell-cycle times to inner cells regardless of whether they gave rise to 32-cell symmetric ICM or asymmetric ICM and TE progenies.
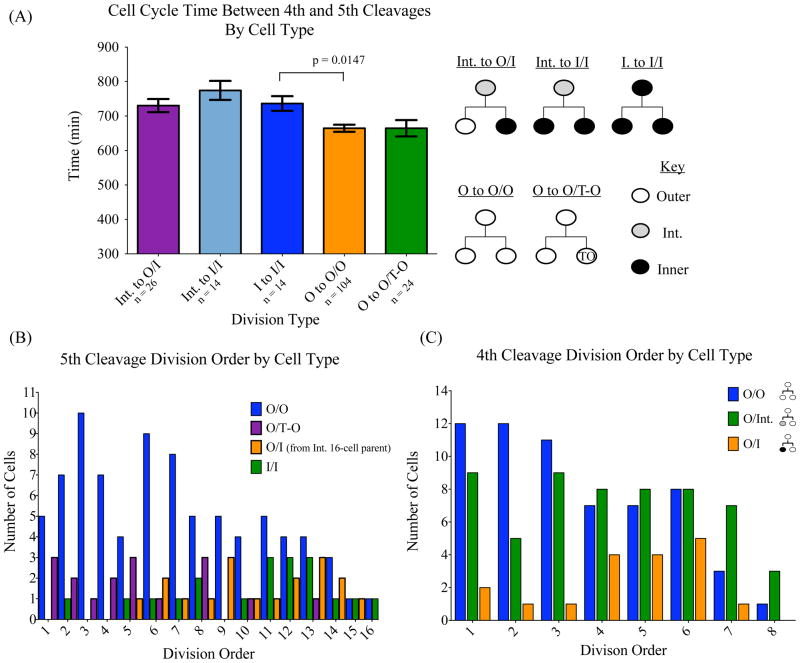
Figure 4.
Cell cycle length and cell lineage. (A) A graph of average cell-cycle length as grouped by cell lineage. For each 16-cell the cell cycle time was calculated as the time between the start frame in which the 16-cell first split from its 8-cell parent during the 4th cleavage and the end frame in which the 16-cell split into two 32-cell daughters during the 5th cleavage. 16-cell stage cells were grouped according to their lineage; Int. to O/I refers to 16-cell intermediate cells that give rise to outer and inner 32-cell progenies. Int. to I/I refers to 16-cell intermediate cells that give rise to two inner 32-cell progenies. I to I/I refers to 16-cell inner cells that give rise to two inner 32-cell progenies. O to O/O refers to 16-cell outer cells that give rise to two outer cell progenies. O to O/T-O refers to 16-cell outer cells that give rise to an outer and a transient outer cell progenies. Lineages for each bar are shown to the right of the graph. n refers to the number of cells analyzed in each category. Error bars represent SEM. p-value = 0.0147. (B) Cell division order as grouped by lineage type at the 5th cell division. 16-cell parents that divided to give rise to two outer 32-cell daughters (O/O) divided early in the 5th cleavage, while divisions from 16-cell parents that resulted in two ICM (I/I) progeny occurred later. 16-cell outer cells that divide into an outer TE cell and a transient-outer cell (O/T-O) that occupies the ICM at the 32-cell stage again behave like outer cells and divide earlier than inner or bi-potent intermediate cells (O/I). (C) Cell division order as grouped by lineage type at the 4th cell division. The cleavage order was determined by following the order in which cells during the 4th and 5th cleavages divided and grouping them according to lineage. During the 4th cleavage 8-cell parents which gave rise to two outer 16-cell daughters (O/O) preferentially divided before other cells in the embryo, while the cleavage order of 8-cell parents which divided to give rise to outer and a bi-potent intermediate daughters (O/Int.) was more evenly distributed. 8-cell parents that would give rise to inner and outer 16-cell daughters (O/I) tended to divide later than other cells in the embryo.
Additionally, inner and intermediate cells showed a strong tendency to divide after their outer 16-cell siblings. During the 5th cleavage, inner and outer 16-cell siblings sharing the same 8-cell parent divided such that the inner cell divided after the outer-cell sibling 86.7% of the time. Similarly, 16- cell intermediate cells divided after their outer-cell siblings 82.2% of the time. Indeed, divisions resulting in two outer cells show a strong preference for occurring early during the 5th cleavage (Fig. 4B) while divisions resulting in two inner progenies trail last in division order. When divisions resulting in asymmetric outer and inner 32-cell daughters are grouped according to whether they give rise to transient-outer cells from outer 16-cell parents or inner cells resulting from intermediate parents, once again the transient-outer cells behave like outer cells and exhibit a tendency for dividing early (Fig. 4B). The intermediate cells, despite being capable of giving rise to asymmetric cell fates, share a similar cell division order with inner cells at the 5th cleavage Fig. 4B).
Similar to the 5th cleavage, during the 4th cleavage division, 8-cell parents that gave rise to two outer daughters showed a preference for dividing early (Fig. 4C), while asymmetric divisions resulting in one outer and one inner daughter occurred late. However, divisions resulting in one outer and one intermediate daughter showed no clear preference for dividing early or late during the 4th cleavage. The above analyses revealed that the intermediate cells share similar cell cycle timing and division order with the inner cells.
Cell cleavage angle is a greater predictor of cell polarity than cell fate
It has been proposed that the cell division angle determines the fate of daughter cells, such that an asymmetric cell division with the axis of chromosome segregation perpendicular to the embryo surface gives rise to an outer and an inner cell with TE and ICM fates, respectively (Bischoff et al., 2008; Zernicka-Goetz, 2005). By contrast, symmetric cell divisions with the chromosome segregation axis tangential to the embryo surface produce daughter cells with the same outer cell fates (Fig. 5A). However, there are studies demonstrating that such division orientations do not correlate with cell fates (Dard et al., 2009b). This discrepancy could be due to the fact that mitotic events are rapid and cells in the early embryo undergo significant movement during divisions, making it challenging to predict an accurate cleavage orientation using imaging conditions that give low temporal resolution. The improved temporal resolution offered by TPLSM allowed us to re-evaluate this issue with much higher accuracy.
Figure 5.
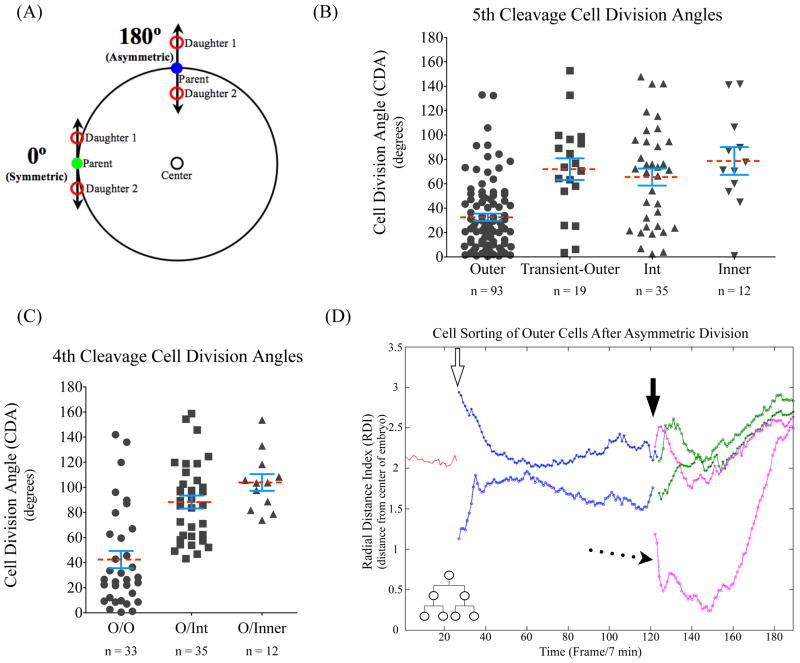
Cell division angle correlates with cell polarity but not cell fates. (A) The cell division angle was determined as described in the Materials and Methods. Briefly, the more symmetric a division (tangential to the surface of the embryo) the closer the cell division angle will be to 0º. An asymmetric division, with one cell moving away from the center of the embryo and one toward, will have a cell division angle closer to 180º. (B) The division angle as grouped by the cell lineage for the 5th cleavage. Degrees in-between 0° and 180° represent all the angles taken by cells dividing during the 16- to 32-cell stage. The division angle of each dividing cell is then grouped based on its lineage. During the 5th cleavage the division angles are shown for 16-cell parents that are outer, outer 16-cells that will give rise to transient-outer, and intermediate and inner 16-cell parents. The outer cells have a tendency of dividing symmetrically as compared to the other three16-cell populations, p-value <0.0001. (C) The division angle as grouped by the cell lineage for the 4th cleavage. During the 4th cleavage 8-cell parents that divide to produce outer and inner (O/Inner), outer and intermediate (O/Int) or two outer 16-cell daughters (O/O). The 8 cells that divide to give two outer cells (O/O) have a tendency to divide symmetrically as compared to the two other types of divisions. p-value < 0.0001. Red dashed lines denote the mean for each group, while SEM bars are in blue. (D) Evidence of cell sorting after asymmetric cell division. Lineage graph of an entirely outer-lineage based on the radial distance index (RDI) of cell nuclei over time from the 8-cell parent to 32-cell progeny. At the 8–16 cell division (open arrow) there is a clear asymmetric division of the 16-cell daughters as indicated by the large difference in RDI between the starting frames of the 16-cell daughters. During the 16-32 cell division, the 32-cell daughters indicated by the pink line again divide asymmetrically (solid arrow). In this case, one 32-cell daughter falls inward close to the center of the embryo (dotted arrow) for nearly 40 frames (~4.5 hours), but then sorts back to the surface of the embryo and occupies a position in the TE post-cavitation. n refers to the number of cells analyzed in each category.
To determine the cell division angle we tracked the chromosome segregation axis and the ensuing nuclei positions (see Materials and Methods). During the 5th cleavage divisions we found that the chromosome segregation axis did not strictly correlate with whether the cell underwent symmetric or asymmetric cell fate division (Fig. 5B). 16-cell inner or intermediate parents show no strong preference for symmetric or asymmetric division angles as defined in Figure 5A. Interestingly, 16-cell parents that produce transient-outer progeny also do not show a preference for symmetric divisions (Fig. 5B). However, outer 16-cell parents that produce only TE progenies did exhibit a preference for division angles parallel to the embryo surface defined as symmetric cleavage (Fig. 5B and C). We found similar results during the 4th cleavage division: 8-cell parents that produced inner or intermediate daughters showed no clear tendency for asymmetric versus symmetric cell division angles, and again the most symmetric division angles were seen in cleavages that produced solely outer progenies (Fig. 5C). Even so, 8-cell and 16-cell parents that contribute only to the outer lineage do occasionally undergo asymmetric cell cleavage. Thus, although an outer, polar cell is more likely to undergo symmetric cleavage than an apolar inner cell, the cell cleavage angle cannot be used as an accurate indicator of cell fate. Instead, the more symmetric the cleavage the higher chance that the dividing cell has a large area of polarized surface exposed on the outside of the embryo. This implies that intermediate cells, like inner cells, though possessing some cell-surface exposure, are inherently apolar in nature.
Cell sorting following cell division re-adjusts cell positions
If cell division angle is independent of cell fate then how do cells of an outer lineage retain their positions and identity when dividing in an asymmetric manner? We observed instances where outer cells divide perpendicular to the embryo surface, in which one daughter cell underwent a dramatic shift in nuclear RDI, occupying and correlating with a very inner position, and remained there for some time. However, these displaced cells would eventually migrate back to the outside of the embryo even after a period of nearly 5 hours on the inside (Fig. 5D). This indicates that cell sorting and cell motility play a role in maintaining cell fates in the pre-implantation embryo (Dietrich and Hiiragi, 2007; Vong et al., 2010).
Discussion
Application of two-photon microscopy for imaging the pre-implantation mouse embryo
The use of two-photon microscopy on a mammalian embryo was first described in 1999 (Squirrell et al., 1999), which showed conclusively that two-photon excitation caused less toxicity to hamster embryos compared to conventional confocal imaging. It has only been recently however, that TPLSM has become more widely appreciated by and available to the scientific community. The commercialization of TPLSM systems and the newest generation of Ti:Sapphire lasers continue to improve their accessibility and ease of use. The long wavelengths of Ti:Sapphire lasers (~670–1100 nm) pass relatively harmlessly through biological specimens (Centonze and White, 1998), and provide high viability with a depth of penetration far superior to that of conventional single-photon confocal microscopy (≳ 600 μm depth versus ~200 μm).
In this study we have demonstrated that TPLSM is indeed a viable, highly effective method for imaging the pre-implantation mouse embryo. While mouse embryos show a short delay in response to imaging over a period of days, they still retain full developmental competency, which indicates that the cellular behaviors observed are likely representative of normal in vivo development. Additionally, by simply reducing the scan time on a similar system we were able to abolish any developmental delay under otherwise identical conditions. Furthermore, the low toxicity provided by TPLSM allowed us to decrease the time intervals between imaging, and to more accurately and completely analyze pre-implantation development. While we chose a time interval of 6–7 minutes as this was the longest time interval to ensure 100% accuracy for the tracking algorithm, and for the purposes of this manuscript only described the stages between 8 and 32-cells, it is important to note that this is not the limit for embryo viability. Embryos can well tolerate shorter intervals, and are easily capable of developing to the 64-cell stage and beyond. The improved temporal and spatial resolution also allowed us to develop a tracking algorithm that is able to track every cell in the embryo and reconstruct their position and behavior with 100% accuracy as verified manually.
Further development of high-resolution imaging and analysis tools will provide a better understanding of how cell-cell contacts and cell surface exposures influence the choice of cell fates in the pre-implantation mouse embryo. Additionally, advances in TPLSM multi-channel imaging will enable the tracking of multiple cell-fate determinants not only in the mouse embryo, but also in a wide variety of tissues and systems. Studies considered previously untenable due to viability concerns using long-term imaging or dimly fluorescent genetic markers may become entirely possible with the application of TPLSM.
Asymmetric cell fate division is performed by a unique population of cells
Many studies have suggested that the mouse embryo maintains the ratio of outer to inner cells either by asymmetric divisions of inner or outer cells or by the inward movement of outer cells. Here we have shown that asymmetric cell fate divisions are performed by a unique cell population, which we refer to as intermediate cells, possessing characteristics of both inner and outer cells. This ambiguity has likely been the source of much confusion regarding the classification of inner or outer cells during the 16-cell stage in previous studies. Depending on the technique used, intermediate cells could be classified as outer cells due to the exposure of some cell-surface membrane or as inner cells based on their cell-cycle timing, behavior, and Cdx2 expression. Indeed, Yamanaka et al. (Yamanaka et al., 2010) described a population of cells during the 16-cell stage that divide asymmetrically to give rise to “secondary inner” and TE progeny. These cells were defined as “outer” during the 16-cell stage based on the injection of a membrane-bound red fluorescent protein. Based on our results we believe that these asymmetrically dividing “outer” cells are in fact the intermediate cells we have characterized in this manuscript.
While the characterization of cellular behavior based on nuclear position is not without its caveats, we have shown that lineages undergoing asymmetric cell fate divisions occupy distinct nuclear positions in the 16-cell stage embryo. Additionally, while it is tempting to classify these cells as outer cells based on their tendency toward surface exposure, they share very little over-lap of nuclear position with those cells of solely outer-lineages. Furthermore, they have many features of inner cells such as lower Cdx2 expression, longer cell-cycle time, and a preference for asymmetric cleavage angles. Therefore it is highly probable that these cells represent a group of inner cells that because of their intermediate position exhibit a unique behavior that affords them their bipotency.
Cell movement and surface exposure contribute to cell fate choices
In addition to describing the intermediate cell population, we observed a group of outer cells that retain all the characteristics and behaviors of outer cells up until cavitation, when they suddenly move inward and appear to assume an ICM fate. Since these cells are positioned next to the blastoceol cavity, we have referred to them as putative PE or transient-outer cells. The observation of these cells supports previously proposed models on the origin of the EPI and PE (Morris et al., 2010; Yamanaka et al., 2006), however it will be important to determine what is prompting these cells to move inside and whether they remain there to contribute to the PE.
The finding that inner and intermediate cells at the 16-cell stage show no preference for symmetric versus asymmetric cleavage is reflective of their apolar nature rather than their eventual cell fates. While intermediate cells exhibit a high tendency toward cell surface exposure, the potential lack of a polarized membrane as compared to outer cells would make it difficult to stabilize mitotic spindle orientation parallel to the embryo surface, leading to randomized spindle orientation and cell cleavage. Outer cells, which do possess a large, polarized outer surface are likely quicker at stabilizing and orienting their spindles parallel to the membrane surface, resulting in symmetric cleavage and a shorter cell-cycle time. It will be important to determine whether the exposed surface of intermediate cells fails to localize apical cell polarity markers as in outer cells. Interestingly, although transient-outer cells are indistinguishable from the other outer cells until cavitation, the 16-cell stage parents that give rise to transient-outer cells do not show the same preference for symmetric cleavage as those solely TE producing parents. It is possible that parents producing transient-outer cells have smaller areas of surface exposure or less polarized surface membranes than parents producing outer TE cells.
Cell-cell contacts and transcriptional regulation of cell fates
Recent studies have suggested that different amounts of cell-cell contact found in inner and outer cells in the pre-implantation embryo result in differential activation of the Hippo pathway, which in turn allows the activation or suppression of Cdx2 in outer and inner cells, respectively (Nishioka et al., 2009). Our imaging analyses have shown that cells in the pre-implantation embryo undergo a substantial amount of displacement during and after cell divisions. Furthermore, we have shown that while intermediate cells do expose their cell surface membrane to the outside they have low levels of Cdx2 expression like inner cells. If indeed intermediate cells exhibit a dynamic cell-surface exposure and constantly exchange between exposed and enclosed positions this would be more in line with their seemingly apolar nature. This implies that mere exposure of cell membranes to the embryo surface is insufficient to inhibit the Hippo pathway and to activate Cdx2.
In addition, the behavior of the transient-outer cells that move inward during or just prior to cavitation warrants further study from the perspective of cell-cell contacts and Cdx2 expression. It will be important to examine whether these cells have less exposed cell surface and weaker Cdx2 expression than those outer cells that only give rise to TE. By following Cdx2 expression live using dual-colored labeling through cavitation and past the 64-cell stage, it should be possible to determine how quickly these internalized cells turn off Cdx2 as well as their final position in the embryo. It is interesting to note that some inner cells have been shown to display weak Cdx2 expression as late as the 32-cell stage (Dietrich and Hiiragi, 2007; Nishioka et al., 2009; Niwa et al., 2005). These weak Cdx2 expressing cells could be the internalized transient-outer cells that are in the process of turning off their Cdx2.
The pre-implantation mouse embryo offers a great opportunity to study how cell polarity, cell movements, and cell-cell contacts influence cell-fate specification. However, limitations of live imaging techniques have caused conflicting results and hindered progress in this area. Our studies reported here demonstrate the utility of TPLSM for the study of pre-implantation development. These studies should stimulate further application of this imaging technique to address many interesting questions regarding the specification of the first lineages.
Materials and Methods
Generation of Histone-2B GFP transgenic mice
Transgenic mice expressing Histone-2B GFP were created in house as described previously (Vong et al., 2010).
Embryo harvesting, culture, imaging, and development
Histone-2B GFP males were crossed with natural, hormone-primed or super-ovulated CD1 wild-type females approximately 6–8 weeks of age. The presence of a vaginal plug the next day indicating copulation was noted as 0.5 d.p.c. Late in the afternoon of 1.5 d.p.c. 2–4 cell embryos were harvested by oviduct flushing with M2 media (MR-015-D, Millipore) and cultured in a droplet of KSOM media (MR-107-D, Millipore) covered with mineral oil (M8410, Sigma) on a World Precision Instruments 35 mm cover-glass Fluorodish (FD35-100, WPI). Embryos were allowed to develop over-night in a 37°C, 5% CO2 incubator before being placed on the microscope stage the next morning at an 8-cell stage.
Embryos were imaged on a Zeiss LSM 510 Confocor3 inverted microscope with a Chameleon Ti:Sapphire laser (Coherent Inc.). The microscope stage and objectives were enclosed by a cage incubator from In Vivo Scientific, and maintained at 37°C and 5% CO2. A 25x LD LCI Plan Apochromat 0.8 W Corr DIC objective used with Immersol (000000-1252-136, Carl Zeiss Microimaging, Inc.) was used at a zoom of x0.7 for all time-lapses. In some experiments multiple-stage positions were utilized to capture a large cluster of embryos. Time-lapses were acquired at 820 nm wavelength at an average laser power of 4–5%, equivalent to 12 mW. Z-stacks were taken at 2 μm intervals, with 51–53 sections for each stack with a scan time of ~981 ms for each section with a line averaging of 2.
To determine the effect of imaging on the potential of embryo development, embryos that were imaged for 48 hours as described above were then transferred into a 2.5 d.p.c. pseudo-pregnant CD1 WT female. Pseudo-pregnancy was induced by mating to vasectomy-treated CD1 male mice. Pups were born naturally allowed to develop to age P12 to ensure developmental competency.
Immunohistochemistry and Reconstruction
CD1 wild-type females were mated with CD1 wild-type males and embryos harvested at 2.5 d.p.c. corresponding to the 16-cell stage. Embryos were fixed in 4% paraformaldehyde in PBS for 10–15 min, permeabilized in a solution of 0.5% Triton X-100 in PBS for 30 min and then blocked over-night in 10% FBS, 0.1% Triton X-100 in PBS. Embryos were then placed in primary antibody diluted in blocking solution for 12 hours or over-night. Primary antibodies used were anti-mouse Cdx2 (1:100, BioGenex, Cdx2-88), and anti-rabbit Oct-4 (1:100, Cell Signaling C30A3). After incubation with the primary antibody embryos were washed three times in PBS and placed in fluorescent secondary and DAPI (1:1000) diluted in blocking solution for 2–4 hours and imaged un-mounted in droplets of PBS on an inverted Leica SP5 confocal microscope. For actin phalloidin staining, in place of the primary antibody step embryos were placed in a 1:500 dilution of Alexa Fluor 568 Phalloidin (Invitrogen, A12380) and 1:1000 of DAPI in blocking solution for 30 min to 1 hour and imaged as before.
Images were acquired in z-stacks every 1–2 μm for each wavelength, and then exported to IMARIS for reconstruction. Nuclei positions were identified using the Spot Identification Function as described previously, and coordinates of each cell nuclei were then exported to MatLab to determine the RDI. For relative Cdx2 levels, the mean intensity was determined by IMARIS for each nuclei in the embryo and corrected for relative background levels. Relative Cdx2 expression was then determined by comparing a cell’s mean intensity with the mean intensity of the closest, outer-most neighbor in the z-plane. This was to control for any loss of intensity from sectioning through the full depth of the embryo.
For phalloidin staining, nuclear positions were obtained as described above, and cells were visualized in 3D in IMARIS to determine cell membrane surface exposure.
Cell-tracking algorithm
To track individual cells accurately, it is necessary to find the correspondence between the cells detected at different time points, according to the positions of their nuclei. More specifically, as the basis of cell lineage analysis, it is necessary to know the history of the trajectory of each cell, which cell was its ancestor, and which cells were its daughters. To achieve this, the positions of all nuclei throughout a movie, each represented by a 3-dimentional vector (x, y, z), were first acquired from IMARIS. These vectors were then sorted according to the time of occurrence, i.e. frame number, and automatically checked to make sure that no nuclei were lost over time. For effective tracking, the first frame of the process was selected such that there were exactly 2N nuclei present, where N is an integer. By doing this, all cells were at the same stage in the first frame, and it is convenient to assign the exact cell-stage numbers and initiate the tracking process.
The algorithm developed is based on the idea that if no division occurs between two frames (the numbers of nuclei are the same), then cells in frame n+1 are assigned to cells in frame n in a one-to-one fashion so as to minimize the total distance summed over all pairs. When division does occur (noted by an increase in the number of nuclei), the cells of frame n+1 are also designated to those in frame n; but in this case, in addition to the one-to-one correspondence, it also happens that two daughter cells in frame n+1 are assigned to the same mother cell in frame n.
Given all the m nuclei positions in the nth frame pn,1 = (xn,1, yn,1, zn,1), ···, pn,m = (xn,m, yn,m, zn,m), and all the k nuclei positions in the (n+1)th frame pn+1,1 = (xn+1,1, yn+1,1, zn+1,1), ···, pn+1,k = (xn+1,k, yn+1,k, zn+1,k), with m ≤ k, the optimal correspondence between nuclei of the two frames was found by minimizing the Total Distance Function (TDF):
In the TDF, Fn,1 and Fn,2 is a 2-partition of the set of all indices for the m nuclei in the nth frame, i.e., Fn,1 ∪ Fn,2 = {1,2, ···, m}, Fn,1 ∩ Fn,2 = Ø. The partition was determined by the forward relationship from the n th frame to the (n +1)th frame: for each i ∈ Fn,1, the cell with nucleus at pn,i does not divide, so that it corresponds to only one nucleus in the (n+1)th frame; on the other hand, for each j ∈ Fn,2, the cell with nucleus at pn,j divides into two daughters in the (n+1)th frame. Similarly, Bn+1,1 and Bn+1,2 is a 2-partition of the set of all indices for the k nuclei in the (n +1)th frame {1,2, ···, k}, but determined by the backward relationship to the nth frame: for each i0 ∈ Bn+1,1, the cell with nucleus at pn+1,i0 corresponds to a cell without division in the previous frame; however, for each pair of j1, j2, the two cells with nuclei at pn+1,j1 and pn+1,j2 are the daughters of the same cell which has divided from the nth to the (n+1)th frame. A combination of these two 2-partitions then uniquely determines a correspondence between nuclei of the two frames. The constants α and β satisfy α +β = 1, weighing the two types of distances. Since the non-dividing cells usually move much less than the displacements from parents to daughters, in our implementations, α was set to be 0.75 and β to be 0.25 so that more tolerance was allowed in the case of division.
The optimization process to find the minimal TDF between each pair of consecutive frames consists of two steps. In the first one, a heuristic search strategy was used to find the initial correspondence between the two groups of points. More specifically, numbers of elements in each subset Fn,1, Fn,2 as well as Bn,1, Bn,2 were first decided by m and k. The single-cell connections were first decided by finding the nearest one available, and the parent-daughter connections were then decided by finding the nearest two available. Clearly, the results from this strategy will be influence by the order of searching. Thus, in the second step of the algorithm, simulated annealing (SA), a strategy developed in genetic algorithms (GA), was used to improve the results closer to the global optimum. More specifically, two pairs of connections were randomly selected from the population and swapped with each other. If the swap decreased the TDF, the results were updated accordingly; otherwise, the swap was discarded and the original results kept unchanged. The total number of random swaps was decided by the size of populations. In our implementation, it was set to be three times of the product of the numbers of points in the two consecutive frames.
Determination of radial distance of cells and division angle
To determine the change of position of cells within each embryo at different time frames, it is necessary to reconstruct the embryos in three dimensions. First, we modeled all cells as 3-dimensional balls, with center positions given by the nuclei, and radii acquired as follows. The radius of a cell in the first frame was set empirically according to the spatial resolution of the imaging process. Assuming that all cells at the same stage have the same volume, and that all division events preserve the total volume of cells, the radius of each cell throughout the movie was thus uniquely determined. It is important to note that the ball-model is not intended as a representation of cell shape, it instead computes relative positions between cells and cells and the embryo.
Next, we reconstructed the surface of the embryo at each time frame as the 3-dimensional convex hull that encompasses all points on the surface of any one of the cells in the same frame. At the same time, the volume of the reconstructed embryo was acquired, and the positions of all points that reside on the outer surface of the convex hull were also recorded (Supplementary Movie 4).
The above 3-dimensional reconstruction of embryos allowed us to determine the radial distance index (RDI) defined as a ratio, for each cell in each frame, between the radial distance from the nucleus to the center of the embryo and the radius of a cell in the initial stage. Thus, it is a normalized distance: the denominator serves as a scaling factor to eliminate the differences in spatial resolution during imaging among different embryos. Since the surface of an embryo is reconstructed as a convex hull, the center of the embryo is approximated accordingly as the center of the 3-dimensional space enclosed by the convex hull. The latter is computed as a weighted average of the centers of all the triangles that contribute to the surface, and the weights being their associated areas.
Calculation of Cell Division Angle
Using the 3-dimentional reconstructed embryos, we also determined the cell division angle (CDA), which measures the difference between the two daughter cells in deviation from their parent cell in each division process. To compute CDA, two angles, θ1 and θ2 are first decided, each representing the deviation of a daughter cell from the parent. For each daughter, this is calculated as the angle between two rays, both emitting from the position of the parent cell, but one points to the center of the embryo (the control direction), the other to the position of the position of the daughter cell. This angle is always between 0° and 180°. CDA is then computed as the absolute value of θ1-θ2. If these two deviations are the same, the CDA for this division process is 0º, representing the division angle that is tangential to the embryo surface. On the other hand, if one daughter cell appears in the control direction, while the other is opposite to the control direction, the CDA is 180º, which represents the division that is perpendicular to the embryo surface (Fig. 4A).
Supplementary Material
Supplementary Movie 1. 3D reconstruction in IMARIS of a mouse embryo expressing Histone-2B GFP starting from an 8-cell stage and ending at a 32-cell stage blastocyst. Each 8-cell stage nuclei (green) is marked with a differently colored spot to allow for easy tracing of each 8-cell lineage by eye. The time interval is every 7 min for 53 2 μm z-sections at 820 nm wavelength for the duration of the movie.
Supplementary Movie 2. Demonstration of transient-outer cell movement during blastocyst formation. Starting from a 16-cell stage, the outer-cell marked by a red spot (arrow) resides on the outer edge of the embryo. After the 5th cleavage division the red cell moves inward at the start of the formation of the blastoceol cavity, and occupies a position in the ICM generally occupied by PE cells.
Supplementary Movie 3. Example of asymmetric cell division in an outer-cell lineage during the 16–32 cell division. The nucleus marked with a red spot undergoes asymmetric division during the 5th cleavage (first arrow), however the two daughter nuclei (also marked by red spots) move outward and occupy a position in the TE after cavitation (final arrows).
Supplementary Movie 4. Reconstruction of a mouse embryo using convex hull projections as described in the Materials and Methods. The embryo shown in this movie is the same as that in Supplementary Movie 1, starting from an 8-cell stage and ending at a 32-cell blastocyst, with the cell surface of each cell modeled as previously described.
Research Highlights.
We utilize two-photon microscopy to image pre-implantation development.
We have created specialized tracking and reconstruction algorithms.
We identify and characterize a third, new cell type in the 16-cell stage embryo.
We provide new insights into the origins of cell fates.
Cdx2 expression is not regulated by cell surface exposure.
Acknowledgments
We would like to acknowledge Michael McCaffery for his infinite patience and the excellent staff and technical assistance from the Johns Hopkins University’s Integrated Imaging Center (IIC). The IIC is funded in part by NIH-NCRR grant #1 S10 RR025505-01 to McCaffery. We would like to thank Chen-Ming Fan and members of the Zheng lab for comments and suggestions to this manuscript. YZ is an investigator of Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bischoff M, Parfitt DE, Zernicka-Goetz M. Formation of the embryonic-abembryonic axis of the mouse blastocyst: relationships between orientation of early cleavage divisions and pattern of symmetric/asymmetric divisions. Development. 2008;135:953–62. doi: 10.1242/dev.014316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centonze VE, White JG. Multiphoton excitation provides optical sections from deeper within scattering specimens than confocal imaging. Biophys J. 1998;75:2015–24. doi: 10.1016/S0006-3495(98)77643-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dard N, Le T, Maro B, Louvet-Vallee S. Inactivation of aPKClambda reveals a context dependent allocation of cell lineages in preimplantation mouse embryos. PLoS One. 2009a;4:e7117. doi: 10.1371/journal.pone.0007117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dard N, Louvet-Vallee S, Maro B. Orientation of mitotic spindles during the 8- to 16-cell stage transition in mouse embryos. PLoS One. 2009b;4:e8171. doi: 10.1371/journal.pone.0008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–31. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Fleming TP. A quantitative analysis of cell allocation to trophectoderm and inner cell mass in the mouse blastocyst. Dev Biol. 1987;119:520–31. doi: 10.1016/0012-1606(87)90055-8. [DOI] [PubMed] [Google Scholar]
- Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, Robson P, Zernicka-Goetz M. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 2008;22:2692–706. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MH, McConnell JM. Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev Biol. 2004;15:583–97. doi: 10.1016/j.semcdb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- MacQueen HA, Johnson MH. The fifth cell cycle of the mouse embryo is longer for smaller cells than for larger cells. J Embryol Exp Morphol. 1983;77:297–308. [PubMed] [Google Scholar]
- Marikawa Y, Alarcon VB. Establishment of trophectoderm and inner cell mass lineages in the mouse embryo. Mol Reprod Dev. 2009;76:1019–32. doi: 10.1002/mrd.21057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Teo RT, Li H, Robson P, Glover DM, Zernicka-Goetz M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc Natl Acad Sci U S A. 2010;107:6364–9. doi: 10.1073/pnas.0915063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, Makita R, Kurihara H, Morin-Kensicki EM, Nojima H, Rossant J, Nakao K, Niwa H, Sasaki H. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–29. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Squirrell JM, Wokosin DL, White JG, Bavister BD. Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability. Nat Biotechnol. 1999;17:763–7. doi: 10.1038/11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Suwinska A, Czolowska R, Ozdzenski W, Tarkowski AK. Blastomeres of the mouse embryo lose totipotency after the fifth cleavage division: expression of Cdx2 and Oct4 and developmental potential of inner and outer blastomeres of 16- and 32-cell embryos. Dev Biol. 2008;322:133–44. doi: 10.1016/j.ydbio.2008.07.019. [DOI] [PubMed] [Google Scholar]
- Vong QP, Liu Z, Yoo JG, Chen R, Xie W, Sharov AA, Fan CM, Liu C, Ko MS, Zheng Y. A role for borg5 during trophectoderm differentiation. Stem Cells. 2010;28:1030–8. doi: 10.1002/stem.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–24. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Ralston A, Stephenson RO, Rossant J. Cell and molecular regulation of the mouse blastocyst. Dev Dyn. 2006;235:2301–14. doi: 10.1002/dvdy.20844. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M. Cleavage pattern and emerging asymmetry of the mouse embryo. Nat Rev Mol Cell Biol. 2005;6:919–28. doi: 10.1038/nrm1782. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M. The first cell-fate decisions in the mouse embryo: destiny is a matter of both chance and choice. Curr Opin Genet Dev. 2006;16:406–12. doi: 10.1016/j.gde.2006.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1. 3D reconstruction in IMARIS of a mouse embryo expressing Histone-2B GFP starting from an 8-cell stage and ending at a 32-cell stage blastocyst. Each 8-cell stage nuclei (green) is marked with a differently colored spot to allow for easy tracing of each 8-cell lineage by eye. The time interval is every 7 min for 53 2 μm z-sections at 820 nm wavelength for the duration of the movie.
Supplementary Movie 2. Demonstration of transient-outer cell movement during blastocyst formation. Starting from a 16-cell stage, the outer-cell marked by a red spot (arrow) resides on the outer edge of the embryo. After the 5th cleavage division the red cell moves inward at the start of the formation of the blastoceol cavity, and occupies a position in the ICM generally occupied by PE cells.
Supplementary Movie 3. Example of asymmetric cell division in an outer-cell lineage during the 16–32 cell division. The nucleus marked with a red spot undergoes asymmetric division during the 5th cleavage (first arrow), however the two daughter nuclei (also marked by red spots) move outward and occupy a position in the TE after cavitation (final arrows).
Supplementary Movie 4. Reconstruction of a mouse embryo using convex hull projections as described in the Materials and Methods. The embryo shown in this movie is the same as that in Supplementary Movie 1, starting from an 8-cell stage and ending at a 32-cell blastocyst, with the cell surface of each cell modeled as previously described.